Abstract
The development and validation of dried sample spots as a method of specimen collection are urgently needed in developing countries for monitoring of human immunodeficiency virus (HIV) infection. Our aim was to test some crucial steps in the use of dried spots, i.e., viral recovery and storage over time. Moreover, we investigated whether dried plasma and blood spots (DPS and DBS, respectively) give comparable viral load (VL) results. Four manual RNA extraction methods from commercial HIV type 1 (HIV-1) VL assays—a QIAamp minikit (Qiagen), the Abbott Molecular sample preparation system, the Nuclisens assay (bioMarieux), and High Pure viral nucleic acid kit (Roche Applied Science)—were compared for VL quantification and PCR amplification for genotypic drug resistance testing on dried spots from spiked plasma and residual samples from HIV-1 patients (n = 47; median VL, 4.13 log10 copies/ml). RNA recovery from DPS was efficient using Nuclisens extraction (median difference, 0.03 log10 copies/ml) and slightly underestimated using the Abbott Molecular sample preparation system (median difference, 0.35 log10 copies/ml). PCR amplification results were in concordance. Measurements from DBS overestimated VL for plasma, with VL results showing <3.7 log10 copies/ml. VL was stable for up to 3 months in spiked DPS stored at 20°C but for only 1 month at 37°C. A faster decline was observed in PCR efficiency: DPS could be stored for 1 week at 37°C and for 1 month at 20°C. In conclusion, the RNA extraction method is an important factor in obtaining reliable RNA quantification and PCR amplification of HIV-1 on DPS/DBS. DBS could be used as an alternative for DPS depending on HIV RNA cutoffs for virological failure. VL measurements remain stable over a longer period than do PCR amplification results.
Due to the efforts of national programs and the support of a wide range of international partners, the number of people receiving antiretroviral therapy (ART) in resource-limited countries has increased significantly over recent years, most notably in sub-Saharan Africa (24). In order to limit the emergence of resistance to antiretroviral drugs, human immunodeficiency virus (HIV) treatment should ideally be accompanied by periodic virological monitoring, such as viral load and resistance testing. However, in resource-limited countries, viral load monitoring and drug resistance tests are not yet available at an affordable cost, and infrastructure requirements limit the scale-up in access to these tests. Treatment initiation and modifications are thus guided mainly by clinical disease progression and by CD4 cell counts, when possible (13, 24). As a consequence, many people with virological failure stay on inadequate drug regimens for long periods. Rapid or uncontrolled emergence of HIV drug resistance is thus feared as a potential consequence of ART scale-up in resource-limited countries. Therefore, ART resistance monitoring is not recommended at the individual patient level in low-income countries, and the WHO has established a system for the surveillance of transmitted acquired resistance to ART (5).
Although plasma is considered optimal for viral load and genotypic drug resistance testing, collection and shipment of plasma are often not feasible in many resource-limited settings, especially in semirural or rural areas, due to cold chain constraints. A major advantage of sampling blood or plasma as dried spots on absorbent paper is that samples can be shipped easily and safely and that no cold chain is required for preservation. Dried blood, plasma, and serum spots (DBS, DPS, and DSS, respectively) have been tested for HIV serology (4, 8, 22), molecular diagnosis (11), CD4+ lymphocyte enumeration (18), and more recently, viral RNA quantification (1-3, 10, 11, 15, 16, 23) and genotypic drug resistance (6, 17, 19, 26). However, the use of dried spots can be recommended only as long as the results are comparable to those obtained with fresh or frozen plasma. Several studies showed the feasibility of viral RNA quantification and genotypic drug resistance testing, although with different performances. The heterogeneity in the methods used for the elution and extraction of viral RNA and for quantification and amplification for genotypic drug resistance testing does not always allow the comparison of different protocols and the description of their advantages and limitations. In addition, the majority of studies focus on the use of dried spots for viral load or resistance testing only. However, adequate monitoring of patients on ART includes viral load but also drug resistance testing, and it is important to examine if both tests can be done on the same spots.
In this study, we compared DBS and DPS with regard to the impact of viral recovery and long-term storage conditions on viral load measurements and PCR amplification for genotypic drug resistance testing. We also studied to what extent DBS can be used for viral load monitoring, because blood is often easier to collect and to use for spots than plasma, especially in small laboratories or health care centers in resource-limited countries with limited infrastructures.
MATERIALS AND METHODS
Samples and preparation of DPS and DBS.
Two sets of samples were studied. First, we evaluated the different assays with DPS prepared from spiked plasma samples. A laboratory strain, HIV type 1 (HIV-1) CRF02-AG (mp642; obtained in 1997 in Cameroon), was diluted in human plasma negative for HIV, and three different 10-fold dilutions were quantified (see Results) and used for spot preparation. In addition, residual plasma and whole blood samples were collected from 47 HIV-infected patients attending the University Hospital of Montpellier, France, between April and May 2008. These samples encompassed viral loads ranging from 0 to 6 log10 copies/ml; 20 of these patient samples had viral loads of <4 log10 copies/ml, 18 had loads between 4 and 5 log10 copies/ml, and 9 had loads of >5 log10 copies/ml.
DPS and DBS were prepared according to a consensus of frequently used protocols described in previous studies (3, 10, 17). Briefly, 50 μl of blood or plasma was spotted on 903 filter paper (Schleicher & Schuell) and dried at room temperature for 3 h. Spots were then placed individually in plastic bags and stored in a hermetic box containing silica desiccant. In order to assess the impact of different storage conditions over time, spiked DPS samples were stored in parallel at 20°C in the laboratory (dry atmosphere) and, to simulate more extreme conditions of temperature and humidity, in a 37°C incubator containing trays of water to maintain a high relative humidity. These two subsets of DPS were analyzed after 1, 2, 4, 8, and 12 weeks.
Viral RNA extraction.
Four different manual RNA extraction methods were compared. The QIAamp viral RNA minikit (Qiagen, Courtaboeuf, France) combines a silica gel-based membrane with the speed of microspin centrifugation. The Abbott sample preparation system (Abbott Molecular, Rungis, France) is an iron particle-based method used for the Abbott RealTime HIV-1 commercial assay for viral load determination. The Nuclisens manual extraction kit (bioMérieux, Craponne, France) is based on the use of silica particles as described by Boom et al. (9); this technology is used for the Nuclisens EasyQ HIV-1 commercial viral load assay. Finally, the High Pure viral nucleic acid kit (Roche Applied Science, Meylan, France), based on column extraction, is to be used with the HIV-1 Cobas TaqMan or Amplicor Monitor commercial assay.
RNAs were extracted from 200-μl liquid plasma samples following the manufacturer's instructions and were eluted in 60 μl of elution buffer. For each extraction method with DPS, two plasma or whole blood spots of 50 μl were extracted according to the instructions of the manufacturer, except for the lysis steps, which were adapted slightly. Elution from spots was performed by cutting spots into two to four pieces that were subsequently incubated with the kit lysis buffer, using 2 ml for QIAamp viral RNA minikit (Qiagen) and High Pure viral nucleic acid kit (Roche) extractions, 3 ml for Abbott sample preparation system extraction, and 9 ml for Nuclisens manual extraction kit (bioMérieux) extraction. After 2 h of incubation at room temperature under gentle agitation, the supernatants were clarified by centrifugation at 1,500 × g for 2 min. RNAs were then extracted according to the instructions of the corresponding extraction method and eluted in 60 μl of elution buffer. Extracted RNAs were stored at −80°C until subsequent use for RNA quantification and PCR amplification. In order to increase the chances of obtaining PCR amplification and viral load measurements after long-term conservation, RNAs were extracted from four spots instead of two spots after 1 month of storage.
Viral load determination.
HIV-1 viral RNA loads were quantified for all samples by use of the same assay, ANRS G2 long terminal repeat-based real-time reverse transcriptase PCR (RT-PCR), which is commercially available under the name “Generic HIV Charge Virale” (Biocentric, Bandol, France) (20, 21). Viral loads were measured following the manufacturer's instructions. Amplification and data collection were carried out using the ABI Prism 7000 sequence detection system (Applied Biosystems). This quantification assay is a real-time TaqMan RT-PCR test with a lower detection limit of 300 copies/ml (2.5 log10 copies/ml). The viral load values were reported as log10-transformed copy numbers of HIV-1 RNA per ml.
PCR amplification of PR and partial RT for genotypic drug resistance testing.
Nested RT-PCR was used to amplify the protease (PR) and RT regions of the pol gene, yielding fragments of 507 and 798 bp, respectively, using published methods, i.e., the ANRS protocol (19; http://www.hivfrenchresistance.org/). Briefly, each RT-PCR was performed with 10 μl of RNA, using the Superscript one-step RT-PCR method for long templates (Invitrogen Life Technologies). Two or 5 μl of the first-round amplification product was then used for nested PCR, using a HotStartTaq master mix kit (Qiagen). Amplification products were visualized by 1% agarose gel electrophoresis with ethidium bromide staining.
Statistical analysis.
For the purpose of viral load analysis, undetectable samples were considered equal to zero, while samples that were detectable but below the detection limit of the assay were recorded as the cutoff value (2.48 log10 copies/ml). The viral load measurements obtained for DPS/DBS extracted with the different extraction kits were compared to the results obtained for liquid plasma (n = 47) by using the Bland-Altman approach (7), in which the differences between individual viral load results from liquid plasmas and spots are plotted against the mean. In addition, a Wilcoxon matched-pair signed-rank test was performed using Stata 10.0 software (Stata Corporation, College Station, TX). The same methods were used to compare viral loads from DBS and DPS of patient samples (n = 39). P values of <0.05 were considered to be significant.
RESULTS
Comparison of different RNA extraction methods for RNA quantification and PCR amplification of HIV-1 for a panel of spiked plasmas and DPS.
Because the recovery of RNA from dried spots is most likely one of the major critical steps in detecting viral RNA, the main objective of this study was to compare the abilities of four different manual viral RNA extraction kits to recover HIV-1 RNA from DPS in comparison to corresponding viral loads in plasma. Samples were also tested for PCR amplification of PR and RT genes for subsequent genotypic drug resistance testing.
(i) Viral load.
We first studied whether the different RNA extraction methods/kits have an impact on viral load quantification by a generic HIV-1 viral load kit. Use of a QIAamp viral RNA mini kit is the extraction method provided with the generic HIV-1 viral load kit. HIV-1 RNA was thus extracted from 200 μl of spiked plasma samples containing three different dilutions of HIV-1, using RNA extraction kits commercially available from Abbott, bioMérieux, and Roche. Viral loads were then determined according to the instructions of the Biocentric assay and compared to the results obtained after extraction with a QIAamp viral RNA mini kit in plasma (Table 1). With the exception of one sample, the differences in viral loads observed with the different extraction methods ranged between −0.02 and 0.31 log10 copies/ml only and corresponded to values observed for inter- and intra-assay variability (20). These results show that the RNA extraction method has no significant influence on the quantification of HIV-1 RNA in plasma with the Biocentric assay.
TABLE 1.
HIV-1 RNA loads in spiked plasma samples for three dilutions of HIV-1, measured in duplicate by a Biocentric kit after RNA extraction with four RNA extraction kits
| Extraction kit (manufacturer) | Mean plasma log10 copies/ml ± SD (log difference compared to Qiagen kit) at indicated dilution
|
||
|---|---|---|---|
| 3.30 ± 0.40 | 4.31 ± 0.11 | 5.32 ± 0.15 | |
| QIAamp viral RNA mini kit (Qiagen) | 3.38 ± 0.02 | 4.44 ± 0.12 | 5.42 ± 0.02 |
| Sample preparation system (Abbott) | 3.40 ± 0.12 (−0.02) | 4.20 ± 0.25 (0.25) | 5.10 ± 0.15 (0.31) |
| Nuclisens manual extraction kit (bioMérieux) | 3.69 ± 0.06 (−0.31) | 4.34 ± 0.17 (0.11) | 5.42 ± 0.06 (−0.01) |
| High Pure viral nucleic acid kit (Roche) | 2.74 ± 0.24 (0.64) | 4.26 ± 0.34 (0.19) | 5.33 ± 0.19 (0.09) |
DPS were then prepared from three different dilutions of spiked plasma samples containing 3.30, 4.31, and 5.32 log10 copies/ml according to the different viral load measurements obtained above (Table 1). For each dilution, viral RNAs were extracted from two DPS (2 × 50 μl) with the four extraction kits (Qiagen, bioMérieux, Abbott, and Roche). The assays were repeated four times for the spots prepared with 3.30 and 5.32 log10 copies/ml and twice for the 4.31 log10 copies/ml dilution. The results are summarized in Table 2. For the lowest viral load, at 3.30 log10 copies/ml, no quantification was possible after extraction with a High Pure viral nucleic acid kit (Roche), and viral RNA was detected in only one of the four extractions with a QIAamp viral RNA mini kit, although with a 0.62-log10 copies/ml decrease compared to the level obtained from plasma. After extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit, viral loads could be measured in 4/4 and 3/4 attempts, respectively. Compared to the corresponding plasmas, the viral loads in DPS samples were slightly decreased after extractions with the Abbott sample preparation system (0.42 log10 copies/ml) and not after those with the Nuclisens manual extraction kit (0.05 log10 copies/ml). The DPS prepared with 4.31 and 5.32 log10 copies/ml could be amplified and quantified, except for one. Importantly, the viral loads in DPS were significantly lower than those in plasma after extractions with the QIAamp viral RNA mini kit (decreases of 1.42 and 1.50 log10 copies/ml, respectively) and the High Pure viral nucleic acid kit (decreases of 1.68 and 2.04 log10 copies/ml, respectively). In contrast, differences were lower for extractions with the Abbott sample preparation system (decreases of 0.70 and 0.74 log10 copies/ml, respectively) and the Nuclisens manual extraction kit (differences of −0.01 and 0.24 log10 copies/ml, respectively).
TABLE 2.
HIV-1 viral loads in DPS after RNA extraction with four different RNA extraction kits
| Spiked plasma dilutiona (log10 copies/ml) | DPS extracted by QIAamp viral RNA mini kit (Qiagen)b
|
DPS extracted by Abbott sample preparation system
|
DPS extracted by Nuclisens manual extraction kit (bioMérieux)b
|
DPS extracted by High Pure viral nucleic acid kit (Roche)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VL | Mean | Plasma − DPSc | VL | Mean | Plasma − DPSc | VL | Mean | Plasma − DPSc | VL | Mean | Plasma − DPSc | |
| 3.30 | — | 2.85 | 2.88 | 0.42 | — | 3.25 | 0.05 | — | ||||
| 2.68 | NA | 0.62 | 3.15 | 2.62 | — | |||||||
| — | 2.77 | 3.57 | — | |||||||||
| — | 2.76 | 3.56 | — | |||||||||
| 4.31 | 2.83 | 2.89 | 1.42 | 3.58 | 3.61 | 0.70 | 4.22 | 4.06 | 0.24 | — | ||
| 2.95 | 3.64 | 3.91 | 2.63 | NA | 1.68 | |||||||
| 5.32 | 3.88 | 3.82 | 1.50 | 4.54 | 4.58 | 0.74 | 5.39 | 5.33 | −0.01 | 3.47 | 3.28 | 2.04 |
| 3.98 | 4.48 | 4.95 | 3.71 | |||||||||
| 3.58 | 4.67 | 5.47 | 2.68 | |||||||||
| 3.84 | 4.64 | 5.51 | 3.25 | |||||||||
DPS were prepared with three different dilutions of human HIV-negative plasma spiked with HIV-1 (Table 1) and were tested in duplicate, and the samples with the lowest and highest viral loads were tested twice in duplicate.
—, undetectable; NA, not applicable.
Difference in log10 copies/ml between the viral loads in plasma and DPS.
(ii) PCR amplification of PR and RT.
Table 3 shows the amplification results for the PR and RT regions of the pol gene for the same DPS extracts as those tested above for viral load. The PR and RT fragments could be amplified successfully from all dilutions after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit, except for one of the two PR amplifications for the lowest dilutions after extraction with the Abbott sample preparation system. After extraction with the High Pure viral nucleic acid kit, amplification was not possible from DPS at 3.30 log10 copies/ml; the highest dilutions were amplified, but without correct reproducibility, for the 4.31 log10 copies/ml dilution. None of the lowest dilutions (3.30 and 4.31 log10 copies/ml) and only one of the two highest dilutions were amplified after extraction with the QIAamp viral RNA mini kit.
TABLE 3.
Amplification of RT and PR regions of the HIV-1 pol gene from DPS prepared with three dilutions of spiked HIV-1 and tested in duplicate after RNA extraction with four RNA extraction kits
| Dilution (log10 copies/ml) | Region amplified | Amplification with RNA extraction kita
|
|||
|---|---|---|---|---|---|
| QIAamp viral RNA mini kit (Qiagen) | Sample preparation system (Abbott) | Nuclisens manual extraction kit (bioMérieux) | High Pure viral nucleic acid kit (Roche) | ||
| 3.30 | RT | − | + | + | − |
| − | + | + | − | ||
| PR | − | − | + | − | |
| − | + | + | − | ||
| 4.31 | RT | − | + | + | − |
| − | + | + | + | ||
| PR | − | + | + | − | |
| − | + | + | + | ||
| 5.32 | RT | + | + | + | + |
| − | + | + | + | ||
| PR | + | + | + | + | |
| − | + | + | + | ||
−, no amplification; +, amplification.
Although the number of tested samples was small, quantification and amplification results obtained with these DPS were concordant.
Comparison of different RNA extraction methods for RNA quantification and PCR amplification of HIV-1 for a panel of patient samples.
In order to confirm to what extent HIV-1 RNA was correctly recovered and quantified from DPS after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit (bioMérieux), we decided to extend our evaluation to real patient samples and used unlinked leftover samples from 47 patients followed at an HIV clinic of the University Hospital of Montpellier. Extraction with the High Pure viral nucleic acid kit (Roche) was excluded from the process because of the limited performance observed in the previous phase. However, we kept the QIAamp viral RNA mini kit (Qiagen) because the Biocentric viral load kit recommends this extraction method.
(i) Viral load.
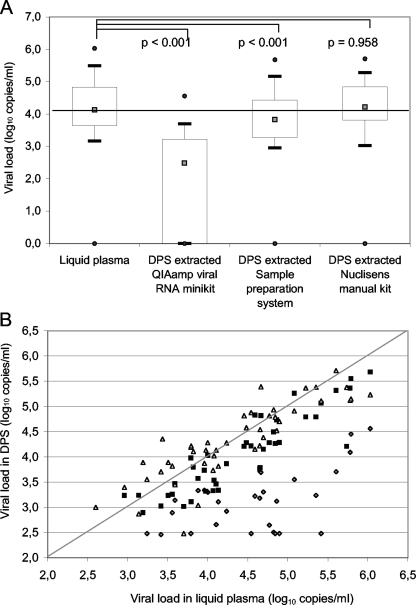
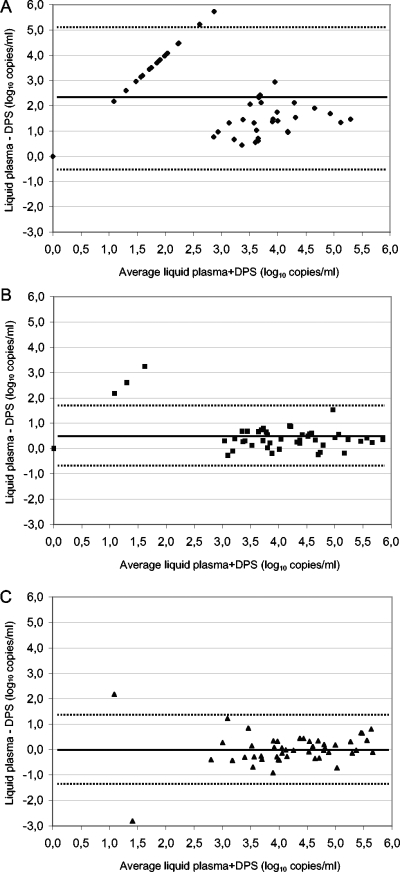
Figure 1 summarizes the results and shows the median viral loads in DPS compared to the viral loads in the corresponding plasmas for the three different extraction methods (Fig. 1A) and the individual results for each method (Fig. 1B). The median viral load in the 47 plasma samples was 4.13 log10 copies/ml (interquartile range, 3.59 to 4.84) and ranged from undetectable (n = 1) to 6.03 log10 copies/ml. The median viral load (2.48 log10 copies/ml) was significantly lower for corresponding DPS samples after extraction with the QIAamp viral RNA mini kit (P < 0.001) (Fig. 1A). Indeed, 16 (34%) of 47 samples were undetectable, and 7 of 47 (15%) were detectable but below the detection limit. These data were concordant with the Bland-Altman analysis (Fig. 2A), where 37 of 47 spot samples had a difference between plasma and DPS of >1 log10, 45 of 47 had a difference of >0.5 log10 copies/ml, and the median difference was 2.06 log10 copies/ml.
FIG. 1.
HIV-1 viral loads in patient samples (n = 47) of plasma and DPS after RNA extractions with a QIAamp viral RNA mini kit (Qiagen), the Abbott sample preparation system, or a Nuclisens manual extraction kit (bioMérieux). (A) Comparison of median viral loads from DPS with those in matched plasma samples. The gray squares represent medians, the boxes represent interquartile ranges, the whiskers represent lower and upper adjacent values, and the black dots represent outside values. (B) Comparison of viral loads in DPS and corresponding plasmas, only for samples with detectable viral loads, after RNA extraction with Qiagen (gray rhombuses), Abbott (black squares), and bioMérieux (white triangles) extraction kits. Viral loads are expressed as log10 copies/ml. P values were calculated by the Wilcoxon test by comparing liquid plasma and spots.
FIG. 2.
Bland-Altman analysis of HIV-1 viral loads in patient samples (n = 47) of plasma versus DPS after RNA extraction with a QIAamp viral RNA mini kit (Qiagen) (A), the Abbott sample preparation system (B), and a Nuclisens manual extraction kit (bioMérieux) (C). Horizontal black lines represent the mean difference, and dotted lines show the standard deviation.
A slight but significant underestimation from DPS after extraction with the Abbott sample preparation system (median viral load, 3.82 log10 copies/ml) was observed compared to plasma (P < 0.001) (Fig. 1A). However, with Bland-Altman analysis, 4 of 47 spot samples (Fig. 2B) had a difference between plasma and DPS of >1 log and 17 of 47 had a difference of >0.5 log10 copies/ml, but the overall median difference was only 0.35 log10 copies/ml (ranging from −0.27 to 3.24 log10 copies/ml).
After RNA extraction from DPS with the Nuclisens manual extraction kit, the different analyses confirmed that the RNA quantification was not different from the corresponding viral loads in plasma. The median viral load in DPS was 4.22 log10 copies/ml and was not different from that in plasma (P = 0.958) (Fig. 1A). The Bland-Altman analysis (Fig. 2C) showed that only 3 of 47 spot samples had a difference between plasma and DPS of >1 log10, 10 of 47 had a difference of >0.5 log10 copies/ml, and the median difference was −0.03 log10 copies/ml (ranging from −2.82 to 2.18). One plasma sample had a viral load below the detection limit, but after extraction from DPS with the Nuclisens manual extraction kit, 657 copies/ml were quantified.
One false-negative result was obtained for DPS extracted with the Nuclisens manual extraction kit, and three were obtained with the Abbott sample preparation system, but these four samples had low plasma viral loads (<2,000 copies/ml in plasma).
(ii) PCR amplification of RT.
Table 4 shows the amplification of the RT region (700 bp) of the pol gene for a subset of 20 of the 47 patients mentioned above, with plasma viral loads ranging between 3.14 and 4.86 log10 copies/ml. The RT fragment could be amplified from a single DPS only after extraction with a QIAamp viral RNA mini kit. All samples with viral loads of ≥4.0 log10 copies/ml could be amplified after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit. With plasma viral loads of <4.0 log10 copies/ml, all except one DPS could be amplified after extraction with the Nuclisens manual extraction kit versus only one with the Abbott sample preparation system.
TABLE 4.
Amplification of RT region of the HIV-1 pol gene after RNA extraction of plasmas and DPS obtained from 20 patients
| Viral load in plasma (log10 copies/ml) | RT region amplificationa
|
|||
|---|---|---|---|---|
| Plasma | DPS extracted by QIAamp viral RNA mini kit (Qiagen) | DPS extracted by Abbott sample preparation system | DPS extracted by Nuclisens manual extraction kit (bioMérieux) | |
| 3.14 | + | − | − | + |
| 3.19 | + | − | + | + |
| 3.42 | − | − | − | − |
| 3.56 | + | − | − | + |
| 3.59 | + | − | − | + |
| 3.70 | + | − | − | + |
| 3.88 | + | + | − | + |
| 4.00 | − | − | + | + |
| 4.07 | + | − | + | + |
| 4.08 | + | − | + | + |
| 4.10 | + | − | + | + |
| 4.23 | + | − | + | + |
| 4.59 | + | − | + | + |
| 4.65 | + | − | + | + |
| 4.65 | + | − | + | + |
| 4.86 | + | − | + | + |
| 5.08 | + | − | + | + |
| 5.78 | + | − | + | + |
| 5.79 | + | − | + | + |
| 6.03 | + | − | + | + |
+, amplification; −, no amplification.
Comparison of viral load measurements from DPS and DBS.
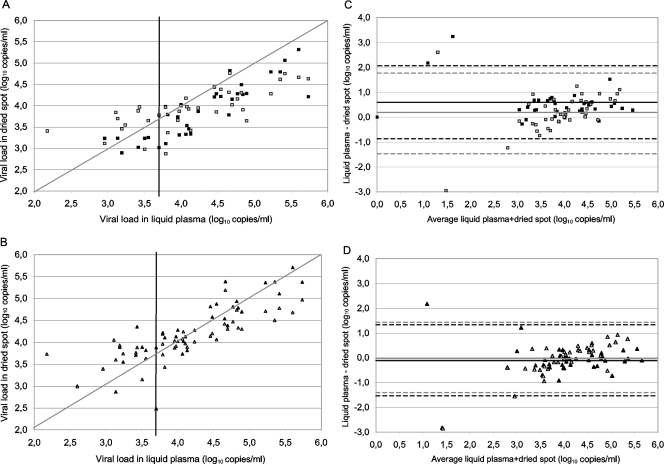
Enough blood was available in 39 of the 47 patient samples to prepare DPS and DBS in parallel to allow for comparison. HIV-1 RNA was extracted from DBS with the Nuclisens manual extraction kit (bioMérieux) and the Abbott sample preparation system only, since extraction with the QIAamp viral RNA mini kit was significantly less efficient, as shown above. The viral loads observed in DBS were then compared to those obtained from corresponding DPS with the same extraction method and with viral loads in the corresponding plasma samples. The results for the 39 patients (viral loads of 0 to 5.73 log10 copies/ml; median viral load = 4.07 log10 copies/ml) are shown in Fig. 3. No false-negative results were obtained from DBS extracted with the Nuclisens manual extraction kit, and only one sample with a low viral load in plasma (400 copies/ml) was negative after extraction with the Abbott sample preparation system, but the viral load value for this sample was close to the detection limit of the assay. Interestingly, the single plasma sample having an undetectable viral load could be quantified from DBS with both extraction methods (889 and 708 copies/ml after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit, respectively), which most likely corresponds to the proviral DNA present in the whole blood.
FIG. 3.
HIV-1 viral loads in patient samples (n = 39) of plasma versus DPS (black) and DBS (gray). HIV-1 RNA was measured in DPS and DBS with detectable viral loads after RNA extraction with the Abbott sample preparation system (A) and a Nuclisens manual extraction kit from bioMérieux (B) and then related to viral loads in plasma. Black vertical lines indicate 3.70 log10 copies/ml in liquid plasma. Bland-Altman analysis was performed for spots after RNA extraction with the RNA extraction kits of Abbott (C) and bioMérieux (D). Horizontal lines represent the mean difference, and dotted lines show the standard deviation (in black for DPS and in gray for DBS).
The trend for RNA quantifications obtained from DBS (Fig. 3) was similar to that observed with DPS. DBS extracted with the Abbott sample preparation system (median viral load = 3.91 log10 copies/ml) (Fig. 3A and C) gave slightly (difference of 0.18) but significantly (P = 0.032) underestimated viral loads, whereas viral loads for DBS extracted with the Nuclisens manual extraction kit (median viral load = 4.02 log10 copies/ml) (Fig. 3B and D) were not statistically different (P = 0.676) from viral loads measured in the corresponding plasmas. The Bland-Altman analysis (Fig. 3C and D) showed that 5 of 39 and 2 of 39 spots had differences between plasma and DBS of >1 log10 and that 16 of 39 and 8 of 39 had differences of >0.5 log10 copies/ml after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit, respectively.
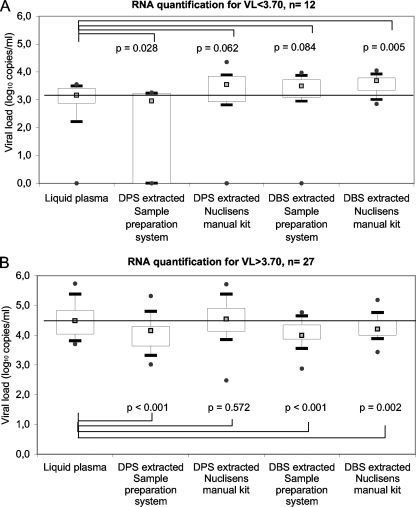
However, in Fig. 3A and B, which show comparisons between viral loads in DBS and DPS versus plasma, viral loads in DBS samples were overestimated when plasma viral loads were below 3.5 to 4.0 log10 copies/ml. This observation suggests that the presence of proviral DNA can influence the viral load results, especially in samples with low viral loads. Therefore, we stratified the samples into two groups according to viral loads in plasma: one group consisted of 12 patient samples with plasma viral loads below 3.70 log10 copies/ml, and the second group consisted of 27 samples with plasma viral loads above 3.70 log10 copies/ml (Fig. 4). We found that the viral loads obtained from DBS prepared from patient samples with viral loads of <3.70 log10 copies/ml and extracted with the Abbott sample preparation system were slightly overestimated (median difference between plasma and DBS = −0.38 log10 copies/ml; P = 0.084) and had, as a consequence, a statistical difference between viral loads measured in DBS and in DPS (P = 0.025). Similarly, quantification from DBS samples extracted with the Nuclisens manual extraction kit was significantly overestimated compared to that from plasma, with a median difference of −0.49 log10 copies/ml (P = 0.005). On the other hand, for samples with plasma viral loads of >3.70 log10 copies/ml, a slight but significant underestimation (median difference = 0.28 log10 copies/ml; P = 0.002) was observed between DBS and plasma after extraction with the Nuclisens manual extraction kit, similar to what was previously observed for extraction with the Abbott sample preparation system.
FIG. 4.
HIV-1 viral loads (log10 copies/ml) in DPS and DBS after RNA extraction with the Abbott sample preparation system or a Nuclisens manual extraction kit from bioMérieux for patient samples stratified according to plasma viral load. (A) Viral loads in plasma of <3.70 log10 copies/ml. (B) Viral loads in plasma of >3.70 log10 copies/ml. P values are calculated by the Wilcoxon test by comparing liquid plasma and spots. The gray squares represent medians, the boxes represent interquartile ranges, the whiskers represent lower and upper adjacent values, and the black dots represent outside values.
Effect of storage conditions on HIV-1 quantification and PCR amplification with DPS.
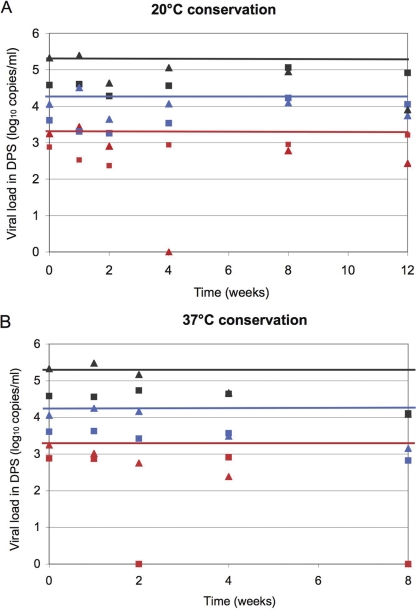
The conservation of viral RNA is another major crucial step for the successful use of dried spots. Two storage conditions of DPS prepared from the three different dilutions of spiked plasma samples, namely, 20°C in a dry atmosphere and 37°C in a humid atmosphere, were tested for conservation for up to 3 months for RNA quantification (Fig. 5) and PCR amplification (Table 5). Samples were tested in duplicate.
FIG. 5.
HIV-1 viral loads over time and under different storage conditions, evaluated from DPS (duplicate mean) prepared from spiked plasma samples with 3.30 (red), 4.31 (blue), and 5.32 (gray) log10 copies/ml and after RNA extraction with the Abbott sample preparation system (squares) and a Nuclisens manual extraction kit (bioMérieux) (triangles). (A) Storage conditions of 20°C and a dry atmosphere. (B) Storage conditions of 37°C and a humid atmosphere.
TABLE 5.
Amplification of RT and PR regions of the HIV-1 pol gene from each DPS, in duplicate, for three dilutions of spiked HIV-1 after RNA extraction over time and at different ambient temperatures and conditions
| Plasma viral load (log10 copies/ml) | Extraction kit | Amplification of indicated regionb
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time 0a
|
1 wk
|
2 wk
|
1 mo
|
2 mo
|
3 mo at 20°C
|
||||||||||||||||
| 20°C
|
37°C
|
20°C
|
37°C
|
20°C
|
37°C
|
20°C
|
37°C
|
||||||||||||||
| RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | RT | PR | ||
| 3.30 | Sample preparation system (Abbott) | + | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Sample preparation system (Abbott) | + | + | − | + | − | − | − | − | − | − | − | + | − | − | − | + | ND | ND | − | − | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | + | − | + | + | − | + | − | − | − | − | − | − | − | − | − | − | |
| 4.31 | Sample preparation system (Abbott) | + | + | + | + | + | + | − | + | − | + | − | + | − | − | − | + | − | − | − | + |
| Sample preparation system (Abbott) | + | + | + | + | + | + | + | − | − | − | + | + | − | − | − | + | − | − | − | + | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | + | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | + | + | + | + | + | + | − | + | − | − | − | + | − | − | + | + | |
| 5.32 | Sample preparation system (Abbott) | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − |
| Sample preparation system (Abbott) | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | + | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | − | − | − | + | |
| Nuclisens manual extraction kit (bioMérieux) | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | − | − | − | + | |
Results are from Table 3.
+, amplification; −, no amplification; ND, not done.
HIV-1 RNA quantification with DPS over time was comparable to initial plasma viral loads when spots were stored at 20°C for 2 months and, to a lesser extent, until 3 months. Median differences, in log10 copies/ml, between plasma and DPS for the three dilutions over time were 0.71 (range, 0.1 to 1.04) and 0.36 (range, 0.1 to 1.41) after extractions with the Abbott sample preparation system and the Nuclisens manual extraction kit, respectively, and are thus similar to the differences observed at time zero for the different dilutions (Table 2). The most important differences were observed for the lowest viral loads. Importantly, the capacity to amplify the RT and PR fragments was not reproducible after 1 week of conservation for the sample with the lowest viral load and decreased significantly after 1 month of conservation for the two other dilutions, in particular for the longest PCR fragment (RT). At 37°C, viral load measurements from DPS started to decrease after 1 month of conservation for both extraction kits. This trend was confirmed by the results after 2 months of conservation; the lowest dilution could not be quantified any more, and median differences compared with plasma viral loads decreased, with 1.48 and 1.21 log10 copies/ml after extraction with the Abbott sample preparation system and 1.15 and 1.23 log10 copies/ml after extraction with the Nuclisens manual extraction kit for the two other dilutions. The capacity to amplify RT fragments from DPS decreased rapidly after 1 week of storage at 37°C.
DISCUSSION
In this study, we showed that the choice of the method used to recover viral RNA from DPS or DBS is critical for viral load measurement and PCR amplification. We also showed the limits of DBS versus DPS for viral load measurements and the difficulties of DPS storage over time under extreme environmental conditions.
Efficiency of viral RNA recovery from DPS.
First of all, we evaluated the impact of RNA extraction methods on the efficiency of HIV-1 RNA recovery. In contrast to previous studies, which generally report on a single commercial HIV viral load assay or compare two viral load assays, we assessed the role of the extraction step in subsequent HIV-1 RNA measurements by using the same HIV viral load assay after each different extraction method. For DPS prepared from spiked plasma samples, we observed a strong decline in HIV-1 RNA recovery after extractions with the QIAamp viral RNA mini kit (Qiagen) and the High Pure viral nucleic acid kit (Roche) but not after extractions with the Abbott sample preparation system or the Nuclisens manual extraction kit (bioMérieux). Moreover, the results for viral load were concordant with the PCR amplification results. Previous studies using Nuclisens commercial viral load assays (nucleic acid sequence-based amplification technology) (1, 3, 15) also reported a good correlation between plasma and DPS or DBS. However, in a previous report on the use of the Amplicor HIV-1 Monitor 1.5 assay (Roche) (2) with DPS, a relatively important decrease in viral load was observed compared to the results for plasma. This observation, in agreement with our results, suggests that this could be related to the extraction method. Studies have reported the use of QIAamp viral RNA extraction kits for genotypic drug resistance testing (12, 19), but never for viral load measurements, with DPS/DBS. However, in these studies, the incubation of the spots was performed in a special buffer before the lysis step with the buffer from the kit to improve the quality of the extracted RNA and subsequent amplification results. The compositions of the lysis buffers in the various extraction kits could explain the differences observed in RNA recovery from DPS. The most important loss in RNA recovery was obtained after extraction using the Qiagen and Roche kits, which use column extraction-based techniques.
The assays of 47 patient samples confirmed our initial observations with spiked DPS for the QIAamp viral RNA mini kit (median difference of 2.06 log10 copies/ml) and allowed a better comparison between the performances of the Abbott and bioMérieux methods. Viral load measurement from DPS after extraction by the Abbott sample preparation system showed a slight underestimation, with a median difference of 0.35 log10 copies/ml. In contrast, the viral load quantifications in DPS extracted by the Nuclisens manual extraction kit were comparable to those in plasma, and the differences between plasma and DPS were uniformly distributed around 0, with the lowest median difference of −0.03 log10 copies/ml. Only one study reported the use of the Abbott sample preparation system until now (C. Garrido, N. Zahonero, V. Soriano, and C. De Mendoza, CROI poster 926, Boston, MA, 2008) and compared the bioMérieux (Nuclisens Easy HIV-1) and Abbott (RealTime HIV-1) commercial assays for viral load quantification. For both techniques, a good correlation was observed between DPS and fresh plasma, but the highest correlation was seen using the bioMérieux assay, which is thus comparable to our results and is thus most likely due to the extraction method used.
In our study, the same trend between the two methods was also observed for PCR amplification. The lower limits for PCR amplification were 3 log10 copies/ml and 4 log10 copies/ml after extractions with the Nuclisens manual extraction kit and the Abbott sample preparation system, respectively. These detection limits are concordant with other published studies using Nuclisens (bioMérieux) and modified Qiagen extraction (6, 14, 16, 17, 19).
Whole blood versus plasma spots for measurement of HIV-1 viral load.
Because under field conditions DBS are easier to prepare than DPS and can be collected by a simple finger prick and spotted directly onto filter paper, we also compared viral load measurements from DBS and DPS with those from plasmas obtained from 39 HIV-1-seropositive patients. Pooling all the results together, HIV-1 viral loads from DBS were not dramatically different from the plasma viral loads. However, a more detailed analysis, taking into account the plasma viral loads, showed a low concordance between DBS and plasma when viral loads were below 5,000 copies/ml. Only few studies have compared viral load measurements from DBS and DPS (1, 15, 23). For 300 DBS samples from patients on ART in Uganda (23), a large number of false-positive results was reported when viral loads were low. Our results confirm these observations, and we interpret this difference as probably related to the presence of proviral DNA, leading to an overestimation compared to the plasma viral load.
Storage of DPS.
The facility to prepare, transport, and store dried spots is the major advantage of this sample support. Different studies have shown that dried spots can be kept for long periods when refrigerated or frozen in hermetic bags with desiccant, such as 1 year at 4°C (25) and 4 years at −20°C (17) for genotypic drug resistance testing and at least 15 days at 4°C (1, 11) and 1 year at −70°C (10) for RNA quantification. These data are important but are only applicable to reference laboratories where the infrastructure for long-term storage is available. However, for application in the field, the limits of DBS or DPS storage under more extreme conditions are also important to know. It was reported in some studies that RNA quantifications from DPS and DBS were still possible after 7 to 15 days at 37°C (1, 11, 15) and after 3 weeks to 1 year at room temperature (10, 23). Results were also reported from DBS for PCR amplification for genotypic drug resistance testing for up to 3 months at 37°C (6) and 5 months at room temperature (26) but were not possible anymore after 5 years at room temperature (17). In resource-poor settings, the time between collection and shipment of dried spots could be several weeks or months, and conservation conditions in the field can vary between different geographical areas, but also in the same setting according to the seasons. In our study, the periods were chosen because under programmatic conditions in some low-income countries, most ART centers are expected to benefit from at least one supervision visit per quarter or should be able to ship the samples to a central laboratory with adequate equipment at one of the proposed periods. Plasma viral loads for a limited number of samples were stable for up to 2 months when samples were stored at 20°C and declined slightly after 3 months. However, at 37°C, viral loads remained stable for only 1 month. Importantly, for PCR amplification, a more rapid decline in PCR efficiency was seen: DPS could be stored for 1 week only at 37°C and for 1 month at 20°C. Differences in results for viral load measurement and PCR as a function of conservation conditions could be related to the different sizes of the amplified fragments in both assays. Our results, especially those for PCR amplification, are somewhat lower than some previously reported, but the storage conditions may not have been completely identical. In our study, only a limited number of samples were analyzed, and these preliminary results need to be confirmed with a larger number of patient samples.
In conclusion, our study shows that the RNA extraction method from DPS or DBS is an important factor in obtaining reliable viral load and PCR amplification results for HIV-1. DBS and DPS give comparable results when viral loads are above 3.70 log10 copies/ml. We identified two extraction methods with relatively good performances, with the Nuclisens manual extraction kit (bioMérieux) being more accurate and having a better sensitivity than the Abbott sample preparation system. However, depending on the field conditions and the equipment available for viral load measurements, each laboratory should evaluate the advantages and limits of the methods they choose as a function of other factors, such as the prices of the tests and the performance of the subsequent viral load assay in detecting circulating HIV-1 variants. DBS could be used as an alternative for DPS if higher HIV RNA cutoffs for virological failure are set, for example, 10,000 versus 1,000 copies/ml, as recommended by WHO in an ART strategy for low-income countries (24). It is important that our study and previous reports compare viral loads in DBS obtained after venipuncture, and these results cannot readily be extrapolated to viral loads in whole blood after a single finger prick. Depending on the storage conditions, the viral load measurements remain stable for a longer time than PCR amplification results, and long-term storage at 37°C in a humid atmosphere must not be advised. Recommendations could be to collect dried spots, store them at ambient temperature with a desiccant, and ship spots rapidly to a laboratory where they could be stored at +4°C or −20°C. Additional studies of on-site storage conditions, transport under various conditions, and subsequent storage at the reference laboratory are needed to determine the stability over time for viral load and genotypic drug resistance testing.
Acknowledgments
This work was financially supported by the IHC Project of the International Union against Tuberculosis and Lung Diseases through a grant from the European Commission (SANTE 2004/078/547/1).
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Alvarez-Munoz, M. T., S. Zaragoza-Rodriguez, O. Rojas-Montes, G. Palacios-Saucedo, G. Vazquez-Rosales, A. Gomez-Delgado, J. Torres, and O. Munoz. 2005. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch. Med. Res. 36382-386. [DOI] [PubMed] [Google Scholar]
- 2.Amellal, B., C. Katlama, and V. Calvez. 2007. Evaluation of the use of dried spots and of different storage conditions of plasma for HIV-1 RNA quantification. HIV Med. 8396-400. [DOI] [PubMed] [Google Scholar]
- 3.Ayele, W., R. Schuurman, T. Messele, W. Dorigo-Zetsma, Y. Mengistu, J. Goudsmit, W. A. Paxton, M. P. de Baar, and G. Pollakis. 2007. Use of dried spots of whole blood, plasma, and mother's milk collected on filter paper for measurement of human immunodeficiency virus type 1 burden. J. Clin. Microbiol. 45891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barin, F., L. Meyer, R. Lancar, C. Deveau, M. Gharib, A. Laporte, J. C. Desenclos, and D. Costagliola. 2005. Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J. Clin. Microbiol. 434441-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertagnolio, S., I. Derdelinckx, M. Parker, J. Fitzgibbon, H. Fleury, M. Peeters, R. Schuurman, D. Pillay, L. Morris, A. Tanuri, G. M. Gershy-Damet, J. Nkengasong, C. F. Gilks, D. Sutherland, and P. Sandstrom. 2008. World Health Organization/HIVResNet drug resistance laboratory strategy. Antivir. Ther. 13(Suppl. 2)49-57. [PubMed] [Google Scholar]
- 6.Bertagnolio, S., L. Soto-Ramirez, R. Pilon, R. Rodriguez, M. Viveros, L. Fuentes, P. R. Harrigan, T. Mo, D. Sutherland, and P. Sandstrom. 2007. HIV-1 drug resistance surveillance using dried whole blood spots. Antivir. Ther. 12107-113. [PubMed] [Google Scholar]
- 7.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i307-310. [PubMed] [Google Scholar]
- 8.Boillot, F., M. Peeters, A. Kosia, and E. Delaporte. 1997. Prevalence of the human immunodeficiency virus among patients with tuberculosis in Sierra Leone, established from dried blood spots on filter paper. Int. J. Tuberc. Lung Dis. 1493-497. [PubMed] [Google Scholar]
- 9.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brambilla, D., C. Jennings, G. Aldrovandi, J. Bremer, A. M. Comeau, S. A. Cassol, R. Dickover, J. B. Jackson, J. Pitt, J. L. Sullivan, A. Butcher, L. Grosso, P. Reichelderfer, and S. A. Fiscus. 2003. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J. Clin. Microbiol. 411888-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassol, S., M. J. Gill, R. Pilon, M. Cormier, R. F. Voigt, B. Willoughby, and J. Forbes. 1997. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J. Clin. Microbiol. 352795-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dachraoui, R., D. Brand, S. Brunet, F. Barin, and J. C. Plantier. 2008. RNA amplification of the HIV-1 Pol and Env regions on dried serum and plasma spots. HIV Med. 9557-561. [DOI] [PubMed] [Google Scholar]
- 13.Gilks, C. F., S. Crowley, R. Ekpini, S. Gove, J. Perriens, Y. Souteyrand, D. Sutherland, M. Vitoria, T. Guerma, and K. De Cock. 2006. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet 368505-510. [DOI] [PubMed] [Google Scholar]
- 14.Hallack, R., L. E. Doherty, J. A. Wethers, and M. M. Parker. 2008. Evaluation of dried blood spot specimens for HIV-1 drug-resistance testing using the Trugene((R)) HIV-1 genotyping assay. J. Clin. Virol. 41283-287. [DOI] [PubMed] [Google Scholar]
- 15.Kane, C. T., H. D. Ndiaye, S. Diallo, I. Ndiaye, A. S. Wade, P. A. Diaw, A. Gaye-Diallo, and S. Mboup. 2008. Quantitation of HIV-1 RNA in dried blood spots by the real-time NucliSENS EasyQ HIV-1 assay in Senegal. J. Virol. Methods 148291-295. [DOI] [PubMed] [Google Scholar]
- 16.Masciotra, S., C. Garrido, A. S. Youngpairoj, A. McNulty, N. Zahonero, A. Corral, W. Heneine, C. de Mendoza, and J. G. Garcia-Lerma. 2007. High concordance between HIV-1 drug resistance genotypes generated from plasma and dried blood spots in antiretroviral-experienced patients. AIDS 212503-2511. [DOI] [PubMed] [Google Scholar]
- 17.McNulty, A., C. Jennings, D. Bennett, J. Fitzgibbon, J. W. Bremer, M. Ussery, M. L. Kalish, W. Heneine, and J. G. Garcia-Lerma. 2007. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J. Clin. Microbiol. 45517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mwaba, P., S. Cassol, R. Pilon, C. Chintu, M. Janes, A. Nunn, and A. Zumla. 2003. Use of dried whole blood spots to measure CD4+ lymphocyte counts in HIV-1-infected patients. Lancet 3621459-1460. [DOI] [PubMed] [Google Scholar]
- 19.Plantier, J. C., R. Dachraoui, V. Lemee, M. Gueudin, F. Borsa-Lebas, F. Caron, and F. Simon. 2005. HIV-1 resistance genotyping on dried serum spots. AIDS 19391-397. [DOI] [PubMed] [Google Scholar]
- 20.Rouet, F., M. L. Chaix, E. Nerrienet, N. Ngo-Giang-Huong, J. C. Plantier, M. Burgard, M. Peeters, F. Damond, D. K. Ekouevi, P. Msellati, L. Ferradini, S. Rukobo, V. Marechal, N. Schvachsa, L. Wakrim, C. Rafalimanana, B. Rakotoambinina, J. P. Viard, J. M. Seigneurin, and C. Rouzioux. 2007. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J. Acquir. Immune Defic. Syndr. 45380-388. [DOI] [PubMed] [Google Scholar]
- 21.Rouet, F., D. K. Ekouevi, M. L. Chaix, M. Burgard, A. Inwoley, T. D. Tony, C. Danel, X. Anglaret, V. Leroy, P. Msellati, F. Dabis, and C. Rouzioux. 2005. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J. Clin. Microbiol. 432709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon, S. S., S. Pulimi, I. I. Rodriguez, S. K. Chaguturu, S. K. Satish Kumar, K. H. Mayer, and S. Solomon. 2004. Dried blood spots are an acceptable and useful HIV surveillance tool in a remote developing world setting. Int. J. STD AIDS 15658-661. [DOI] [PubMed] [Google Scholar]
- 23.Waters, L., A. Kambugu, H. Tibenderana, D. Meya, L. John, S. Mandalia, M. Nabankema, I. Namugga, T. C. Quinn, B. Gazzard, S. J. Reynolds, and M. Nelson. 2007. Evaluation of filter paper transfer of whole-blood and plasma samples for quantifying HIV RNA in subjects on antiretroviral therapy in Uganda. J. Acquir. Immune Defic. Syndr. 46590-593. [DOI] [PubMed] [Google Scholar]
- 24.WHO. 2006. HIV/AIDS programme. Antiretroviral therapy for HIV infection in adults and adolescents: recommendations for a public health approach. WHO, Geneva, Switzerland. www.who.int/hiv/pub/guidelines/adult/en/index.html. [PubMed]
- 25.Youngpairoj, A. S., S. Masciotra, C. Garrido, N. Zahonero, C. de Mendoza, and J. G. Garcia-Lerma. 2008. HIV-1 drug resistance genotyping from dried blood spots stored for 1 year at 4°C. J. Antimicrob. Chemother. 611217-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziemniak, C., A. George-Agwu, W. J. Moss, S. C. Ray, and D. Persaud. 2006. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. J. Virol. Methods 136238-247. [DOI] [PubMed] [Google Scholar]