Abstract
The Mycobacterium avium complex (MAC) comprises genomically similar but phenotypically divergent bacteria that inhabit diverse environments and that cause disease in different hosts. In this study, a whole-genome approach was used to examine the polymorphic PE (Pro-Glu) and PPE (Pro-Pro-Glu) gene families, implicated in immunostimulation and virulence. The four major groups of MAC organisms were examined, including the newly sequenced type strains of M. intracellulare and M. avium subsp. avium, plus M. avium subsp. paratuberculosis and M. avium subsp. hominissuis, for the purpose of finding genetic differences that could be exploited to design diagnostic tests specific to these groups and that could help explain their divergence in pathogenesis and host specificity. Unique and missing PPE genes were found in all MAC members except M. avium subsp. avium. Only M. intracellulare had a unique PE gene. Apart from this, most PE and PPE sequences were conserved, with average nucleotide sequence identities of 99.1 and 98.1%, respectively, among the M. avium subspecies, but only 82.9 and 79.7% identities with the PE and PPE sequences of M. intracellulare, respectively. A detailed analysis of the amino acid sequences was performed between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis. Most differences were detected in the PPE proteins, with amino acid substitutions and frame shifts leading to unique amino acid sequences. In conclusion, several unique PPE proteins were identified in MAC organisms next to numerous polymorphisms in both the PE and PPE gene families. These substantial differences could help explain the divergence in phenotypes within the MAC and could lead to diagnostic tests with better discriminatory abilities.
Mycobacterium avium and Mycobacterium intracellulare are collectively known as the Mycobacterium avium complex (MAC). For genotypic, phenotypic, and historical reasons, multiple subspecies of M. avium are recognized; these include Mycobacterium avium subsp. paratuberculosis, M. avium subsp. hominissuis, Mycobacterium avium subsp. avium, and M. avium subsp. silvaticum (50). All four M. avium subspecies and M. intracellulare possess a high degree of genetic similarity but are capable of infecting a diverse range of host species.
M. avium subsp. paratuberculosis is the causative organism of Johne's disease (paratuberculosis), a debilitating chronic enteritis in ruminants (49), and has been implicated in Crohn's disease in humans (18). M. avium subsp. avium and M. avium subsp. silvaticum are limited almost exclusively to avian species (53), in which they cause tuberculosis. M. avium subsp. hominissuis is a recent designation and was added to reflect the distinction of human and porcine isolates from bird-type strains when genotypic methods showed that M. avium isolates from humans in particular but also pigs rarely shared the genetic profiles of organisms found in birds (38, 53). M. avium subsp. hominissuis and M. intracellulare are ubiquitous, saprophytic mycobacteria commonly found in soil and water (12, 17, 28). Best known for causing disseminated infection in patients infected with human immunodeficiency virus, M. intracellulare and M. avium subsp. hominissuis are increasingly recognized as emerging pathogens of immunocompetent hosts (9, 26, 29) and as the etiologic agents of chronic pulmonary infections.
Despite their divergent phenotypes and the diseases that they cause, the genomes of M. avium subsp. hominissuis and M. avium subsp. paratuberculosis share greater than 97% nucleotide identity over large regions of their genomes (5). This genetic similarity between M. avium subsp. paratuberculosis and other members of the MAC confounds the diagnosis of M. avium subsp. paratuberculosis infections by serological and PCR-based tests. In the current serological tests used to diagnose M. avium subsp. paratuberculosis infection, such as enzyme-linked immunosorbent assay, the gamma interferon release assay, and the agarose gel immunodiffusion assay, many of the antigens used are not specific for M. avium subsp. paratuberculosis (31). Most tests use complex, ill-defined mixtures of proteins derived from whole-cell or fractionated extracts of M. avium subsp. paratuberculosis, which allows cross-reactivity with antibodies generated against other mycobacterial species (56). The tests could be improved by using multiple, specific, well-defined antigens which would increase the consistencies, specificities, and sensitivities of the tests. The selection of such M. avium subsp. paratuberculosis-specific protein antigens requires a thorough comparison of the protein-coding sequences of all MAC members to allow differentiation between these closely related organisms to address clinical and epidemiologic needs.
For the detection of M. avium subsp. paratuberculosis by PCR, a few specific genetic sequences have previously been identified, such as the insertion sequence IS900 (21), the F57 element (41), and the hspX gene (15); but the value of these sequences for use in diagnostic assays for M. avium subsp. paratuberculosis infection remains unclear because of concerns about their specificity (13, 16, 42) or a lack of rigorous evaluations with large samples of clinical isolates. In fact, false-positive PCR results have been reported for suspected paucibacillary M. avium subsp. paratuberculosis infections in patients with Crohn's disease (30). All of these facts demonstrate that both in veterinary settings in which M. avium subsp. paratuberculosis is an established pathogen and in human investigations studying the putative link between M. avium subsp. paratuberculosis and Crohn's disease, there is a need to identify true M. avium subsp. paratuberculosis-specific sequences to develop new diagnostic tests.
A recent comparative study of the genomes of different mycobacterial species has indicated that the major differences among these species are in the gene products constituting the cell wall and the polymorphic gene families encoding the PE and the PPE proteins (36), which are unique to mycobacteria. The names PE and PPE are derived from the motifs Pro-Glu and Pro-Pro-Glu, respectively, found in conserved domains near the N termini of these proteins. The PE and PPE gene families are highly expanded in the pathogenic members of this genus but show a conspicuous paucity in the nonpathogenic species. Although no precise function is known for any member of these families, members of the PE and PPE families have been linked to virulence (34, 43) or have at least been shown to influence interactions with other cells (7). Some PPE proteins are thought to be expressed on the cell surface (7, 14) and have been found to be immunodominant antigens (8).
The first aim of the present study was to identify all PE and PPE orthologues in the major groups of the MAC. Although M. avium subsp. silvaticum is not included in the MAC, isolates of this subspecies are largely indistinguishable from M. avium subsp. avium (53). The second aim was to identify PE and PPE orthologues that are missing in some members of the complex. The final aim was to report how unique PPE genes, insertions or deletions (indels), and frame shifts in orthologue genes result in unique protein fragments in M. avium subsp. paratuberculosis that could be exploited as M. avium subsp. paratuberculosis-specific targets in the development of more specific diagnostic tests.
MATERIALS AND METHODS
Source material.
A Roche 454 pyrosequencer was used for genome sequencing. The genome data for M. avium subsp. avium strain TMC 25291T was collected and kindly provided by Vivek Kapur. Genome data for Mycobacterium intracellulare strain ATCC 13950T were generated at the Genome Quebec Innovation Centre, McGill University, and are deposited in GenBank as genome project 27955.
The genome sequences of M. avium subsp. paratuberculosis K-10 and M. avium subsp. hominissuis 104 were previously determined in the laboratory of V. Kapur (33) and at The Institute for Genomic Research (www.tigr.org), respectively.
Identification of orthologous PE and PPE genes.
The BLAST algorithm was used to find orthologues of all M. avium subsp. paratuberculosis and M. avium subsp. hominissuis PE and PPE genes in the fully sequenced genome of M. avium subsp. avium and M. intracellulare and vice versa. The corresponding genes and proteins, the latter only between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis, were aligned in a pairwise manner; and the identity and gap were calculated by using a Ktuple of 2 and gap penalties of 7 and 4, respectively, and the DNAMAN (version 5.2.9) program (Lynnon Bio-Soft, Quebec, Canada).
Orthologues in other mycobacterial subspecies were identified by the BLASTp algorithm in the nonredundant GenBank coding sequence translations, RefSeq Proteins, and SwissProt databases and were subsequently compared by pairwise alignment.
Gap closing.
Alignment of the PE and PPE genes of M. avium subsp. hominissuis, M. avium subsp. avium, and M. avium subsp. paratuberculosis by the Vector NTI program (Invitrogen) revealed gaps in the sequences of the PPE genes (MaaPPE2, MaaPPE30, MaaPPE24, MaaPPE20, MaaPPE17, MaaPPE14, MaaPPE10, MaaPPE9, MaaPPE31, MaaPPE32, MaaPPE36, MaaPPE38, MaaPPE39) and the PE genes (MaaPE5 and MaaPE9) of the newly sequenced M. avium subsp. avium genome. Primers were designed around these gaps and were used to amplify the affected sequences from the genomic DNA of M. avium subsp. avium strain TMC 25291T. PCRs were performed with the Roche Expand high-fidelity PCR system; and the reaction mixtures (50 μl) contained 2 μl template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, 20 pmol of each primer, 5 μl of the manufacturer's PCR buffer containing MgCl2 (final MgCl2 concentration, 1.5 mM), and 1.75 U of Taq polymerase. The PCR conditions were denaturation at 94°C for 5 min, followed by 30 cycles of PCR with denaturation at 94°C for 45 s, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The final extension time was 10 min at 72°C.
Identification of unique PE and PPE genes and proteins.
PE or PPE genes were considered unique when they had less than 65% DNA identity with their closest orthologue in all of the other subspecies. PE and PPE genes were defined to be missing from a member of the MAC complex if no orthologue with an identity higher than 65% with the PE and PPE genes of any of the other members could be found. PE and PPE protein fragments were identified as divergent when frame shifts were discovered or when stretches of more than 80 amino acids (aa) were <30% identical to the homologous protein.
Amino acid sequence variations in the PE and PPE proteins.
The indels were identified in the DNA sequences, and the conservative and mismatched amino acid substitutions in the protein sequences were analyzed manually on the basis of the amino acid physicochemical grouping described by Lim (35).
Categorization of PPE proteins in sublineages.
The PPE proteins were categorized in sublineages on the basis of the presence of the sublineage-specific motifs and PPE domain structure, as described previously (54).
Phylogenetic analysis of PPE genes.
PPE orthologues that were present in all four members of the M. avium complex and that had limited gaps in DNA sequence alignment were selected to be used for phylogenetic analysis of the MAC. The sequences of 14 PPE genes (MapPPE29, MapPPE28, MapPPE23, MapPPE19, MapPPE18, MapPPE16, MapPPE13, MapPPE8, MapPPE7, MapPPE31, MapPPE32, MapPPE33, MapPPE34, MapPPE35, or orthologues) were combined and concatenated, generating 16,851-bp sequences, and were aligned and compared by use of the ClustalW program (www.ebi.ac.uk/clustalw).
Single-nucleotide polymorphisms (SNPs) in the orthologues of MapPPE23 and MapPPE24 in M. avium subsp. avium, M. avium subsp. hominissuis, M. avium subsp. paratuberculosis, and M. intracellulare were analyzed in detail; and a phylogram for both genes was created by use of the ClustalW program.
Nucleotide sequence accession numbers.
All M. avium subsp. avium PE and PPE gene locus sequences were deposited in GenBank under accession numbers EU854954 to EU854962 and EU864963 to EU854996 (Tables 1 and 2), respectively. All M. intracellulare PE and PPE gene locus sequences were deposited in GenBank under accession numbers EU854997 to EU855007 and EU855008 to EU855045, respectively.
TABLE 1.
Similarity of DNA sequences of PPE gene family orthologues in the MAC
| MAC PPE locus name | M. tuberculosis orthologue | Locus name in:
|
Sub- lineage | % Identity between the following pairs by nucleotide sequence alignment of PPE gene orthologues:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. avium subsp. paratuberculosis | M. avium subsp. hominissuis | M. avium subsp. avium | M. intracellulare | M. avium subsp. paratuberculosis-M. avium subsp. hominissuis | M. avium subsp. paratuberculosis- M. avium subsp. avium | M. avium subsp. paratuberculosis- M. intracellulare | M. avium subsp. hominissuis- M. avium subsp. avium | M. avium subsp. hominissuis-M. intracellulare | M. avium subsp. avium-M. intracellulare | |||
| MACPPE1 | PPE20 | MAP0123 | Mav_0118 | MaaPPE1 | MiPPE1 | II | 99.7 | 99.3 | 83.4 | 97.9 | 81.4 | 81.1 |
| MACPPE1 | PPE20 | MAP0124 | Mav_0118 | MaaPPE1 | MiPPE1 | (II)* | 98.3 | 95.5 | 74.7 | 97.9 | 81.4 | 81.1 |
| MACPPE2 | PPE69 | MAP0158 | Mav_0152 | MaaPPE2 | MiPPE2 | III | 99.2 | 99.3 | 76.7 | 100.0 | 80.3 | 76.5 |
| MACPPE3 | PPE65 | MAP0442 | Mav_0535 | MaaPPE3 | MiPPE3 | IV | 99.0 | 98.8 | 77.3 | 99.0 | 80.7 | 77.3 |
| MACPPE4 | Aa | Mav_0790c | I | |||||||||
| MACPPE5 | A | MAP0966c | MaaPPE5 | MiPPE5 | V | 98.6 | 83.0 | 82.8 | ||||
| MACPPE6 | PPE15 | MAP1003c | Mav_1179 | MaaPPE6 | MiPPE6 | IV | 99.4 | 99.6 | 84.4 | 99.5 | 84.7 | 84.5 |
| MACPPE7 | A | MAP2601 | Mav_1322c | MaaPPE7 | MiPPE7 | IV | 99.1 | 99.0 | 77.0 | 99.9 | 76.9 | 76.8 |
| MACPPE8 | A | MAP2600 | Mav_1324c | MaaPPE8 | MiPPE8 | IV | 99.3 | 99.3 | 80.3 | 100.0 | 79.7 | 79.7 |
| MACPPE9 | A | MAP2595 | Mav_1329c | MaaPPE9 | MiPPE9 | II | 99.0 | 99.1 | 82.6 | 100.0 | 81.3 | 82.4 |
| MACPPE10 | PPE18 | MAP2575c | Mav_1347 | MaaPPE10 | MiPPE10 | IV | 99.0 | 99.1 | 81.4 | 99.9 | 81.0 | 81.1 |
| MACPPE11 | A | Mav_1998 | MaaPPE11 | MiPPE11 | IV | 73.1 | 73.1 | 90.3 | ||||
| MACPPE12 | A | Mav_2006 | IV | |||||||||
| MACPPE13 | A | MAP2136c | Mav_2039 | MaaPPE13 | MiPPE13 | IV | 99.0 | 99.1 | 80.8 | 99.9 | 80.8 | 80.8 |
| MACPPE14 | A | MAP1813c | Mav_2429 | MaaPPE14 | MiPPE14 | II | 97.5 | 97.5 | 83.2 | 99.0 | 83.4 | 83.2 |
| MACPPE15 | A | NT03MA1810 | Mav_2514c | MaaPPE15 | II | 98.3 | 98.1 | 98.6 | ||||
| MACPPE16 | A | MAP1675 | Mav_2746c | MaaPPE16 | MiPPE16 | (IV)* | 100.0 | 98.8 | 89.2 | 100.0 | 76.7 | 89.6 |
| MACPPE16 | A | MAP1676 | Mav_2746c | MaaPPE16 | MiPPE16 | IV | 99.4 | 98.6 | 72.3 | 99.5 | 76.7 | 70.5 |
| MACPPE17 | PPE33 | MAP1522 | Mav_2905c | MaaPPE17 | MiPPE17 | IV | 98.9 | 98.6 | 86.0 | 98.9 | 85.3 | 85.3 |
| MACPPE18 | PPE29, PPE32 | MAP1521 | Mav_2906c | MaaPPE18 | MiPPE18 | IV | 98.9 | 99.0 | 88.4 | 99.4 | 88.7 | 89.0 |
| MACPPE19 | PPE30, PPE33 | MAP1519 | Mav_2909c | MaaPPE19 | MiPPE19 | IV | 99.2 | 99.4 | 87.5 | 99.2 | 87.8 | 87.5 |
| MACPPE20 | PPE32 | MAP1518 | Mav_2910c | MaaPPE20 | MiPPE20 | IV | 98.4 | 98.3 | 86.2 | 99.9 | 86.0 | 86.0 |
| MACPPE21 | PPE31 | MAP1516 | Mav_2913c | MaaPPE21 | IV | 98.6 | 98.6 | 100.0 | ||||
| MACPPE22 | PPE31 | MAP1515 | Mav_2914c | MaaPPE22 | IV | 98.2 | 98.1 | 99.9 | ||||
| MACPPE23 | PPE26 | MAP1506 | Mav_2924c | MaaPPE23 | MiPPE23 | IV | 97.8 | 98.9 | 72.7 | 98.3 | 71.6 | 72.0 |
| MACPPE24 | PPE25 | MAP1505 | Mav_2925c | MaaPPE24 | MiPPE24 | IV | 93.9 | 97.6 | 74.7 | 93.4 | 74.3 | 74.6 |
| MACPPE24 | PPE26 | MAP1506 | Mav_2926c | MaaPPE23 | MiPPE23 | IV | 98.9 | 98.9 | 72.7 | 100.0 | 72.1 | 72.0 |
| MACPPE24 | PPE25 | MAP1505 | Mav_2928c | MaaPPE24 | MiPPE24 | IV | 97.6 | 97.6 | 74.7 | 99.8 | 74.6 | 74.6 |
| MACPPE27 | A | MAP1155 | Mav_3353c | MaaPPE27 | IV | 98.2 | 98.2 | 99.4 | ||||
| MACPPE28 | A | MAP1153 | Mav_3355c | MaaPPE28 | MiPPE28 | IV | 98.8 | 98.7 | 76.9 | 99.7 | 76.6 | 76.5 |
| MACPPE29 | A | MAP1152 | Mav_3356c | MaaPPE29 | MiPPE29 | IV | 99.1 | 99.1 | 70.0 | 100.0 | 70.2 | 70.2 |
| MACPPE30 | A | MAP1144c | MaaPPE30 | MiPPE30 | II | 99.1 | 76.1 | 75.7 | ||||
| MACPPE31 | A | MAP2927 | Mav_3715 | MaaPPE31 | MiPPE31 | IV | 98.3 | 98.6 | 68.6 | 98.6 | 68.6 | 68.3 |
| MACPPE32 | PPE50 | MAP3184 | Mav_4014 | MaaPPE32 | MiPPE32 | IV | 98.9 | 99.5 | 81.9 | 99.4 | 82.0 | 82.1 |
| MACPPE33 | PPE51 | MAP3185 | Mav_4015 | MaaPPE33 | MiPPE33 | IV | 98.7 | 98.6 | 76.6 | 99.0 | 76.7 | 76.9 |
| MACPPE34 | A | MAP3419c | Mav_4273c | MaaPPE34 | MiPPE34 | IV | 99.5 | 99.3 | 77.8 | 99.5 | 77.9 | 78.0 |
| MACPPE35 | A | MAP3420c | Mav_4274c | MaaPPE35 | MiPPE35 | IV | 99.5 | 99.3 | 83.8 | 99.5 | 83.8 | 83.6 |
| MACPPE36 | A | MAP3490 | Mav_4349 | MaaPPE36 | MiPPE36 | II | 99.1 | 99.3 | 82.7 | 98.9 | 81.5 | 81.7 |
| MACPPE37 | PPE10 | MAP3939c | Mav_4704 | MaaPPE37 | MiPPE37 | V | 99.7 | 99.5 | 89.6 | 99.5 | 89.5 | 89.8 |
| MACPPE38 | PPE4 | MAP3782 | Mav_4867c | MaaPPE38 | MiPPE38 | II | 98.7 | 98.6 | 84.3 | 99.1 | 85.4 | 84.8 |
| MACPPE39 | PPE2 | Mav_4872c | MaaPPE39 | II | 99.6 | |||||||
| MACPPE40 | A | MAP3765 | Mav_4879c | MaaPPE40 | MiPPE40 | II | 74.9 | 74.4 | 77.1 | 98.7 | 83.3 | 82.9 |
| MACPPE41 | PPE3 | MAP3725 | II | |||||||||
| MACPPE42 | A | MAP3737 | II | |||||||||
| MACPPE43 | PPE65 | MiPPE43 | IV | |||||||||
| MACPPE44 | A | MiPPE44 | IV | |||||||||
| MACPPE45 | A | MiPPE45 | IV | |||||||||
| MACPPE46 | PPE32 | MiPPE46 | IV | |||||||||
| MACPPE47 | A | MiPPE47 | IV | |||||||||
| MACPPE48 | PPE31 | MiPPE48 | IV | |||||||||
| MACPPE49 | A | MAP1152 | Mav_3356c | MiPPE49 | IV | 99.1 | 99.1 | 70.5 | 100.0 | 70.7 | 70.7 | |
| Mean | 98.1 | 98.2 | 79.6 | 98.6 | 79.6 | 80.0 | ||||||
| SD | 4.1 | 4.0 | 5.5 | 4.4 | 5.6 | 6.1 | ||||||
A, absent; *, truncated gene.
TABLE 2.
Similarity of DNA sequences of PE gene family orthologues in the MAC
| MAC PE locus name | M. tuberculosis orthologue | Locus name in:
|
% Identity by nucleotide sequence alignment of PE gene orthologues from the following pairs:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. avium subsp. paratuberculosis | M. avium subsp. hominissuis | M. avium subsp. avium | M. intracellulare | M. avium subsp. paratuberculosis-M. avium subsp. hominissuis | M. avium subsp. paratuberculosis-M. avium subsp. avium | M. avium subsp. paratuberculosis-M. intracellulare | M. avium subsp. hominissuis-M. avium subsp. avium | M. avium subsp. hominissuis-M. intracellulare | M. avium subsp. avium-M. intracellulare | ||
| MACPE1 | Rv1386 | Map0122 | Mav_0117 | MaaPE1 | MiPE1 | 99.4 | 99.0 | 81.2 | 98.4 | 80.9 | 80.9 |
| MACPE2 | Rv3893 | Map0157 | Mav_0151 | MaaPE2 | MiPE2 | 100.0 | 100.0 | 77.6 | 100.0 | 79.2 | 77.6 |
| MACPE3 | Rv3622 | Map0441 | Mav_0534 | MaaPE3 | MiPE3 | 98.0 | 97.7 | 81.3 | 97.0 | 81.0 | 82.3 |
| MACPE4 | Rv1040 | Map1003c | Mav_1179c | MaaPE4 | MiPE4 | 99.5 | 99.3 | 83.9 | 99.2 | 83.8 | 84.8 |
| MACPE5 | Rv1195 | Map2576c | Mav_1346 | MaaPE5 | MiPE5 | 99.5 | 99.5 | 83.6 | 100.0 | 83.6 | 83.6 |
| MACPE6 | Rv1788 | Map1514 | Mav_2915c | MaaPE6 | MiPE7 | 99.3 | 99.3 | 69.0 | 100.0 | 68.3 | 68.3 |
| MACPE7 | Rv1791 | Map1507 | Mav_2923 | MaaPE7 | MiPE7 | 97.9 | 98.7 | 92.0 | 99.5 | 89.3 | 92.7 |
| MACPE8 | Rv1791 | Map1507 | Mav_2923 | MaaPE7 | MiPE8 | 97.9 | 98.7 | 71.3 | 99.5 | 73.1 | 72.0 |
| MACPE9 | Map4144 | Mav_4488 | MaaPE9 | MiPE9 | 99.7 | 99.3 | 68.8 | 98.9 | 84.6 | 86.1 | |
| MACPE10 | Rv0287 | Map3783 | Mav_4866 | MaaPE10 | MiPE10 | 99.7 | 99.0 | 91.5 | 99.7 | 92.0 | 91.8 |
| MACPE11 | Rv0285 | Map3781 | Mav_4868c | MaaPE11 | MiPE11 | 98.4 | 99.0 | 88.0 | 98.7 | 89.6 | 88.7 |
| MACPE12 | MiPE12 | ||||||||||
| Mean | 99.1 | 99.1 | 81.7 | 99.1 | 83.2 | 83.7 | |||||
| Stdev | 0.8 | 0.6 | 8.1 | 0.9 | 6.7 | 7.2 | |||||
RESULTS
Identification of orthologous and unique sequences.
A total of 28 PPE genes were conserved in all four members of the complex (Table 1). Although most orthologues were intact, pseudogenes, frame-shifted genes, or remaining fragments of former complete PPE genes (MapPPE1, MapPPE16) were also observed. Map0123 and Map0124 (MapPPE1) were identified as nonoverlapping partial orthologues of MahPPE1, caused by an early stop codon in Map0123.
Map1675 and Map1676 (MapPPE16) were identified as nonoverlapping partial orthologues of MahPPE16, caused by an early stop codon in Map1675.
Over the whole complex, 49 different PPE paralogues existed, with 36 M. avium subsp. paratuberculosis, 38 M. avium subsp. hominissuis, 36 M. avium subsp. avium, and 38 M. intracellulare paralogues being detected. Consequently, there were almost identical duplicate genes (namely, the repeating pairs MahPPE23-MahPPE25, MahPPE24-MahPPE26, MapPPE21-MapPPE22, MahPPE21-MahPPE22, and MaaPPE21-MaaPPE22) and subspecies-specific paralogues in M. avium subsp. paratuberculosis (MapPPE41, MapPPE42), M. avium subsp. hominissuis (MahPPE4, MahPPE12), and M. intracellulare (MiPPE43, MiPPE44, MiPPE45, MiPPE46, MiPPE47, MiPPE48). The homologies of these loci and their closest orthologues are given in Table 3, as are other mycobacterial species with which the closest orthologues have homology. Additionally, missing paralogues, defined as missing from one subspecies but present in at least two members of the complex, were found in M. avium subsp. paratuberculosis (n = 2), M. avium subsp. hominissuis (n = 2), and M. intracellulare (n = 6) (Table 4). This comparison has allowed the definition of new PPE locus names for the MAC.
TABLE 3.
Similarity in DNA sequence between PE and PPE genes that are unique to a member of the MAC and their closest orthologue in other members of the complex or other mycobacterial species
| Gene | Unique PPE genes and closest orthologuesa
|
% Nucleotide sequence identity between genes from the following pairs:
|
Locus of non-MAC mycobacterial spp. with which homology exists | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. avium subsp. paratuberculosis | M. avium subsp. hominissuis | M. avium subsp. avium | M. intracellulare | M. avium subsp. paratuberculosis-M. avium subsp. hominissuis | M. avium subsp. paratuberculosis-M. avium subsp. avium | M. avium subsp. paratuberculosis-M. intracellulare | M. avium subsp. hominissui-M. avium subsp. avium | M. avium subsp. hominissui- M. intracellulare | M. avium subsp. avium-M. intracellulare | ||
| PPE | PPE33 | PPE4 | PPE33 | PPE18 | 53.8 | 53.7 | 50.2 | Rv1548c | |||
| PPE20 | PPE12 | PPE20 | PPE43 | 57.9 | 59.5 | 58.8 | Mt1850 | ||||
| PPE41 | PPE36 | PPE36 | PPE40 | 58.2 | 55.6 | 59.5 | Mt0269 | ||||
| PPE42 | PPE39 | PPE39 | PPE40 | 60.8 | 60.1 | 56.4 | Mt0269 | ||||
| PPE6 | PPE6 | PPE6 | PPE45 | 55.4 | 42.3 | 42.7 | Mt1745 | ||||
| PPE18 | PPE18 | PPE18 | PPE46 | 58.4 | 58.5 | 57.6 | Mt1850 | ||||
| PPE31 | PPE31 | PPE31 | PPE47 | 60.4 | 60.2 | 60.1 | Mul_2096 | ||||
| PPE18 | PPE18 | PPE18 | PPE44 | 58.4 | 58.4 | 58.5 | Mt1851 | ||||
| PPE18 | PPE18 | PPE18 | PPE43 | 58.3 | 58.3 | 58.1 | Mt1850 | ||||
| PPE21 | PPE21 | PPE21 | PPE48 | 61.9 | 61.9 | 61.9 | Mul_0947 | ||||
| PE | PE7 | PE7 | PE7 | PE11 | 63.0 | 64.0 | 64.0 | Rv1791 | |||
Unique PE and PPE genes are indicated in boldface.
TABLE 4.
Similarity in DNA sequence between PE and PPE genes that are missing from members of the MAC and their closest orthologues in other members of the complex
| Orthologues of the missing PPE genesa
|
% Nucleotide sequence identity between genes from the following pairs:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| M. avium subsp. paratuberculosis | M. avium subsp. hominissuis | M. avium subsp. avium | M. intracellulare | M. avium subsp. paratuberculosis-M. avium subsp. hominissuis | M. avium subsp. paratuberculosis-M. avium subsp. avium | M. avium subsp. paratuberculosis-M. intracellulare | M. avium subsp. hominissuis-M. avium subsp. avium | M. avium subsp. hominissuis-M. intracellulare | M. avium subsp. avium-M. intracellulare |
| PPE5 | PPE26 | PPE5 | PPE5 | 58.3 | 55.6 | 52.7 | |||
| PPE30 | PPE39 | PPE30 | PPE30 | 59.4 | 59.6 | 61.8 | |||
| PPE27 | PPE27 | PPE27 | PPE47 | 59.6 | 59.9 | 59.8 | |||
| PPE15 | PPE15 | PPE15 | PPE36 | 55.6 | 54.6 | 54.3 | |||
| PPE22 | PPE11 | PPE11 | PPE11 | 57.0 | 56.2 | 54.2 | |||
| PPE36 | PPE39 | PPE39 | PPE36 | 61.8 | 65.3 | 62.0 | 62.7 | ||
| PPE22 | PPE22 | PPE22 | PPE48 | 61.1 | 60.6 | 60.5 | |||
| PPE21 | PPE21 | PPE21 | PP448 | 61.9 | 61.9 | 61.9 | |||
The closest orthologues in other members of the complex are indicated in boldface.
Close orthologues were found for the remaining PPE genes and for all M. avium subsp. hominissuis and M. avium subsp. paratuberculosis PE genes (Tables 1 and 2), including orthologues for the PE and the PPE gene fragments of the natural PE-PPE fusion in Map1003, both of which are found in the annotated gene Mav_1179 in two different reading frames with an 8-bp overlap. By performing BLASTn searches of the M. avium subsp. paratuberculosis genome with M. avium subsp. hominissuis PE sequences, a previously nonrecognized M. avium subsp. paratuberculosis PE gene, MapPE10, was discovered.
Deletions averaging 170 bp in size (size range, 38 bp to 464 bp) in 14 M. avium subsp. avium PPE genes compared to the sequences of their orthologues in M. avium subsp. paratuberculosis and M. avium subsp. hominissuis were initially found after compilation of the contigs. The affected regions in the M. avium subsp. avium genome were amplified by PCR, and subsequent sequencing confirmed that the gaps were due to insufficient overlap of the contigs. Several small indels between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis still existed, and these are extensively covered in the next section. The numerous indels in the M. intracellulare PE and PPE genes were not investigated further.
With respect to the PE genes, 12 different PE paralogues were found (Table 2). M. avium subsp. paratuberculosis, M. avium subsp. hominissuis, and M. avium subsp. avium each had orthologues for 10 PE genes. M. intracellulare was found to have two MapPE7 orthologues (MiPE7 and MiPE8), with MiPE7 also being the closest orthologue to MapPE6 (Table 4). Again, the new names for all PE genes in MAC are defined in Table 2.
Sequence similarity.
Among the M. avium subspecies, the level of DNA identity was high for all conserved PE and PPE genes, with average levels of identity of 99.1% ± 0.8% and 98.3% ± 4.1%, respectively, for M. avium subsp. paratuberculosis, M. avium subsp. hominissuis, and M. avium subsp. avium. The only outliers were MapPPE24 and its first orthologue, MahPPE25; MapPPE40 and MahPPE40; and MahPPE11 and MaaPPE11. The level of identity between M. avium subsp. hominissuis and M. avium subsp. paratuberculosis was high for all PE proteins, with an average of 98.0% ± 2.8%, but there was greater variability in the level of identity for the PPE proteins, with an average of 90.2% ± 21.2%; the variability was mainly caused by frame shifts.
The average levels of nucleotide sequence identity of the M. intracellulare PE and PPE genes with the M. avium PE and PPE genes were 82.0% ± 7.3% and 79.7% ± 5.7%, respectively.
Detailed comparison of M. avium subsp. paratuberculosis and M. avium subsp. hominissuis PE and PPE proteins and genes.
Orthologues of M. avium subsp. avium and M. avium subsp. hominissuis were found to be very similar, with an average level of nucleotide sequence identity of 98.6%, such that only M. avium subsp. hominissuis and M. avium subsp. paratuberculosis were selected for use in a more detailed amino acid sequence comparison (see Tables S1 and S2 in the supplemental material). While BLAST searches determined high levels of nucleotide sequence identity between sequences coding for the PE and PPE proteins and the flanking regions in M. avium subsp. paratuberculosis and M. avium subsp. hominissuis, large amino acid sequence differences caused by one or numerous frame shifts were observed in some PE and PPE loci. Frame shifts have created some unique M. avium subsp. paratuberculosis protein fragments in MapPE6 (aa 195 to 314), MapPPE1 (aa 185 to 301), and MapPPE15 (aa 62 to 368). Additional frame shifts in M. avium subsp. hominissuis genes MapPPE2 (aa 153 to 396) and MapPPE13 (aa 314 to 410) create differences in protein sequences between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis but are not M. avium subsp. paratuberculosis specific because those frame shifts do not occur in M. avium subsp. avium.
Categorization of sublineages.
Of the 36 M. avium subsp. paratuberculosis PPE proteins, 10 belonged to sublineage II, 1 to sublineage III, 23 to sublineage IV, and 2 to sublineage V. Of the 38 M. avium subsp. hominissuis PPE proteins, 8 belonged to sublineage II, 1 to sublineage III, 28 to sublineage IV, and 1 to sublineage V. Of the 36 M. avium subsp. avium PPE proteins, 9 belonged to sublineage II, 1 to sublineage III, 24 to sublineage IV, and 2 to sublineage V. Three unique M. avium subsp. hominissuis PPE proteins belonged to sublineage IV, while the other two belonged to sublineage II. One unique M. avium subsp. paratuberculosis PPE protein belonged to the sublineage IV, while the other four belonged to sublineage II.
SNP analysis of orthologues of duplicated PPE genes.
Some PPE genes have duplicate paralogues in one of the members of the complex. MahPPE24 and MahPPE26 in M. avium subsp. hominissuis are both orthologues of MapPPE24, just as MahPPE23 and MahPPE25 are both orthologues of the single gene MapPPE23. The paralogues MahPPE23 and MahPPE25 have 93.3% amino acid identity, while the paralogues MahPPE22 and MahPPE24 have 98.3% amino acid identity. The order of the genes in M. avium subsp. hominissuis suggests duplication of both neighboring genes together, although an extra gene, Mav_2927, is inserted between the MapPPE24 and MapPPE23 orthologues MahPPE26 and MahPPE25, respectively.
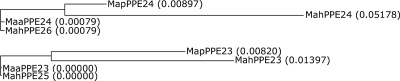
SNP analysis of these orthologues (Tables 5 and 6) within the complex indicates that M. avium subsp. avium and M. avium subsp. paratuberculosis share SNPs with different orthologues in M. avium subsp. hominissuis. This suggests that the duplication of the neighboring PPE proteins was present in an ancestor of these members of the complex (M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis) and that M. avium subsp. avium and M. avium subsp. paratuberculosis have retained different paralogues of the duplicate. This is also illustrated by the phylograms of the MapPPE23 and MapPPE24 orthologues (Fig. 1).
TABLE 5.
SNPs in the orthologues of MACPPE23 in the MAC indicating genetic relation between the members
| Orthologue | SNP at base pair:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 15 | 16 | 150 | 293 | 328 | 344 | 411 | 483 | 452 | 675 | 789 | 864 | 944 | 945 | 946 | 1039 | 1080 | 1125 | |
| MiPPE23 | C | G | G | G | A | C | G | G | G | C | G | T | G | A | |||||
| MahPPE25 | C | A | A | C | A | T | G | G | G | C | G | C | C | A | C | C | |||
| MaaPPE23 | C | A | A | C | A | T | G | G | G | C | G | C | C | A | C | C | |||
| MahPPE23 | T | G | G | C | A | T | G | G | G | C | G | C | G | A | T | C | |||
| MapPPE23 type II | T | G | G | T | T | G | G | T | C | T | C | T | C | G | T | G | G | T | T |
| MapPPE23 type III | T | G | G | T | A | T | A | G | C | C | C | T | C | G | T | T | |||
| MapPPE23 type I | T | G | G | T | A | T | G | G | C | C | C | T | C | G | T | T | |||
TABLE 6.
SNPs in the orthologues of MACPPE24 in the MAC indicating genetic relation between the members
| Orthologue | SNP at base pair:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 381 | 562 | 563 | 564 | 571 | 572 | 577 | 579 | 582 | 585 | 588 | 590 | 593 | 597 | |
| MiPPE24 | C | G | C | G | G | T | G | G | G | G | C | A | C | G |
| MahPPE26 | T | A | G | C | A | T | C | G | A | G | T | G | C | G |
| MaaPPE24 | T | A | G | C | A | T | C | G | A | G | T | G | C | G |
| MahPPE24 | T | G | C | T | G | C | G | A | G | C | C | A | G | A |
| MapPPE24 | C | G | C | T | G | C | G | A | G | C | C | A | G | A |
FIG. 1.
Phylograms of the MapPPE23 and MapPPE24 orthologues in the MAC based on their nucleotide sequences, as calculated by the use of ClustalW (EMBL-EBI) software. The relative branch lengths are given for all the orthologues.
Phylogenetic analysis.
A phylogram was drawn from the distances calculated with the ClustalW program (Fig. 2) for the concatenated nucleotide sequences of orthologues of 15 PPE genes of M. avium subsp. avium, M. avium subsp. hominissuis, M. avium subsp. paratuberculosis, and M. intracellulare. M. intracellulare is the more distant relative within the MAC. M. avium subsp. avium and M. avium subsp. hominissuis are the closest relatives.
FIG. 2.
Phylogram of the concatenated nucleotide sequences of orthologues of 15 PPE genes of members of the MAC, as calculated by the use of ClustalW (EMBL-EBI) software. The relative branch lengths are given for all the orthologues.
DISCUSSION
Studies involving comparisons of complete microbial genomes can readily reveal commonalities in gene content and genome organization between closely related bacteria and the major differences that distinguish them. While it is clear that bioinformatics has its limits in predicting the influences of gene content and genome organization on virulence and immunity, an important first step in determining the basis for differences in pathogenicity is the documentation of the differences between closely related organisms. Because of the previously reported link between PE and PPE proteins and pathogenicity for organisms of the M. tuberculosis complex, this protein family from phenotypically different MAC organisms represented a reasonable candidate for comparative analysis.
Previously, the PE and PPE genes in the fully sequenced and annotated genomes of M. tuberculosis H37Rv and CDC1551 were analyzed by comparative genomics (54). Although the genome of M. avium subsp. paratuberculosis strain K-10 was also analyzed, complete sequences for additional MAC organisms were not yet available, and it was not possible to detail the divergence of the PE and PPE proteins within the MAC. The genomes of several MAC organisms, including the type strains of M. avium subsp. avium and M. intracellulare, have now been sequenced. This provided an opportunity to reexamine the relationships between and evolutionary history of the members of the subfamilies of the PE and PPE protein families, to identify subfamily-specific characteristics, and to determine the extent of PE and PPE sequence similarity and variation.
In the present study, new uniform PPE and PE locus names for all members of the MAC were proposed. These uniform orthologue names will simplify the reporting of the findings of future studies of these polymorphic gene families in MAC. The names of the MAC PPE genes were rooted on the M. avium subsp. hominissuis genome because the sequence of M. avium subsp. paratuberculosis strain K-10 has previously been described to have small and large genomic inversions and, thus, a gene order that differs from those in M. avium subsp. hominissuis, M. avium subsp. avium, and other M. avium subsp. paratuberculosis isolates (5, 57).
This is the first time that the sequences of multiple genes that are potentially associated with virulence were compared between four members of the MAC. Previously, single genes (20, 46, 48, 52) and insertion sequences (10, 23, 32) were targeted to characterize MAC isolates. The study of multiple related genomes also provides insight into the distribution and evolution of these genes. The unique M. avium subsp. paratuberculosis genes are located in a subspecies-specific large sequence polymorphism and were likely acquired via horizontal gene transfer. Similarly, the unique M. avium subsp. hominissuis genes are found in genomic regions conserved among a subset of M. avium subsp. hominissuis isolates (44, 57). The missing orthologues of MACPPE05 and MACPPE11 also correspond to earlier deletions identified in M. avium subsp. hominissuis and M. avium subsp. paratuberculosis, respectively (44).
The present study demonstrated on average 98.6% nucleotide sequence similarity between the M. avium subsp. avium and the M. avium subsp. hominissuis PPE genes. This is in agreement with the high average similarities of 99.1 and 98.1% between the conserved PE and PPE genes of M. avium subsp. hominissuis and M. avium subsp. paratuberculosis, respectively. Indeed, very few genetic differences were previously found when the genomes of M. avium subsp. paratuberculosis and M. avium subsp. hominissuis were compared, with greater than 97% nucleotide sequence identity over large genomic regions and 100% nucleotide sequence identity of the 16S rRNA genes from these two subspecies being identified (5). In another study, the comparison of genes orthologous between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis has revealed 98 to 99% nucleotide sequence identity (33). However, the high degree of nucleotide sequence identity observed among the M. avium subspecies does not hold true for the complete MAC, with only an average of 79.7% similarity between M. intracellulare and the M. avium subspecies. A previously observed mean similarity between M. intracellulare and M. avium was 91% (51).
From an analysis of 48% of the genome, only 27 predicted coding sequences were found to be absent in M. avium subsp. avium and M. avium subsp. hominissuis (2). In another report, 39 genes of M. avium subsp. paratuberculosis were found to be unique to M. avium subsp. paratuberculosis and were thus missing from M. avium subsp. hominissuis (33). Other reports indicate that M. avium subsp. paratuberculosis coding sequences are absent from M. avium subsp. hominissuis (3, 40). The unique PPE proteins mentioned above are not included in these lists since they were all recognized as members of the polymorphic PPE protein family, and the various other differences between M. avium subsp. paratuberculosis and M. avium subsp. hominissuis PPE proteins at the protein level were also not previously described. They are, however, equally valuable for consideration as targets for new discriminative diagnostic tests.
The present analysis indicates that all of the PE proteins and most of the PPE proteins of M. avium subsp. paratuberculosis are highly homologous to the corresponding proteins of M. avium subsp. hominissuis. This is in agreement with a comparison of other proteins between M. avium subsp. hominissuis and M. avium subsp. paratuberculosis by Bannantine et al. (5), who described levels of similarity that ranged from 94 to 100%. However, one PE protein and six PPE proteins with high levels of nucleotide sequence identity were found to be very different at the protein level due to frame shifts that led to unique protein fragments in the M. avium subsp. paratuberculosis and the M. avium subsp. hominissuis PPE proteins. Other studies have shown that unique sequences have considerable potential for use in the development of more specific and sensitive diagnostic assays for the detection of M. avium subsp. paratuberculosis infection by both molecular assay- and immunoassay-based approaches (2-4, 40). It is also possible that these different amino acid sequences have significant impacts on the protein functions. Even the single amino acid substitutions identified could potentially have an influence on the immunogenicity and virulence of the PE and PPE proteins (25).
While unique and missing genes were identified in the M. avium subsp. hominissuis, M. avium subsp. paratuberculosis, and M. intracellulare genomes, most genes in all four members of the complex also contained numerous SNPs. These SNPs can be used for subspecies differentiation. SNPs are frequently used in epidemiological and evolutionary studies to differentiate between closely related species, subspecies, and strains of bacteria without knowledge of what effect the SNP may have on gene function or protein activity (1, 19, 24, 27, 47).
A study by Turenne et al. (52), who used a PCR- and sequencing-based strategy to investigate the complete hsp65 gene in the MAC, identified 10 SNPs that differentiated M. avium subsp. hominissuis and M. avium subsp. paratuberculosis. Other studies (37) showed that SNPs are a major source of genotypic variation within the MAC, and as demonstrated by Semret et al. (45), SNPs can be used in conjunction with large sequence polymorphisms to identify possible evolutionary paths within the MAC.
The existence of unique PPE proteins as well as subtle variations in the PE and PPE gene families, such as SNPs, might be the cause of the differences in pathogenesis between mycobacterial species, as previously suggested by Marri et al. (36). For example, if these PE and PPE genes are expressed in vivo, they could potentially cause the differential responses of bovine macrophages to M. avium subsp. avium/M. avium subsp. hominissuis and M. avium subsp. paratuberculosis, as observed previously (55). This previous suggestion is supported by previous observations that a specific M. avium subsp. hominissuis PPE protein is associated with the ability to grow in macrophages and is involved in virulence in mice (34). Other SNPs, such as the ones in the rpoV and mma3 genes of M. bovis, have been shown to have a marked impact on virulence and cellular functions (6, 11), and SNPs in M. tuberculosis (6, 11) have been shown to be responsible for altered phenotypes. The identification of SNPs in genes that are linked to virulence is the first step in explaining the differences in virulence that are caused by these SNPs.
The findings of the present analysis suggest that the duplication of the neighboring PPE genes (MapPPE23 and MapPPE24) was present in an ancestor of these members of the complex (M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis) and that M. avium subsp. avium and M. avium subsp. paratuberculosis have retained different paralogues of the duplicate, while M. avium subsp. hominissuis still contains both duplicates. This corresponds to the findings described in an earlier report that M. avium subsp. hominissuis is a diverse group of organisms from which two pathogenic clones (M. avium subsp. paratuberculosis and M. avium subsp. avium) have evolved independently (51).
The present study is based on the full genomic sequences of single isolates of M. avium subsp. paratuberculosis, M. avium subsp. hominissuis, M. avium subsp. avium, and M. intracellulare and thus will not identify sequence variations between subtypes (e.g., the bovine, ovine, and intermediate type variants of M. avium subsp. paratuberculosis) within a subspecies. Recently, SNPs have been found in the PPE gene sequence of MapPPE23. The sequences of the bovine strains appeared to consistently differ at 8 nucleotides from the sequences of the intermediate type strains and 7 nucleotides from the sequences of the ovine type strains and the homologous genes in M. avium subsp. hominissuis (MahPPE23 and MahPPE25, respectively) (22).
On the basis of the DNA sequences of the PPE genes, M. avium subsp. avium seems to be more closely related to M. avium subsp. hominissuis than to M. avium subsp. paratuberculosis. However, M. avium subsp. avium shares two PPE proteins (MACPPE5 and MACPPE30 with M. avium subsp. paratuberculosis that are absent in M. avium subsp. hominissuis. This could also indicate a function for these gene products in host adaptation because both M. avium subsp. avium and M. avium subsp. paratuberculosis are believed to be adapted to ruminant and bird hosts, respectively (53), whereas M. avium subsp. hominissuis has a more ubiquitous host distribution.
Two M. avium subsp. hominissuis-specific genes (MACPPE4 and MACPPE11) were found in this study. The corresponding gene products could be used to identify immune responses against this M. avium subspecies, and misinterpretations due to cross-reactivity in current diagnostics for Johne's disease would thereby be avoided. This subspecies is a heterogeneous group of strains with at least six distinct hsp65 sequences (hsp65 sequevars) (52). This has previously led to the suggestion that human isolates simply reflect what is found in their environment (44). It will be intriguing to know whether the M. avium subsp. hominissuis-specific MAC PPE genes are present in all sequevars and all environmental isolates or whether those genes are unique to isolates causing disease in humans.
After the demonstration of immune responses against one PPE protein in M. avium subsp. paratuberculosis-infected cows (39), studies are now under way to heterologously express the unique PPE M. avium subsp. paratuberculosis genes as well as the PPE genes coding for unique protein fragments for the construction of a partial protein array to evaluate the humoral and cell-mediated immunostimulatory capabilities of these recently discovered unique proteins. The combination of genomic information, molecular tools, and immunological assays will thus provide key insights into the host immune response to M. avium subsp. paratuberculosis infection. Overall, the elucidation of all of the unique sequences as well as those that may be associated with the cell surface of M. avium subsp. paratuberculosis provides a strong foundation on which to develop the next generation of specific and sensitive diagnostic assays for M. avium subsp. paratuberculosis.
In conclusion, the existence of several unique PPE proteins in both M. avium subsp. hominissuis and M. avium subsp. paratuberculosis was demonstrated, as were differences created by frame shifts and indels in both the PE and the PPE gene families. These substantial differences could help explain the important differences in phenotypes between members of the MAC.
Supplementary Material
Acknowledgments
We thank Vivek Kapur from the Department of Veterinary and Biomedical Sciences at Pennsylvania State University for providing us with the genome sequence data for M. avium subsp. avium.
This work has been supported by the Margaret Gunn Endowment for Animal Research (to J.M.D.B.).
Footnotes
Published ahead of print on 14 January 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 1853392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., E. Baechler, Q. Zhang, L. L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 401303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., J. K. Hansen, M. L. Paustian, A. Amonsin, L. L. Li, J. R. Stabel, and V. Kapur. 2004. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannantine, J. P., J. F. J. Huntley, E. Miltner, J. R. Stabel, and L. E. Bermudez. 2003. The Mycobacterium avium subsp. paratuberculosis 35 kDa protein plays a role in invasion of bovine epithelial cells. Microbiology 1492061-2069. [DOI] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., Q. Zhang, L. L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr, M. A., B. G. Schroeder, J. N. Brinkman, R. A. Slayden, and C. E. Barry. 2000. A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 1823394-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, M. J., G. Delogu, Y. P. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 697326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. R. Murthy, V. M. Katoch, and S. E. Hasnain. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 716338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coker, R. J., T. J. Hellyer, I. N. Brown, and J. N. Weber. 1992. Clinical aspects of mycobacterial infections in HIV-infection. Res. Microbiol. 143377-381. [DOI] [PubMed] [Google Scholar]
- 10.Collins, D. M., D. M. Gabric, and G. W. deLisle. 1989. Identification of a repetitive DNA-sequence specific to Mycobacterium paratuberculosis. FEMS Microbiol. Lett. 60175-178. [DOI] [PubMed] [Google Scholar]
- 11.Collins, D. M., R. P. Kawakami, G. W. Delisle, L. Pascopella, B. R. Bloom, and W. R. Jacobs. 1995. Mutation of the principal sigma-factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 928036-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, F. M. 1989. Mycobacterial disease, immunosuppression, and acquired immunodeficiency syndrome. Clin. Microbiol. Rev. 2360-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13431-442. [DOI] [PubMed] [Google Scholar]
- 14.Delogu, G., C. Pusceddu, A. Bua, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52725-733. [DOI] [PubMed] [Google Scholar]
- 15.Ellingson, J. L. E., C. A. Bolin, and J. R. Stabel. 1998. Identification of a gene unique to Mycobacterium avium subspecies paratuberculosis and application to diagnosis of paratuberculosis. Mol. Cell. Probes 12133-142. [DOI] [PubMed] [Google Scholar]
- 16.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209267-271. [DOI] [PubMed] [Google Scholar]
- 17.Falkinham, J. O. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feller, M., K. Huwiler, R. Stephan, E. Altpeter, A. Shang, H. Furrer, G. E. Pfyffer, T. Jemmi, A. Baumgartner, and M. Egger. 2007. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect. Dis. 7607-613. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. Deboy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 1845479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frothingham, R., and K. H. Wilson. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 1752818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, E. P., M. L. V. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 179063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths, T. A., K. Rioux, and J. De Buck. 2008. Sequence polymorphisms in a surface PPE protein distinguish types I, II, and III of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 461207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutacker, M. M., J. C. Smoot, C. A. L. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 1621533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, D. P., M. Hill, H. M. Vordermeier, M. Jones, G. Hewinson, H. Thangaraj, and J. Ivanyi. 1997. Mutagenesis of an immunodominant T cell epitope can affect recognition of different T and B determinants within the same antigen. Mol. Immunol. 34315-322. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh, C. R. 1991. Current concepts—Mycobacterium avium complex infection in the acquired-immunodeficiency-syndrome. N. Engl. J. Med. 3241332-1338. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, A. L., R. Friedman, and M. Murray. 2002. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg. Infect. Dis. 81342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inderlied, C. B., C. A. Kemper, and L. E. M. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson, M. A., P. C. Hopewell, D. M. Yajko, W. K. Hadley, E. Lazarus, P. K. Mohanty, G. W. Modin, D. W. Feigal, P. S. Cusick, and M. A. Sande. 1991. Natural-history of disseminated Mycobacterium avium complex infection in AIDS. J. Infect. Dis. 164994-998. [DOI] [PubMed] [Google Scholar]
- 30.Jeyanathan, M., D. C. Alexander, C. Y. Turenne, C. Girard, and M. A. Behr. 2006. Evaluation of in situ methods used to detect Mycobacterium avium subsp. paratuberculosis in samples from patients with Crohn's disease. J. Clin. Microbiol. 442942-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalis, C. H. J., M. T. Collins, J. W. Hesselink, and H. W. Barkema. 2003. Specificity of two tests for the early diagnosis of bovine paratuberculosis based on cell-mediated immunity: the Johnin skin test and the gamma interferon assay. Vet. Microbiol. 9773-86. [DOI] [PubMed] [Google Scholar]
- 32.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. McFadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 52265-2272. [DOI] [PubMed] [Google Scholar]
- 33.Li, L. L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 10212344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Y. J., E. Miltner, M. Wu, M. Petrofsky, and L. E. Bermudez. 2005. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell. Microbiol. 7539-548. [DOI] [PubMed] [Google Scholar]
- 35.Lim, D. V. 1989. Microbiology. West Publishing Company, St. Paul, MN.
- 36.Marri, P. R., J. P. Bannantine, and G. B. Golding. 2006. Comparative genomics of metabolic pathways in Mycobacterium species: gene duplication, gene decay and lateral gene transfer. FEMS Microbiol. Rev. 30906-925. [DOI] [PubMed] [Google Scholar]
- 37.Marsh, I. B., and R. J. Whittington. 2007. Genomic diversity in Mycobacterium avium: single nucleotide polymorphisms between the S and C strains of M. avium subsp. paratuberculosis and with M. a. avium. Mol. Cell. Probes 2166-75. [DOI] [PubMed] [Google Scholar]
- 38.Mijs, W., P. de Haas, R. Rossau, T. van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 521505-1518. [DOI] [PubMed] [Google Scholar]
- 39.Newton, V., S. L. B. Mckenna, and J. De Buck. 24 September 2008. Presence of PPE proteins in Mycobacterium avium subsp. paratuberculosis isolates and their immunogenicity in cattle. Vet. Microbiol. [Epub ahead of print.] [DOI] [PubMed]
- 40.Paustian, M. L., A. Amonsin, V. Kapur, and J. P. Bannantine. 2004. Characterization of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis: implications for diagnosis of Johne's disease. J. Clin. Microbiol. 422675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poupart, P., M. Coene, H. Vanheuverswyn, and C. Cocito. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 311601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puyang, X. L., K. Lee, C. Pawlichuk, and D. Y. Kunimoto. 1999. IS1626, a new IS900-related Mycobacterium avium insertion sequence. Microbiology 1453163-3168. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 2881436-1439. [DOI] [PubMed] [Google Scholar]
- 44.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. Van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 433704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semret, M., C. Y. Turenne, P. de Haas, D. M. Collins, and M. A. Behr. 2006. Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms. J. Clin. Microbiol. 44881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smole, S. C., F. McAleese, J. Ngampasutadol, C. F. von Reyn, and R. D. Arbeit. 2002. Clinical and epidemiological correlates of genotypes within the Mycobacterium avium complex defined by restriction and sequence analysis of hsp65. J. Clin. Microbiol. 403374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 401798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, D. S., V. Kapur, K. Stockbauer, X. Pan, R. Frothingham, and J. M. Musser. 1997. Subspecific differentiation of Mycobacterium avium complex strains by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. Int. J. Syst. Bacteriol. 47414-419. [DOI] [PubMed] [Google Scholar]
- 49.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 12305-312. [DOI] [PubMed] [Google Scholar]
- 50.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40254-260. [DOI] [PubMed] [Google Scholar]
- 51.Turenne, C. Y., D. M. Collins, D. C. Alexander, and M. A. Behr. 2008. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J. Bacteriol. 1902479-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turenne, C. Y., M. Semret, D. V. Cousins, D. M. Collins, and M. A. Behr. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turenne, C. Y., R. Wallace, Jr., and M. A. Behr. 2007. Mycobacterium avium in the postgenomic era. Clin. Microbiol. Rev. 20205-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Pittius, N. C. G., S. L. Sampson, H. Lee, Y. Kim, P. D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss, D. J., O. A. Evanson, A. Moritz, M. Q. Deng, and M. S. Abrahamsen. 2002. Differential responses of bovine macrophages to Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium. Infect. Immun. 705556-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiley, E. L., T. J. Mulhollan, B. Beck, J. A. Tyndall, and R. G. Freeman. 1990. Polyclonal antibodies raised against Bacillus Calmette-Guerin, Mycobacterium duvalii, and Mycobacterium paratuberculosis used to detect mycobacteria in tissue with the use of immunohistochemical techniques. Am. J. Clin. Pathol. 94307-312. [DOI] [PubMed] [Google Scholar]
- 57.Wu, C. W., J. Glasner, M. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.