Abstract
In this study we report on the development of a multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) method for the molecular typing of Mycoplasma pneumoniae. The genomic content of M. pneumoniae M129 was analyzed for VNTRs, and 5 of the 17 VNTRs identified were selected for use in an MLVA assay. The method was based on a GeneScan analysis of VNTR loci labeled with fluorescent dyes by multiplex PCR and capillary electrophoresis. This approach was applied to a collection of 265 isolates from various European countries, Japan, and Tunisia; and 26 distinct VNTR types were found. The VNTR assay was compared to the P1 adhesin PCR-restriction fragment length polymorphism (RFLP) typing method and showed a far better resolution than the P1 PCR-RFLP method. The discriminatory power of MLVA (Hunter-Gaston diversity index [HGDI], 0.915) for the 265 isolates was significantly higher than that of the P1 PCR-RFLP method (HGDI, 0.511). However, there was a correlation between the typing results obtained by MLVA and the P1 gene PCR-RFLP method. The potential value of MLVA of M. pneumoniae as an epidemiological tool is discussed, and the use of the VNTR markers in further investigations of the potential use of MLVA in outbreaks of M. pneumoniae infections is proposed.
Mycoplasma pneumoniae represents one of the most common etiological agents of community-acquired respiratory tract infections, with the clinical courses ranging from mild forms of pharyngitis and tracheobronchitis to severe cases of interstitial pneumonia. M. pneumoniae infections are responsible for 20% or more of all cases of community-acquired pneumonia (22) and occur endemically, with epidemic peaks occurring every 4 to 7 years. School-age children and young adults are the populations that are the most affected.
Strain subtyping by molecular methods is a powerful tool for surveillance and outbreak investigation. However, molecular typing is hampered by the fact that M. pneumoniae is a genetically homogeneous species and isolates are poorly differentiated by PCR-restriction fragment length polymorphism (RFLP) analysis of the P1 gene, the most common genotyping method. PCR-RFLP analysis of the gene encoding the P1 protein, a major adhesin that induces a strong humoral immune response during infection, enables the separation of isolates into two types, types 1 and 2 (1, 2, 17). More recent studies used repetitive regions, RepMp2/3 and RepMp4, which can be found in the P1 gene, for molecular typing. Besides the subtype classification, those methods allow the identification of variants of subtypes 1 and 2 (3, 5, 8).
In addition to these molecular typing methods that target only one gene, other methods based on the study of the whole genome have been adapted to M. pneumoniae, such as pulsed-field electrophoresis (1) and multilocus sequence typing (MLST) (4). Pulsed-field electrophoresis is able to differentiate strains into two groups (groups 1 and 2), like P1 gene PCR-RFLP analysis can, and can subdivide group 2 into two subgroups, subgroups 2a and 2b (1). Due to the homogeneity of the M. pneumoniae species, very little polymorphism can be found in M. pneumoniae housekeeping genes. Thus, the attempt to use MLST with housekeeping and structural genes was not useful for molecular typing (4). Among the new genotyping methods, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) has successfully been tested with many bacterial species (20). MLVA is a molecular typing method based on the variation in the copy number of tandemly repeated sequences, called VNTRs, found at different loci on the genome. The variation of the copy number of these tandem repeats (TRs) depends on the isolate tested. Recently, MLVA has proven its suitability for the subtyping of isolates of one animal Mycoplasma species, Mycoplasma mycoides subsp. mycoides SC (12). Furthermore, a multilocus genotyping system has recently been developed for the human species Mycoplasma genitalium, based in part on the analysis of short TR sequences (10).
As current genotyping methods have low discriminatory powers, an alternative method with a high discriminatory ability is needed to differentiate M. pneumoniae isolates according to geographical origin, specimen type, isolation date, and patient age or to differentiate a reinfection from a relapse. The present study aims to develop a new epidemiological tool based on whole-genome analysis, an MLVA scheme, by using VNTR loci selected from the sequence of the sole published genome of M. pneumoniae, that of strain M129, and to apply it to a large collection of isolates.
MATERIALS AND METHODS
Bacterial strains, culture, and DNA preparation.
A total of 265 M. pneumoniae strains were used in this study, including the 2 reference strains of types 1 and 2, M. pneumoniae M129 (ATCC 29342) and FH (ATCC 15531), respectively. Among the 263 clinical strains, 7 were isolated from extra-respiratory tract specimens. The isolates were from France (n = 210), Germany (n = 15), Denmark (n = 11), Japan (n = 9), Belgium (n = 8), Tunisia (n = 8), and Spain (n = 2) and were collected between 1962 and 2008. All except two of the isolates were single patient isolates. The strain collection included 253 fully macrolide-susceptible and 12 macrolide-resistant isolates (11, 14). The characteristics of the strains analyzed by the MLVA typing method are shown Table 1. The growth conditions used for the M. pneumoniae strains have been described previously (21). DNA was isolated with a MagNa Pure LC DNA isolation kit I (Roche Diagnostics, France), according to the manufacturer's instructions.
TABLE 1.
Characteristics of the 263 M. pneumoniae isolates used in this study
| Isolatea | Origin
|
Yr of isolation | P1 gene PCR-RFLP type | MLVA type | |
|---|---|---|---|---|---|
| Country | Source | ||||
| J7 | Germany | Respiratory tract | 1991 | 2 | B |
| J4 | Germany | Respiratory tract | 1991 | 2 | O |
| Ho | Germany | Respiratory tract | 1991 | 2a | B |
| J17 | Germany | Respiratory tract | 1992 | 1 | P |
| J19 | Germany | Respiratory tract | 1992 | 1 | P |
| J16 | Germany | Respiratory tract | 1992 | 1 | P |
| J13 | Germany | Respiratory tract | 1992 | 1 | P |
| J20 | Germany | Respiratory tract | 1992 | 1 | U |
| J11 | Germany | Respiratory tract | 1992 | 1 | X |
| J22 | Germany | Respiratory tract | 1993 | 1 | Z |
| Sta | Germany | Respiratory tract | 1995 | 2a | M |
| B2146 | Germany | Respiratory tract | 1996 | 2 | B |
| 79692 | Germany | Respiratory tract | 2000 | 2a | M |
| 79809 | Germany | Respiratory tract | 2000 | 2a | M |
| F59 | Germany | Respiratory tract | Unknown | 2a | V |
| M5 | Denmark | Respiratory tract | 1962 | 1 | P |
| M40 | Denmark | Respiratory tract | 1963 | 1 | P |
| M38 | Denmark | Respiratory tract | 1963 | 1 | Z |
| M62 | Denmark | Respiratory tract | 1964 | 1 | X |
| M547 | Denmark | Respiratory tract | 1967 | 2 | T |
| M1121 | Denmark | Respiratory tract | 1977 | 1 | J |
| M1873 | Denmark | Respiratory tract | 1987 | 2 | C |
| M2018b | Denmark | Respiratory tract | 1988 | 1 | P |
| M2155 | Denmark | Respiratory tract | 1990 | 1 | P |
| M4350 | Denmark | Respiratory tract | 1991 | 2 | V |
| 4817 | Denmark | Respiratory tract | 1993 | 1a | J |
| J173b | Japan | Unknown | Unknown | 1 | A |
| J186b | Japan | Unknown | Unknown | 1 | A |
| J380b | Japan | Respiratory tract | 2000-2003 | 1 | J |
| J381b | Japan | Respiratory tract | 2000-2003 | 1 | K |
| J378b | Japan | Respiratory tract | 2000-2003 | 2 | M |
| J377b | Japan | Respiratory tract | 2000-2003 | 2 | C |
| J376b | Japan | Respiratory tract | 2000-2003 | 2 | C |
| J382b | Japan | Respiratory tract | 2000-2003 | 1 | P |
| J379b | Japan | Respiratory tract | 2000-2003 | 2 | S |
| AV1 | Belgium | Unknown | 1992 | 2 | C |
| AV2 | Belgium | Unknown | 1992 | 1 | E |
| AV4 | Belgium | Unknown | 1992 | 1 | E |
| AV3 | Belgium | Unknown | 1992 | 1 | P |
| AV8 | Belgium | Unknown | 1993 | 1 | E |
| AV9 | Belgium | Unknown | 1993 | 1 | E |
| AV6 | Belgium | Unknown | 1993 | 1 | J |
| AV7 | Belgium | Unknown | 1993 | 1 | J |
| A131 | Tunisia | Respiratory tract | 2006 | 1 | P |
| A138 | Tunisia | Respiratory tract | 2006 | 1 | P |
| A167 | Tunisia | Respiratory tract | 2006 | 1 | P |
| A168 | Tunisia | Respiratory tract | 2006 | 1 | P |
| A195 | Tunisia | Respiratory tract | 2007 | 1 | P |
| A251 | Tunisia | Respiratory tract | 2007 | 1 | P |
| A340 | Tunisia | Respiratory tract | 2008 | 1 | P |
| A355 | Tunisia | Respiratory tract | 2008 | 1 | P |
| B2285 | Madrid, Spain | Pericardial fluid | 1995 | 1 | E |
| B2283 | Madrid, Spain | Pericardial fluid | 1995 | 1 | E |
| Pn19 | Bordeaux, France | Respiratory tract | 1992 | 1 | E |
| Pn25 | Bordeaux, France | Respiratory tract | 1992 | 1 | E |
| Pn18 | Bordeaux, France | Respiratory tract | 1992 | 1 | E |
| Pn15 | Bordeaux, France | Respiratory tract | 1992 | 1 | J |
| B699 | Bordeaux, France | Respiratory tract | 1992 | 1 | P |
| Pn22 | Bordeaux, France | Respiratory tract | 1992 | 1 | R |
| Pn21 | Bordeaux, France | Respiratory tract | 1992 | 1 | U |
| B718 | Bordeaux, France | Respiratory tract | 1993 | 1 | D |
| B833 | Bordeaux, France | Respiratory tract | 1993 | 1 | J |
| B773 | Bordeaux, France | Respiratory tract | 1993 | 2 | O |
| B4812 | Bordeaux, France | Respiratory tract | 1993 | 1 | P |
| B767 | Bordeaux, France | Respiratory tract | 1993 | 1 | X |
| B936 | Bordeaux, France | Respiratory tract | 1994 | 1 | J |
| B932 | Bordeaux, France | Cervix | 1994 | 1 | X |
| B2036 | Bordeaux, France | Respiratory tract | 1995 | 1 | E |
| B2329 | Bordeaux, France | Respiratory tract | 1996 | 1 | P |
| B2365 | Bordeaux, France | Respiratory tract | 1996 | 1 | U |
| B2359 | Bordeaux, France | Respiratory tract | 1996 | 1 | U |
| B2085 | Bordeaux, France | Respiratory tract | 1996 | 2 | W |
| B2483 | Bordeaux, France | Respiratory tract | 1997 | 1 | P |
| B2759 | Bordeaux, France | Respiratory tract | 1999 | 2 | H |
| B2882 | Bordeaux, France | Synovial fluid | 1999 | 2 | O |
| B2832 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2818 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2823 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2852 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2892 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2864 | Bordeaux, France | Respiratory tract | 1999 | 2 | O |
| B2854 | Bordeaux, France | Respiratory tract | 1999 | 1 | U |
| B2829 | Bordeaux, France | Respiratory tract | 1999 | 1 | U |
| B3063 | Bordeaux, France | Respiratory tract | 2000 | 1 | E |
| B2912 | Bordeaux, France | Respiratory tract | 2000 | 1 | J |
| B3041 | Bordeaux, France | Respiratory tract | 2000 | 2 | O |
| B3049 | Bordeaux, France | Respiratory tract | 2000 | 1 | P |
| B2918 | Bordeaux, France | Respiratory tract | 2000 | 1 | Q |
| B2934 | Bordeaux, France | Respiratory tract | 2000 | 2 | T |
| B3140 | Bordeaux, France | Respiratory tract | 2001 | 2 | C |
| B3163 | Bordeaux, France | Sternal wound | 2001 | 2 | C |
| B3188 | Bordeaux, France | Respiratory tract | 2001 | 1 | E |
| B3174 | Bordeaux, France | Respiratory tract | 2001 | 1 | E |
| B3148 | Bordeaux, France | Respiratory tract | 2001 | 2 | H |
| B3179 | Bordeaux, France | Respiratory tract | 2001 | 1 | U |
| B3168 | Bordeaux, France | Respiratory tract | 2001 | 1 | U |
| B3119 | Bordeaux, France | Respiratory tract | 2001 | 2 | V |
| B3357 | Bordeaux, France | Respiratory tract | 2002 | 2 | C |
| B3487 | Bordeaux, France | Respiratory tract | 2003 | 1 | A |
| B3568 | Bordeaux, France | Respiratory tract | 2003 | 1 | A |
| B3448 | Bordeaux, France | Respiratory tract | 2003 | 2 | C |
| B3570 | Bordeaux, France | Respiratory tract | 2003 | 1 | E |
| B3573 | Bordeaux, France | Respiratory tract | 2003 | 2 | H |
| B3436 | Bordeaux, France | Respiratory tract | 2003 | 2 | T |
| B3816 | Bordeaux, France | Respiratory tract | 2004 | 2 | B |
| B3786 | Bordeaux, France | Respiratory tract | 2004 | 2 | C |
| B3836 | Bordeaux, France | Respiratory tract | 2004 | 2 | C |
| B3887 | Bordeaux, France | Respiratory tract | 2004 | 1 | G |
| B3803 | Bordeaux, France | Respiratory tract | 2004 | 2 | H |
| B3648 | Bordeaux, France | Respiratory tract | 2004 | 2 | O |
| B3805 | Bordeaux, France | Respiratory tract | 2004 | 2 | O |
| B3676 | Bordeaux, France | Respiratory tract | 2004 | 2 | O |
| B3654 | Bordeaux, France | Respiratory tract | 2004 | 2 | O |
| B3627 | Bordeaux, France | Respiratory tract | 2004 | 2 | O |
| B3766 | Bordeaux, France | Respiratory tract | 2004 | 1 | U |
| B3722 | Bordeaux, France | Respiratory tract | 2004 | 1 | U |
| B3834 | Bordeaux, France | Respiratory tract | 2004 | 1 | U |
| B3737 | Bordeaux, France | Respiratory tract | 2004 | 1 | X |
| B4051 | Bordeaux, France | Respiratory tract | 2005 | 1 | J |
| B3996 | Bordeaux, France | Respiratory tract | 2005 | 1 | N |
| B3858 | Bordeaux, France | Respiratory tract | 2005 | 2 | O |
| B3912 | Bordeaux, France | Synovial fluid | 2005 | 1 | P |
| B3885 | Bordeaux, France | Respiratory tract | 2005 | 1 | P |
| B3891 | Bordeaux, France | Respiratory tract | 2005 | 2 | T |
| B3896 | Bordeaux, France | Respiratory tract | 2005 | NTc | S |
| B3881 | Bordeaux, France | Respiratory tract | 2005 | 1 | U |
| B3869 | Bordeaux, France | Respiratory tract | 2005 | 1 | U |
| B3851 | Bordeaux, France | Respiratory tract | 2005 | 1 | U |
| B4027 | Bordeaux, France | Respiratory tract | 2005 | 2 | V |
| B4079 | Bordeaux, France | Respiratory tract | 2005 | 2 | W |
| B3944 | Bordeaux, France | Respiratory tract | 2005 | 2 | V |
| B4100 | Bordeaux, France | Respiratory tract | 2006 | 1 | E |
| B4391 | Bordeaux, France | Respiratory tract | 2006 | 1 | I |
| B4291 | Bordeaux, France | Respiratory tract | 2006 | 2 | H |
| B4254 | Bordeaux, France | Respiratory tract | 2006 | 2 | H |
| B4223 | Bordeaux, France | Respiratory tract | 2006 | 1 | J |
| B4112 | Bordeaux, France | Respiratory tract | 2006 | 2 | M |
| B4381 | Bordeaux, France | Respiratory tract | 2006 | 2 | O |
| B4074 | Bordeaux, France | Respiratory tract | 2006 | 1 | P |
| B4104 | Bordeaux, France | Respiratory tract | 2006 | 2 | S |
| B4288 | Bordeaux, France | Respiratory tract | 2006 | 2 | T |
| B4209 | Bordeaux, France | Respiratory tract | 2006 | 1 | U |
| B4229 | Bordeaux, France | Respiratory tract | 2006 | 1 | U |
| B4098 | Bordeaux, France | Respiratory tract | 2006 | 2 | V |
| B4628 | Bordeaux, France | Respiratory tract | 2007 | 2 | B |
| B4578 | Bordeaux, France | Respiratory tract | 2007 | 1 | E |
| B4550 | Bordeaux, France | Respiratory tract | 2007 | 1 | J |
| B4581 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4470 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4495 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4543 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4567 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4675 | Bordeaux, France | Respiratory tract | 2007 | 2 | O |
| B4449 | Bordeaux, France | Respiratory tract | 2007 | 2 | T |
| B4466 | Bordeaux, France | Respiratory tract | 2007 | 2 | S |
| B4594 | Bordeaux, France | Respiratory tract | 2007 | 2 | T |
| B4608 | Bordeaux, France | Respiratory tract | 2007 | 2 | T |
| L20730 | Lyon, France | Respiratory tract | 1992 | 1 | X |
| L25861 | Lyon, France | Respiratory tract | 1993 | 2 | T |
| L1 | Lyon, France | Respiratory tract | 1994 | 1 | J |
| L2 | Lyon, France | Respiratory tract | 1995 | 1 | E |
| L3 | Lyon, France | Respiratory tract | 1995 | 1 | X |
| L8 | Lyon, France | Respiratory tract | 1996 | 2 | B |
| L33 | Lyon, France | Respiratory tract | 1996 | 1 | E |
| L11 | Lyon, France | Respiratory tract | 1996 | 1 | J |
| L5 | Lyon, France | Respiratory tract | 1996 | 1 | J |
| L10 | Lyon, France | Respiratory tract | 1996 | 1 | Q |
| L4 | Lyon, France | Respiratory tract | 1996 | 1 | P |
| L12 | Lyon, France | Respiratory tract | 1996 | 1 | U |
| L13 | Lyon, France | Respiratory tract | 1996 | 1 | U |
| L6 | Lyon, France | Respiratory tract | 1996 | 1 | U |
| L9 | Lyon, France | Respiratory tract | 1996 | 1 | U |
| L7 | Lyon, France | Respiratory tract | 1996 | 1 | X |
| L18 | Lyon, France | Respiratory tract | 1997 | 2 | C |
| L17 | Lyon, France | Respiratory tract | 1997 | 1 | E |
| L20 | Lyon, France | Respiratory tract | 1997 | 1 | J |
| L22 | Lyon, France | Respiratory tract | 1997 | 1 | J |
| L35 | Lyon, France | Respiratory tract | 1997 | 1 | J |
| L19 | Lyon, France | Respiratory tract | 1997 | 2 | L |
| L14 | Lyon, France | Respiratory tract | 1997 | 2 | M |
| L34 | Lyon, France | Respiratory tract | 1997 | 1 | P |
| L16 | Lyon, France | Respiratory tract | 1997 | 1 | P |
| L21 | Lyon, France | Respiratory tract | 1997 | 1 | U |
| L36 | Lyon, France | Respiratory tract | 1997 | 1 | U |
| L37 | Lyon, France | Respiratory tract | 1997 | 1 | U |
| L15 | Lyon, France | Respiratory tract | 1997 | 2 | W |
| L24 | Lyon, France | Respiratory tract | 1998 | 1 | E |
| L38 | Lyon, France | Respiratory tract | 1998 | 2 | O |
| L39 | Lyon, France | Respiratory tract | 1998 | 1 | P |
| L23 | Lyon, France | Respiratory tract | 1998 | 2 | S |
| L26 | Lyon, France | Respiratory tract | 1998 | 1 | U |
| L27 | Lyon, France | Respiratory tract | 1998 | 1 | U |
| L25 | Lyon, France | Respiratory tract | 1998 | 2 | V |
| L28 | Lyon, France | Respiratory tract | 1998 | 1 | X |
| L53 | Lyon, France | Respiratory tract | 1999 | 1 | E |
| L54 | Lyon, France | Respiratory tract | 1999 | 2 | H |
| L40 | Lyon, France | Respiratory tract | 1999 | 2 | H |
| L47 | Lyon, France | Respiratory tract | 1999 | 2 | G |
| L43 | Lyon, France | Respiratory tract | 1999 | 1 | J |
| L56 | Lyon, France | Respiratory tract | 1999 | 1 | J |
| L42 | Lyon, France | Respiratory tract | 1999 | 2 | O |
| L49b | Lyon, France | Respiratory tract | 1999 | 2 | O |
| L50 | Lyon, France | Respiratory tract | 1999 | 2 | M |
| L41 | Lyon, France | Respiratory tract | 1999 | 1 | P |
| L45 | Lyon, France | Respiratory tract | 1999 | 1 | P |
| L48 | Lyon, France | Respiratory tract | 1999 | 1 | P |
| L55 | Lyon, France | Respiratory tract | 1999 | 1 | P |
| L52b | Lyon, France | Respiratory tract | 1999 | 2 | T |
| L51 | Lyon, France | Respiratory tract | 1999 | 2 | T |
| L32 | Lyon, France | Respiratory tract | 2000 | 1 | J |
| L31 | Lyon, France | Respiratory tract | 2000 | 2 | O |
| L57 | Lyon, France | Respiratory tract | 2000 | 2 | O |
| L59 | Lyon, France | Respiratory tract | 2000 | 2 | N |
| L62 | Lyon, France | Respiratory tract | 2000 | 2 | M |
| L63 | Lyon, France | Respiratory tract | 2000 | 2 | O |
| L61 | Lyon, France | Respiratory tract | 2000 | 2 | T |
| L58 | Lyon, France | Respiratory tract | 2000 | 1 | U |
| C60 | Caen, France | Respiratory tract | 2005 | 2 | H |
| C66 | Caen, France | Respiratory tract | 2005 | 1 | U |
| C69 | Caen, France | Respiratory tract | 2005 | 2 | S |
| C62 | Caen, France | Respiratory tract | 2005 | 1 | X |
| C71 | Caen, France | Respiratory tract | 2005 | 1 | Z |
| C72 | Caen, France | Respiratory tract | 2005 | 1 | Z |
| C77 | Caen, France | Respiratory tract | 2006 | 2 | C |
| C91 | Caen, France | Respiratory tract | 2006 | 2 | B |
| C84 | Caen, France | Respiratory tract | 2006 | 1 | J |
| C89 | Caen, France | Respiratory tract | 2006 | 2 | H |
| C78 | Caen, France | Respiratory tract | 2006 | 1 | P |
| C87 | Caen, France | Respiratory tract | 2006 | 1 | P |
| C75 | Caen, France | Respiratory tract | 2006 | 1 | U |
| C83 | Caen, France | Respiratory tract | 2006 | 2 | T |
| C76 | Caen, France | Respiratory tract | 2006 | 2 | W |
| C90 | Caen, France | Respiratory tract | 2006 | 2 | V |
| C82 | Caen, France | Respiratory tract | 2006 | 2 | Y |
| C95 | Caen, France | Respiratory tract | 2007 | 2 | H |
| C98 | Caen, France | Respiratory tract | 2007 | 2 | H |
| C93 | Caen, France | Respiratory tract | 2007 | 1 | P |
| C94 | Caen, France | Respiratory tract | 2007 | 1 | P |
| C96 | Caen, France | Respiratory tract | 2007 | 1 | P |
| C92 | Caen, France | Respiratory tract | 2007 | 1 | Z |
| FG8 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | E |
| FG12 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | J |
| FG6 | Saint-Etienne, France | Respiratory tract | 1997 | 2 | O |
| FG9 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | P |
| FG11 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | P |
| FG7 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | U |
| FG2 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | U |
| FG5 | Saint-Etienne, France | Respiratory tract | 1997 | 1 | U |
| FG10 | Saint-Etienne, France | Respiratory tract | 1997 | 2 | U |
| FG13 | Saint-Etienne, France | Respiratory tract | 1998 | 1 | E |
| FG14 | Saint-Etienne, France | Respiratory tract | 1998 | 1 | P |
| B4560 | Bayonne, France | Respiratory tract | 2007 | 1 | E |
| B4602 | Bayonne, France | Respiratory tract | 2007 | 1 | J |
| B4625 | Bayonne, France | Respiratory tract | 2007 | 1 | P |
| B4561 | Bayonne, France | Respiratory tract | 2007 | 2 | S |
| N35 | Nantes, France | Respiratory tract | 1986 | 2 | V |
| N37 | Nantes, France | Respiratory tract | 1988 | 2 | V |
| N41 | Nantes, France | Respiratory tract | 1989 | 1 | P |
| B2362 | Poitiers, France | Respiratory tract | 1996 | 1 | U |
| B2644 | Poitiers, France | Respiratory tract | 1998 | 2 | B |
| B1145 | Brest, France | Cerebrospinal fluid | 1994 | 1 | P |
| B3098 | Argenteuil, France | Respiratory tract | 2000 | 1 | F |
| B4574 | Grenoble, France | Respiratory tract | 2007 | 2 | G |
P1 gene typing.
The type of P1 adhesin gene of 155 clinical isolates collected in France between 1994 and 2006 was previously determined (14). For the 102 remaining clinical isolates, typing by P1 gene PCR-RFLP analysis was performed as described previously (1, 14). Among six clinical isolates characterized previously, one isolate was a type 1a variant (3) and the five other isolates were type 2a variants (5) (Table 1).
Identification of TRs.
VNTR markers were identified in the sequenced genome of M. pneumoniae M129 (6) by using the Tandem Repeats Finder program and the Microorganisms Tandem Repeats database, available at http://minisatellites.u-psud.fr. Loci were chosen on the basis of matches of more than 80% between the DNA sequences of the repeat units. The different TR loci were designated by the letters Mpn, followed by a number corresponding to the order of detection and the position on the genome.
To screen for variability in the number of TRs, PCR primers targeting the regions flanking TR loci were designed. As a preliminary step, a total of 17 selected VNTRs were tested with specific primers to amplify DNA from a set of 13 diverse M. pneumoniae strains, including the 2 reference strains, 5 type 1 clinical isolates, 5 type 2 clinical isolates, and 1 isolate that could not be classified by P1 gene PCR-RFLP analysis. These strains were collected over a long time frame and from diverse geographical origins. Each locus was amplified individually and was analyzed by conventional agarose gel electrophoresis. To confirm that length polymorphisms were the result of repeat copy number variations, the PCR products were purified with a Wizard PCR preps DNA purification system (Promega) and were sequenced by using the fluorescence-labeled dideoxynucleotide technology, according to the manufacturer's recommendations (Applied Biosystems). By this approach, five VNTR loci were selected and taken forward for full assessment.
PCR amplification for MLVA.
The five loci ultimately selected for MLVA were multiplexed in two solutions, solutions M1 and M2, and the amplifications were performed with a Mastercycler epgradient S apparatus (Eppendorf, Hamburg, Germany) in a final volume of 25 μl. The reaction mixtures contained 1× Qiagen PCR buffer containing 1.5 mM MgCl2; 0.2 mM deoxynucleotide triphosphates; 2 mM and 3 mM MgCl2 for solutions M1 and M2, respectively; 1.25 U of HotStar Taq polymerase (Qiagen, Hilden, Germany); 0.5 μM each primer pairs Mpn1-F-Mpn1-R, Mpn14-F-Mpn14-R, and Mpn16-F-Mpn16-R for solution M1 and primer pairs Mpn13-F-Mpn13-R and Mpn15-F-Mpn15-R for solution M2; and 1 μl of template DNA. Forward or reverse primers were fluorescently labeled at the 5′ end with 4,4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein (HEX), 6-carboxyfluorescein (FAM; Eurogentec, France), or NED (2′-chloro-5′-fluoro-7′,8′-fused phenyl-1,4-dichloro-6-carboxyfluorescein; Applied Biosystems), depending on the locus to be amplified (Table 2). Both multiplex solutions were run with the same cycling conditions of 95°C for 15 min, followed by 25 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min and with a final extension of 72°C for 10 min. Prior to GeneScan analysis, the amplicons were diluted 1:50 in distilled water, and 1 μl of the diluted samples was mixed with 10 μl of Hi-Di formamide (Applied Biosystems) and 0.5 μl of GeneScan ROX 500 size standard (Applied Biosystems). After heat denaturation for 5 min at 95°C and rapid cooling on ice, the fragments were separated on an ABI 3130 genetic analyzer (Applied Biosystems) and were electrophoresed at 15,000 V for 20 min at 60°C. The GeneScan data were analyzed with GeneMapper software (version 4.0; Applied Biosystems) to perform sizing and to calculate the number of repeats in the PCR fragments. Each locus was identified according to color. An allele number string, based on the number of repeats at each locus, was assigned to all isolates. The number of repeats was rounded up to an integer value.
TABLE 2.
Oligonucleotide primers used for MLVA
| Primer namea | Dye sequence (5′ → 3′) |
|---|---|
| Mpn1-F | GTTGAAGTTATGCCGGTAGC |
| Mpn1-R | HEX-CGCGATAGAAGGCATACTGC |
| Mpn13-F | GACCAGCATTAGATTGCTATG |
| Mpn13-R | NED-AACAAATTAAGCAGCTCACG |
| Mpn14-F | CTCAGGGCGAAACCTTAAAG |
| Mpn14-R | 6-FAM-GCAATGGCTTTCAGCACAAC |
| Mpn15-F | HEX-CAACAGCACCACATCTTTAG |
| Mpn15-R | GCTAATCTTGCAAACGCTGC |
| Mpn16-F | NED-GACGCGTTCGCTAAAAGAG |
| Mpn16-R | CAGGCTCAACCAAATAATGG |
F, forward primer; R, reverse primer.
Data analysis.
The data with the calculated number of repeats were imported into the Bionumerics (version 5.0) software package (Applied Maths) and a minimum-spanning tree (MST) was generated for visualization of the relationships between large numbers of isolates in a single compact image. The MST was created on the basis of the categorical coefficient and a priority rule consisting of the highest number of single-locus variants (when two types were equal distances from the tree, the type with the highest number of single-locus variants was linked first). The creation of hypothetical types was allowed. A detailed description of the analysis of MLVA by MST can be found in the study of Schouls et al. (18). The polymorphism index of individual or combined VNTR loci was calculated by using the Hunter-Gaston diversity index (HGDI) (7).
Stability determination.
Five M. pneumoniae isolates, including reference strains M129 and FH, were chosen for use in the stability study. Each strain was passaged 10 times in Hayflick modified broth medium supplemented with glucose to determine the stability of each locus before and after the 10 passages. DNA was extracted and subjected to multiplexing PCR.
RESULTS
Identification of VNTRs for typing by MLVA and stability.
Seventeen regions with TRs spread over the genome of M. pneumoniae M129 were identified by using the Microorganisms Tandem Repeats database (http://minisatellites.u-psud.fr). Amplification was achieved by the use of primer pairs complementary to the sequences in DNA flanking each direct-repeat locus (data not shown). Preliminary amplification of these 17 loci, named Mpn1 to Mpn17, from both reference strains M. pneumoniae M129 and FH and 11 clinical strains showed that only six loci were polymorphic with different allele sizes. Eight markers turned out to be monomorphic, while for two loci, specific primers failed to amplify the DNA satisfactorily. One marker was eliminated from this study since the sequence variability observed between strains was not related to a variation in repeat number but was due to insertions.
The stability of the six polymorphic VNTR markers was studied for five strains after 10 passages in Hayflick broth medium. Analysis of the five strains resulted in identical MLVA profiles for all markers except one, Mpn6, which harbored a variation in the amplification size between the 1st and the 10th passages of one strain.
Thus, an MLVA scheme that included five markers (Mpn1, Mpn13, Mpn14, Mpn15, and Mpn16) and that allowed unambiguous type assignment by using agarose gels was proposed. The PCR products ranging in size from 219 to 419 bp. The five loci were distributed around the genome from base pair positions 111920 to 736123. Four of the five VNTRs were located in open reading frames, and three of them encoded hypothetical proteins. Marker Mpn1 was located in the hsdS gene, which encodes the S subunit of a type I restriction-modification enzyme. The characteristics of each VNTR marker are shown in Table 3. The sizes of the unit repeats ranged from 12 bp for Mpn1 to 47 bp for Mpn16. Sequencing of the PCR products of different sizes at each of the five loci from each of the 13 strains confirmed the sizes and sequences of the individual VNTR loci.
TABLE 3.
Characteristics of VNTR markers
| Name | Genome positiona (bp) | Locus (protein no. in the genome sequence) | Repeat size (bp) | % Identity between TRs | HGDIb (265 strains) |
|---|---|---|---|---|---|
| Mpn1 | 111920 | HsdS (MPN 089) | 12 | 100 | 0.784 |
| Mpn13 | 596741 | Intergenic | 16 | 83 | 0.495 |
| Mpn14 | 608651 | Hypothetical protein (MPN 501) | 21 | 84 | 0.404 |
| Mpn15 | 645561 | Hypothetical protein (MPN 524) | 21 | 81 | 0.494 |
| Mpn16 | 736123 | Hypothetical protein (MPN 613) | 47 | 100 | 0.015 |
Position (5′ end) on the M. pneumoniae M129 genome sequence (6).
HGDI, Hunter Gaston diversity index.
Typing of clinical strains.
The set of five VNTR markers was used to type a large collection of 265 M. pneumoniae strains isolated from several countries, including France, Germany, Denmark, Japan, Belgium, Tunisia, and Spain. This study mostly included strains isolated from the respiratory tract, and 12 of them were resistant to macrolides; but the study also included some extra-respiratory tract isolates. The use of fluorescently labeled primers in a multiplex PCR associating Mpn1-Mpn14-Mpn16 and Mpn13-Mpn15 and adapted to capillary electrophoresis allowed the easier interpretation of the results compared to that required for agarose gel electrophoresis. All loci were clearly identified on electropherograms according to their size ranges and colors, and the amplicon sizes were extracted and turned into allele numbers by the Genetic Analyzer software (Applied Biosystems).
The five VNTRs were very efficiently amplified from all 265 strains, and the size variations of the amplicons were exact multiples of the repeats. The results are shown in Table 4. Marker Mpn1 was the most discriminatory VNTR, displaying seven different alleles with repeat copy numbers ranging from 1 to 7, depending on the isolate. Markers Mpn13 and Mpn15 showed three different allele sizes and had repeat units that ranged in number from 2 to 4 and 6 to 8, respectively. Markers Mpn14 and Mpn16 gave two different-sized PCR products. Marker Mpn16 was the most homogeneous marker, with all except two isolates harboring 2 repeat units of 47 bp. This was also reflected by the diversity index of each VNTR, estimated from the HGDI, with a value of 0.784 for the most discriminatory marker (Mpn1) and a value of 0.015 for the less discriminatory one (Mpn16) (Table 3). The overall discriminatory index of the MLVA method was 0.915.
TABLE 4.
Number of repeat units of the VNTR loci in 265 isolates of M. pneumoniae
| MLVA type | No. of isolates by MLVA type | No. of repeats at the following VNTR loci:
|
P1 gene PCR-RFLP type(s) | ||||
|---|---|---|---|---|---|---|---|
| Mpn1 | Mpn13 | Mpn14 | Mpn15 | Mpn16 | |||
| A | 4 | 1 | 4 | 5 | 7 | 2 | 1 |
| B | 8 | 2 | 3 | 5 | 6 | 2 | 2 |
| C | 12 | 2 | 3 | 6 | 6 | 2 | 2 |
| D | 1 | 2 | 4 | 5 | 6 | 2 | 1 |
| E | 24 | 2 | 4 | 5 | 7 | 2 | 1 |
| F | 1 | 2 | 4 | 5 | 8 | 2 | 1 |
| G | 3 | 3 | 3 | 5 | 6 | 2 | 1 and 2 |
| H | 12 | 3 | 3 | 6 | 6 | 2 | 2 |
| I | 1 | 3 | 3 | 6 | 7 | 2 | 1 |
| J | 24 | 3 | 4 | 5 | 7 | 2 | 1 |
| K | 1 | 3 | 4 | 5 | 6 | 2 | 1 |
| L | 1 | 4 | 2 | 6 | 6 | 2 | 2 |
| M | 8 | 4 | 3 | 5 | 6 | 2 | 2 |
| N | 2 | 4 | 3 | 5 | 7 | 2 | 1 and 2 |
| O | 30 | 4 | 3 | 6 | 6 | 2 | 2 |
| P | 46 | 4 | 4 | 5 | 7 | 2 | 1 |
| Q | 2 | 4 | 4 | 5 | 7 | 1 | 1 |
| R | 1 | 5 | 3 | 5 | 7 | 2 | 1 |
| S | 7 | 5 | 3 | 5 | 6 | 2 | 2 and NTa |
| T | 14 | 5 | 3 | 6 | 6 | 2 | 2 |
| U | 33 | 5 | 4 | 5 | 7 | 2 | 1 and 2 |
| V | 10 | 6 | 3 | 5 | 6 | 2 | 2 |
| W | 4 | 6 | 3 | 6 | 6 | 2 | 2 |
| X | 10 | 6 | 4 | 5 | 7 | 2 | 1 |
| Y | 1 | 7 | 3 | 5 | 6 | 2 | 2 |
| Z | 5 | 7 | 4 | 5 | 7 | 2 | 1 |
NT, Mycoplasma pneumoniae isolate not classified by PCR-RFLP analysis of P1 adhesin gene.
Analysis of the combination of the five VNTR loci for the 265 M. pneumoniae isolates revealed 26 MLVA types, types A to Z (Table 4), while analysis of only the 210 French strains showed 25 types. The 26th MLVA type was represented by only one Japanese strain, strain J381. Between one and eight MLVA profiles were found for the isolates that originated from countries other than France. Reference strains M. pneumoniae M129 and FH harbored two different MLVA profiles and belonged to types P and T, respectively. It should be noted that 59% of the 265 isolates were of five MLVA types and that each of these five types contained at least 20 strains.
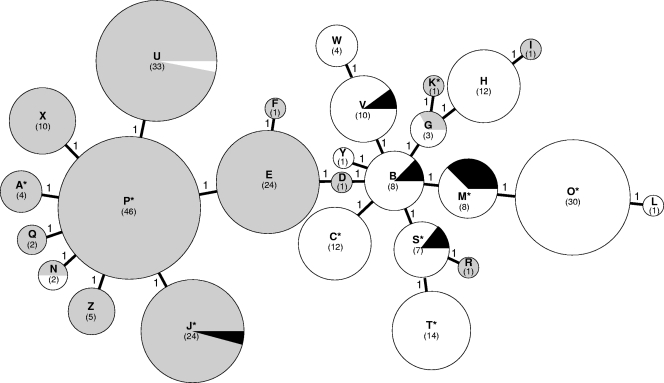
As a complementary analysis, the MLVA profiles were imported into the Bionumerics software package (Applied Maths), and the genetic relationships of the 265 isolates were deduced by construction of an MST (Fig. 1). This population modeling highlighted the diversity among the isolates tested, but each type was closely related to each other, differing by no more than one locus and emphasizing the lack of distinct clusters. There was no obvious link between the MLVA type and the year when the strains were collected, the patient's age, or resistance to macrolides. Indeed, the 12 macrolide-resistant isolates were dispersed into nine MLVA types (Fig. 1). No association could be established between the 26 unique allele strings and the geographical origin, except for the Tunisian and Spanish isolates. The eight Tunisian isolates were obtained between 2006 and 2008 from different geographical areas and belonged to the same MLVA type, type P, the most important type. The two Spanish strains isolated from two different patients in 1995, both of which were from pericardial fluid or tissue, belonged to MLVA type E (Fig. 1). Furthermore, a clustering was observed on the basis of the P1 subtype, although there were a few exceptions (Fig. 1). P1 variant 1a (strain 4817) belonged to MLVA type J, which included only type 1 isolates, while the five 2a variants (strains F59, Ho, Sta, 79809, and 79692) belonged to MLVA types B, M, and V, which included only type 2 isolates.
FIG. 1.
MST of the MLVA profiles of the 265 M. pneumoniae isolates. Each circle denotes a particular MLVA type, as indicated by a letter and with the number of isolates corresponding to this genotype given in parentheses. The size of the circle is proportional to the number of isolates belonging to the indicated MLVA genotype. The distance between neighboring genotypes is expressed as the number of allelic changes and is outlined by short bold lines for one change. The color of the circles indicates the P1 adhesin type: gray for type 1, white for type 2, and black for characterized variants. Asterisks in the MLVA type indicate the presence of macrolide-resistant isolates.
DISCUSSION
MLVA has been used to genotype several species of bacteria, including recently two Mycoplasma species, M. mycoides subsp. mycoides SC (12) and M. genitalium (10). In the study presented here, we evaluated the applicability of MLVA to the molecular typing of M. pneumoniae. We performed MLVA with a collection of isolates originating from European countries, Japan, and Tunisia. A collection of five VNTR markers which can be used to genotype M. pneumoniae isolates by simple PCR and agarose gel electrophoresis was identified. To improve the assay, two multiplex PCRs with fluorescently labeled primers were performed; one PCR amplified the Mpn1, Mpn14, and Mpn16 loci and one PCR amplified the Mpn13 and Mpn15 loci. The application of multiplex PCR and capillary electrophoresis on a gene analyzer enabled a high-throughput version of the analysis to be performed and allowed the easier interpretation of the results compared to that required for gel agarose electrophoresis, in particular, for VNTRs with small numbers of repeat units.
The stability of the VNTRs was evaluated in vitro and was confirmed before and after 10 repeated passages of five strains in broth medium. For one patient, two distinct respiratory tract specimens (one sputum specimen and one bronchoalveolar lavage fluid specimen) that were positive for M. pneumoniae were available at the same time. Both isolates were genotyped and were found to harbor the same MLVA type, which also suggested the in vivo stability of the VNTRs chosen (data not shown).
The discriminatory power of this MLVA scheme with the five markers was 0.915 for the collection of 265 isolates. The discriminatory index should ideally be 1.00, but in practice, it should at least be on the order of 0.90 to 0.95 for a typing system to be considered more or less ideal (19). Thus, the global HGDI of this MLVA scheme in our study was very satisfactory. This value must be interpreted in comparison with the discriminatory powers of the other available typing methods. In comparison, the HGDI of the P1 PCR-RFLP typing method for the 265 isolates was only 0.511. The allelic diversity of the VNTR loci varied significantly at each locus, with the Mpn1, Mpn13, Mpn14, and Mpn15 loci being more discriminatory than the Mpn16 locus. The association of this set of markers with variable HGDIs makes this MLVA, in theory, the optimal MLVA scheme for epidemiological studies. Of the five VNTRs tested, only one was located in an intergenic region. Interestingly, marker Mpn1 was located in a coding region containing the hsdS gene, which encodes the S subunit of a type I restriction-modification enzyme involved in protection against foreign DNA (15). The recognition sequence of this enzyme depends on the S subunit, which is known to be variable (16) and which could explain the importance of the allelic variation of the Mpn1 marker.
No clustering was emphasized on the basis of geographical location or specimen origin for any of the isolates except the Spanish and Tunisian isolates. It is interesting to note that two clinical strains, isolated in Spain in 1995 from the pericardial site from two different patients, were of the same MLVA type (13), suggesting that these strains may have a particular virulence mechanism. However, 22 other strains belonged to the same MLVA type as the 2 clinical strains isolated in Spain in 1995. The eight isolates from Tunisia that were studied clustered in the main MLVA type, suggesting a very low level of genetic diversity and the diffusion of a homogeneous population of M. pneumoniae in that country. The 12 macrolide-resistant isolates of M. pneumoniae, which comprised 9 Japanese isolates, 2 French isolates, and 1 Danish isolate, were not related and clustered into nine MLVA types, suggesting the absence of a link between this typing method and macrolide resistance. Nevertheless, this observation could also be explained by the absence of a particular emerging macrolide-resistant clone.
Furthermore, our MLVA scheme was used to study the course of infection in a single patient. Indeed, two clinical isolates obtained 2 months apart from the same patient, isolates B2829 and B2892, showed two different MLVA profiles, as well as two different P1 PCR-RFLP subtypes, suggesting more a M. pneumoniae reinfection than a relapse in this case.
The P1 gene PCR-RFLP typing method (1, 17) divided the clinical isolates only into P1 types 1 and 2. One or the other of the two types tended to predominate among the clinical isolates over time. These changes in the P1 adhesin type may play a role in the development of outbreaks. However, in disagreement with this theory, clear shifts between the M. pneumoniae types did not appear to be correlated with M. pneumoniae epidemic cycles (17). In France, since 1998, both types were present in about the same proportion without a clear dominance of one type over the other one (14). Compared to the findings of the P1 gene PCR-RFLP typing method, which is usually performed to subtype M. pneumoniae, MLVA analysis showed a better power to discriminate among the different isolates, as shown by comparison of the HGDIs between the two methods (see above). Furthermore, while P1 PCR-RFLP typing gave only three patterns (type 1, type 2, and not classified), 26 MLVA types were obtained for the 265 isolates tested. Interestingly, there was a correlation between the typing results obtained by MLVA and PCR-RFLP analysis of the P1 gene. As shown by the MST modeling, M. pneumoniae strains belonging to type 1 or 2 were well separated into two parts, with the exception of a few strains. Beside subtypes 1 and 2, one 1a variant (3) and five 2a variants characterized previously (5) belonged to MLVA types which included only type 1 and type 2 isolates, respectively. Furthermore, one isolate, isolate B3896, was previously found to be different and could not be classified into type 1, type 2, or one of the different subtype variants already described (3, 5, 8, 14). This isolate belonged to MLVA type S, in which only type 2 isolates clustered (Fig. 1), suggesting that isolate B3896 was more related to type 2 than to type 1. These data are in agreement with the sequence analysis of the P1 gene of this isolate, which was more homologous to that of type 2 than to that of type 1 (14).
MST modeling illustrates the relationship of subtypes on the basis of the number of VNTR loci that differ between two MLVA genotypes. Analysis of this population modeling revealed that the 26 MLVA types identified did not differ by more than one locus. Furthermore, no cluster could be individualized, highlighting the homogeneous character of the M. pneumoniae species, despite the high HGDI of the typing method developed in this study. As M. pneumoniae is a relatively homogeneous species, few typing methods have previously been able to detect significant differences between strains (2, 5). One attempt to subtype M. pneumoniae by MLST with housekeeping and structural genes was not sufficiently discriminatory to be used for epidemiological purposes (4).
Although we have studied an important set of clinical M. pneumoniae strains that originated from various countries and that were isolated in different years, unfortunately, no isolates were obtained from an epidemic outbreak of M. pneumoniae disease. Further community outbreaks of acute respiratory tract infections due to M. pneumoniae have been described worldwide (9, 22), and it would be very interesting to evaluate our MLVA scheme in such epidemic situations. The level of discrimination of this typing method should be confirmed by comparing outbreak-related strains to a set of control strains that were isolated from a similar time period and geographical area but that are not epidemiologically related.
In conclusion, this is, to our knowledge, the first attempt to perform the molecular typing of M. pneumoniae by the MLVA method with a consequential number of clinical isolates. Other markers could be identified to improve the MLVA scheme proposed in this study and to find a better correlation between strain information and typing by VNTR.
One of the advantages of MLVA typing is that it is PCR based and does not require the growth of bacteria. Therefore, another important issue will be to adapt the method directly for clinical specimens, since the value of M. pneumoniae culture is far from being satisfactory, is much more fastidious, and is less sensitive than detection by PCR. Furthermore, the data obtained by MLVA could be shared in a web-based database for M. pneumoniae, as has already been done for other bacterial species.
Acknowledgments
We thank M. P. Milon, F. Grattard, and J. Petitjean for providing M. pneumoniae clinical isolates from the Hospices Civils de Lyon, St. Etienne Hospital, and Caen Hospital, respectively. We are greatly indebted to J. S. Jensen for providing the Danish isolates, E. Jacobs and R. Dumke for the German isolates, T. Sasaki for the Japanese isolates, A. Ben Hassem and A. Touati for the Tunisian isolates, M. Messeguer for the Spanish isolates, and H. Goossens for the Belgian isolates.
Footnotes
Published ahead of print on 9 February 2009.
REFERENCES
- 1.Cousin-Allery, A., A. Charron, B. de Barbeyrac, G. Fremy, J. Skov Jensen, H. Renaudin, and C. Bébéar. 2000. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 124103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorigo-Zetsma, J. W., J. Dankert, and S. A. Zaat. 2000. Genotyping of Mycoplasma pneumoniae clinical isolates reveals eight P1 subtypes within two genomic groups. J. Clin. Microbiol. 38965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorigo-Zetsma, J. W., B. Wilbrink, J. Dankert, and S. A. Zaat. 2001. Mycoplasma pneumoniae P1 type 1- and type 2-specific sequences within the P1 cytadhesin gene of individual strains. Infect. Immun. 695612-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumke, R., I. Catrein, E. Pirkil, R. Herrmann, and E. Jacobs. 2003. Subtyping of Mycoplasma pneumoniae isolates based on extended genome sequencing and on expression profiles. Int. J. Med. Microbiol. 292513-525. [DOI] [PubMed] [Google Scholar]
- 5.Dumke, R., P. C. Luck, C. Noppen, C. Schaefer, H. von Baum, R. Marre, and E. Jacobs. 2006. Culture-independent molecular subtyping of Mycoplasma pneumoniae in clinical samples. J. Clin. Microbiol. 442567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 244420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenri, T., R. Taniguchi, Y. Sasaki, N. Okazaki, M. Narita, K. Izumikawa, M. Umetsu, and T. Sasaki. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 674557-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klement, E., D. F. Talkington, O. Wasserzug, R. Kayouf, N. Davidovitch, R. Dumke, Y. Bar-Zeev, M. Ron, J. Boxman, W. Lanier Thacker, D. Wolf, T. Lazarovich, Y. Shemer-Avni, D. Glikman, E. Jacobs, I. Grotto, C. Block, and R. Nir-Paz. 2006. Identification of risk factors for infection in an outbreak of Mycoplasma pneumoniae respiratory tract disease. Clin. Infect. Dis. 431239-1245. [DOI] [PubMed] [Google Scholar]
- 10.Ma, L., S. Taylor, J. S. Jensen, L. Myers, R. Lillis, and D. H. Martin. 2008. Short tandem repeat sequences in the Mycoplasma genitalium genome and their use in a multilocus genotyping system. BMC Microbiol. 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuoka, M., M. Narita, N. Okazaki, H. Ohya, T. Yamazaki, K. Ouchi, I. Suzuki, T. Andoh, T. Kenri, Y. Sasaki, A. Horino, M. Shintani, Y. Arakawa, and T. Sasaki. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob. Agents Chemother. 484624-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAuliffe, L., R. D. Ayling, and R. A. Nicholas. 2007. Identification and characterization of variable-number tandem-repeat markers for the molecular epidemiological analysis of Mycoplasma mycoides subspecies mycoides SC. FEMS Microbiol. Lett. 276181-188. [DOI] [PubMed] [Google Scholar]
- 13.Meseguer, M. A., S. Garcia-Rull, J. Picher, J. Ortiz-Saracho, L. Maiz, and F. Baquero. 1995. Isolation of Mycoplasma pneumoniae from pericardial tissue. Eur. J. Clin. Microbiol. Infect. Dis. 14825-826. [DOI] [PubMed] [Google Scholar]
- 14.Pereyre, S., A. Charron, H. Renaudin, C. Bébéar, and C. M. Bébéar. 2007. First report of macrolide-resistant strains and description of a novel nucleotide sequence variation in the P1 adhesin gene in Mycoplasma pneumoniae clinical strains isolated in France over 12 years. J. Clin. Microbiol. 453534-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 621094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha, E. P., and A. Blanchard. 2002. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 302031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki, T., T. Kenri, N. Okazaki, M. Iseki, R. Yamashita, M. Shintani, Y. Sasaki, and M. Yayoshi. 1996. Epidemiological study of Mycoplasma pneumoniae infections in Japan based on PCR-restriction fragment length polymorphism of the P1 cytadhesin gene. J. Clin. Microbiol. 34447-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 1865496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3)1-46. [DOI] [PubMed] [Google Scholar]
- 20.Vergnaud, G., and C. Pourcel. 2006. Multiple locus VNTR (variable number of tandem repeat) analysis (MLVA), p. 83-104. In E. Stackebrandt (ed.), Molecular identification, systematics and population structure of prokaryotes. Springer-Verlag, Berlin, Germany.
- 21.Waites, K. B., C. M. Bébéar, J. A. Roberston, D. F. Talkington, and G. E. Kenny (ed.). 2001. Cumitech 34, Laboratory diagnosis of mycoplasmal infections. Coordinating ed., F. S. Nolte. American Society for Microbiology, Washington, DC.
- 22.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]