Abstract
Spirochetes of the genus Treponema were cultured from 7 of 10 cases of digital dermatitis in sheep. Two cultures comprised Treponema phagedenis-like and Treponema medium/Treponema vincentii-like spirochetes, respectively, while the remaining cultures comprised mixed populations of Treponema medium/Treponema vincentii-like, Treponema phagedenis-like, and Treponema denticola/Treponema putidum-like organisms.
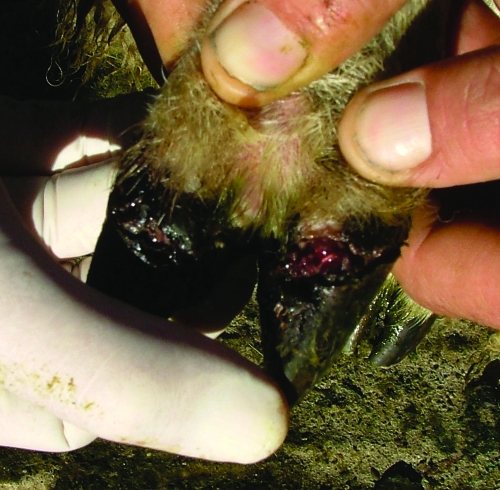
Contagious ovine digital dermatitis (CODD) is a disease of the ovine hoof which results in acute, severe lameness (Fig. 1). In contrast to virulent footrot, which is characterized clinically by lesions involving the heel and the interdigital area, CODD is characterized by ulcerative lesions of the coronary band which progress and result in disruption of the abaxial wall lining the hoof and loss of the horn case in untreated cases (1, 9, 11). The causative agent of CODD is unknown; however, spirochetes have been associated with clinical cases of CODD (2, 3, 9). While evidence of CODD in Ireland is sparse, with only one documented case (3), anecdotal evidence from shepherds highlighting persistent “incurable” footrot and ineffective vaccine strategies suggest that CODD may be prevalent and being incorrectly diagnosed as virulent footrot.
FIG. 1.
An acute lesion of CODD on the coronary band. If untreated, the infection will lead to loss of the hoof case.
Two geographically distinct lowland sheep farms with persistent flock lameness problems were identified. Charcoal anaerobic swabs were used to take samples from 10 crossbred sheep with acute lesions of the coronary band. Swab buds were buried deep in the lesion within the broken skin-horn junction and transported immediately to the laboratory. Samples were dipped vigorously in 10 ml fastidious anaerobic broth (Lab M Limited, United Kingdom) containing 20% fetal calf serum (Sigma, United Kingdom) and 10 μg/ml each rifampin (rifampicin), enrofloxacin, and marbofloxacin (Sigma, United Kingdom) and incubated at 37°C in anaerobic conditions.
Following 12 days of incubation, dark-field microscopy confirmed spirochetes in 7 of the 10 cultures from clinical cases of CODD. Contamination from other bacterial sources was confirmed negative in all cases by aerobic incubation on blood agar. Further morphological analysis was performed by scanning electron microscopy (SEM) and transmission EM (TEM) (Fig. 2 A, B, and C). For SEM, an aliquot of spirochetes containing 2.5% glutaraldehyde in 0.1 M Sorensen phosphate buffer (pH 7.3) was incubated at room temperature for 1 h and centrifuged and the pellet postfixed with 1% osmium tetroxide in 0.1 M Sorensen phosphate buffer (pH 7.4) for 1 h at room temperature and washed twice. The pellet was dispersed in water and applied to a poly-l-lysine-coated glass microscope slide (Menzel-Glaser, Germany), air dried, and coated in gold (SEM coating unit Polaron E5100). Samples were examined by using a JEOL JSM 5410LV (JEOL, United Kingdom) SEM at 15 kV. For TEM, an aliquot of spirochetes containing 2.5% glutaraldehyde in 0.1 M Sorensen phosphate buffer (pH 7.3) was incubated at room temperature for 1 h, centrifuged, and postfixed with 1% osmium tetroxide in 0.1 M Sorensen phosphate buffer (pH 7.3) for 1 h at room temperature. Samples were embedded in Epon resin using standard methods, and ultrathin (80 nm) sections were cut using a diamond knife and a Leica UC6 ultramicrotome, picked up on 200-mesh copper grids, and contrasted with uranyl acetate (20 min) and leaf citrate (10 min). Sections were examined in a Tecnai 12 BioTwin TEM (FEI Electron Optics, The Netherlands) using an acceleration voltage of 120 kV and an objective aperture of 20 μm. Digital images at various magnifications were acquired with a MegaView 3 camera (Soft Imaging Systems, Germany). The basic defining features of treponemes were evident: a helical shape, outer and inner membranes, and approximately five flagellar filaments located in the periplasmic space (Fig. 2A, B, and C) (7).
FIG. 2.
EM results. (A) Scanning electron micrograph of a spirochete cultured from the CODD lesion shown in Fig. 1. The spirochete was identified as Treponema phagedenis-like based on 16S rRNA gene phylogenetic analysis. (B) Transmission electron micrograph of a cultured Treponema phagedenis-like spirochete. 1, Outer membrane; 2, inner membrane; 3, periplasmic flagella. Bar, 200 nm. (C) Cross section of a cultured Treponema phagedenis-like organism (TEM). Arrows indicate evidence of periplasmic flagella. Bar, 200 nm.
Cultured spirochete species were also typed at the molecular level. Briefly, genomic DNA was extracted from 250 μl of cultured broth using a DNeasy blood and tissue kit (Qiagen, United Kingdom) and stored at −20°C until use. The 16S rRNA genes and 16S-23S rRNA gene intergenic spacer region 2 region were amplified, sequenced, and analyzed for species identification (10). Typing of mixed cultures was implemented with a PCR method used to identify the association of Treponema medium/Treponema vincentii-like, Treponema phagedenis-like, and Treponema denticola/Treponema putidum-like DD treponemes with bovine DD lesions, which uses species-specific primers located within the 16S rRNA gene (6, 6a). All seven cultures of spirochetes were typed within the genus Treponema (Table 1). Two of seven samples were pure cultures of T. phagedenis-like (100% homology to the sequence deposited in GenBank under accession number EF057411) and T. medium/T. vincentii-like (99.4% homology to the sequence deposited in GenBank under accession number EF061252) DD treponemes (data not shown). Mixed cultures included various permutations of the T. medium/T. vincentii-like, T. phagedenis-like, and T. denticola/T. putidum-like DD treponemes, as shown in Table 1.
TABLE 1.
Genetic analysis of cultured spirochetesa
| Sample | Group-specificb PCR result
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Mayo A | − | + | − |
| Wicklow 2 | + | + | + |
| Wicklow 3 | + | + | + |
| Wicklow 4 | − | + | + |
| Wicklow 5 | + | − | + |
| Wicklow 6 | + | − | + |
| Wicklow 8 | + | − | − |
Sample Mayo A was 100% homologous to T. phagedenis-like spirochetes (comparable to the sequence deposited in GenBank under accession no. EF057411), and Wicklow 8 was 99.4% homologous to T. medium/Treponema vincentii-like spirochetes (comparative to the sequence deposited in GenBank under accession no. EF061252). The remaining samples were mixed cultures.
Group 1, Treponema medium/Treponema vincentii-like spirochetes; group 2, Treponema phagedenis-like spirochetes; group 3, Treponema denticola/Treponema putidum-like spirochetes. +, present; −, absent.
In this study, treponemes were identified in 70% of CODD lesions, similar to the results of a study by Moore et al. (8). Negative culture results should be interpreted carefully in light of the fastidious nature of treponemes in culture and the variability in swab sampling techniques, leading to potential false-negative results. T. phagedenis-like and T. medium/T. vincentii-like species have also been previously associated with DD in cattle (4, 10; N. J. Evans et al., submitted). This suggests that these treponemes are associated with DD in both cattle and sheep, highlighting the potential for interspecies transmission, as noted in previous studies (2, 5). This transmission potential raises specific disease control and biosecurity issues for multienterprise farming and may account for the inability of some enterprises to achieve lameness-free status in cases where alternate housing units and grazing of paddocks were used for sheep and cattle as a means of disease control. The two farms investigated in this study were combined cattle and sheep enterprises.
This study has identified spirochetes in clinical cases of DD in sheep and has characterized these spirochetes as belonging to the genus Treponema. A definitive population of culturable spirochetes from DD lesions in cattle and sheep is emerging, and an immune response to spirochete infection in sheep has been observed (5). In order to develop potential immunoprophylactic control measures for DD, the nature, extent, and specificity of this immune response will need to be determined in both cattle and sheep.
Nucleotide sequence accession numbers.
Partial 16S rRNA gene sequences for isolates designated Mayo A and Wicklow 8 (Table 1) have been deposited in GenBank under accession numbers FM210038 and FM210039, respectively.
Acknowledgments
This work was funded by the UCD School of Agriculture, Food Science, and Veterinary Medicine, University College Dublin.
We are grateful to Märit Pringle for advice on culturing techniques, David Cottell for assistance with EM, Jacques Izard for guidance on interpretation of electron micrographs, and Yvonne Abbot of the UCD Veterinary Hospital bacteriology laboratory for technical assistance.
Footnotes
Published ahead of print on 9 February 2009.
REFERENCES
- 1.Abbott, K. A., and C. J. Lewis. 2005. Current approaches to the management of ovine footrot. Vet. J. 16928-41. [DOI] [PubMed] [Google Scholar]
- 2.Collighan, R. J., R. D. Naylor, P. K. Martin, B. A. Cooley, N. Buller, and M. J. Woodward. 2000. A spirochete isolated from a case of severe virulent ovine foot disease is closely related to a treponeme isolated from human periodontitis and bovine digital dermatitis. Vet. Microbiol. 74249-257. [DOI] [PubMed] [Google Scholar]
- 3.Demirkan, I., S. D. Carter, C. Winstanley, K. D. Bruce, N. M. McNair, M. Woodside, and C. A. Hart. 2001. Isolation and characterisation of a novel spirochaete from severe virulent ovine foot rot. J. Med. Microbiol. 501061-1068. [DOI] [PubMed] [Google Scholar]
- 4.Demirkan, I., H. F. Williams, A. Dhawi, S. D. Carter, C. Winstanley, K. D. Bruce, and C. A. Hart. 2006. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 101948-955. [DOI] [PubMed] [Google Scholar]
- 5.Dhawi, A., C. A. Hart, I. Demirkan, I. H. Davies, and S. D. Carter. 2005. Bovine digital dermatitis and severe virulent ovine foot rot: a common spirochaetal pathogenesis. Vet. J. 169232-241. [DOI] [PubMed] [Google Scholar]
- 6.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, W. D. Vink, R. W. Blowey, C. A. Hart, and S. D. Carter. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet. Microbiol. 130141-150. [DOI] [PubMed] [Google Scholar]
- 6a.Evans, N. J., J. M. Brown, I. Demirkan, P. Singh, B. Getty, D. Timofte, W. D. Vink, R. D. Murray, R. W. Blowey, R. J. Birtles, C. A. Hart, and S. D. Carter. 2009. Association of unique, isolated treponemes with bovine digital dermatitis lesions. J. Clin. Microbiol. 47689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izard, J., and A. Limberger. 2006. Structural and genomic features of treponemal architecture, p. 39-60. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponema: molecular and cellular biology, Caister Academic Press, Norfolk, United Kingdom.
- 8.Moore, L. J., M. J. Woodward, and R. Grogono-Thomas. 2005. The occurrence of treponemes in contagious ovine digital dermatitis and the characterisation of associated Dichelobacter nodosus. Vet. Microbiol. 111199-209. [DOI] [PubMed] [Google Scholar]
- 9.Naylor, R. D., P. K. Martin, J. R. Jones, and M. C. Burnell. 1998. Isolation of spirochaetes from an incident of severe virulent ovine footrot. Vet. Rec. 143690-691. [PubMed] [Google Scholar]
- 10.Stamm, L. V., H. L. Bergen, and R. L. Walker. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J. Clin. Microbiol. 403463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter, A. 1997. Virulent foot rot in sheep. Vet. Rec. 14127. [PubMed] [Google Scholar]