Abstract
Systematic sequence analysis of human immunodeficiency virus type 1 (HIV-1) variants with RNA levels underestimated by the Cobas TaqMan HIV-1 assay demonstrated that mutations at a single position of the downstream primer can lead to the underestimation of HIV-1 RNA levels by more than 2 log and to false-negative results in minipool screening of blood donors. Mutations at this position are found in about 2% of all HIV-1 M gag sequences.
Quantification of human immunodeficiency virus type 1 (HIV-1) RNA in human plasma (determination of the viral load) is one of the cornerstones in the management of care for HIV-infected patients. HIV-1 RNA detection also shortens the diagnostic window in blood donor screening. In recent years, the use of real-time PCR has substantially improved the performance of quantitative nucleic acid detection. The major advantages of real-time PCR over assays with end point detection are the extension of the linear range, the reduction of test variability, faster turnaround times, and easier automation. For viruses with high degrees of genetic variability, however, it can be more difficult to design appropriate primer and probe sets for real-time PCR that are able to equally quantify all variants. We identified several samples with falsely low or undetectable viral loads as determined by the Cobas TaqMan (CTM) HIV-1 assay (Roche Diagnostics, Mannheim, Germany). In an attempt to determine the reasons for the underquantitation, we sequenced parts of the gag region from five underquantified viruses as well as from viruses of two control patients that were accurately quantified by the CTM assay.
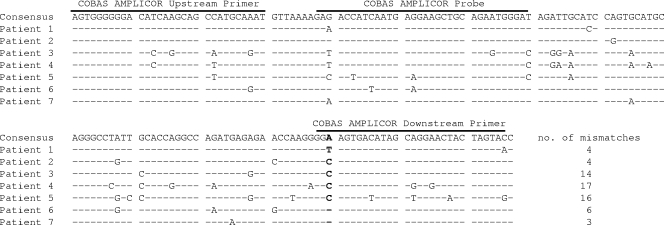
All patients except patient 3 were untreated at the time when the underestimation was observed. For patient 1, the sample for comparative testing of the viral load and determination of the gag sequence was obtained about 5 months after infection via a contaminated erythrocyte concentrate. For the other patients, no information on the duration of infection and the mode of transmission was available. Table 1 shows the viral load and subtype data for the five patients with viruses substantially underquantified by the CTM assay and for two control patients. For patients 1 to 5, the HIV-1 RNA concentrations determined with the CTM assay were between 30- and 780-fold lower than those determined with other viral load assays targeting either the gag gene (Cobas Amplicor Monitor version 1.5 [v1.5]) or the pol gene (RealTime HIV-1 assay [Abbott Diagnostics, Wiesbaden, Germany] and VERSANT branched-DNA assay 3.0 [Siemens Healthcare Diagnostics, Fernwald, Germany]) of HIV-1. In contrast, for the control patients, the values from the CTM assay were equal to or even higher than those from the other assays. For only one of the patients (patient 5), the Cobas Amplicor Monitor HIV-1 v1.5 assay, which targets the same HIV-1 gene as the CTM assay but is based on competitive PCR, also failed to detect the virus correctly, whereas in all other cases, it compared well to the assays targeting the pol gene. Sequence analysis revealed that these underquantified viruses belonged to four different HIV-1 subtypes. Two of them belonged to subtype B, which is most prevalent in Western Europe and North America. Unfortunately, a straightforward sequence comparison to identify mutations relevant to these failures was impossible, because the manufacturer was unwilling to disclose the primer and probe sequences of the CTM assay. Therefore, the PCR product of a CTM strongly positive control reaction was cloned into a pGEM-T Easy vector (Promega) and sequenced. The sequence was very similar to the sequences of the Cobas Amplicor Monitor v1.5 assay primers and probe, which have been published earlier (3). Figure 1 shows the 157-bp sequence of the cloned PCR product from the CTM assay (corresponding to nucleotides 1359 to 1515 of the HXB2 reference sequence; GenBank accession no. K03455) and the localization of the Cobas Amplicor Monitor v1.5 primers and probe. The 5′ ends of the sequences from the two assays are identical, whereas at the 3′ end, the CTM PCR product is 2 nucleotides longer than the Cobas Amplicor Monitor v1.5 downstream primer. A comparison of the sequences of the five viruses that were underquantified by the CTM assay with the two control sequences revealed that only one position in all underquantified viruses is mutated but that the nucleotide at this position in the two control samples is identical to that in the wild-type sequence. This position corresponds to nucleotide 1488 in the HXB2 reference sequence and is highlighted in bold in Fig. 1. It is located close to the 3′ end of the Cobas Amplicor Monitor downstream primer. A notice released from the manufacturer to the Paul-Ehrlich-Institute in the case of patient 1 (who was infected via a blood transfusion) suggests that this position 1488 is also located at the −3 position of the downstream primer in the CTM assay. The importance of position 1488 is also supported by the fact that the degrees of underestimation of the two subtype B virus loads are comparable to those of the three non-B virus loads, although the non-B viruses contain a much higher number of mismatches throughout the CTM amplicon (14 to 17, compared to 3 to 6 for the subtype B viruses). Intriguingly, quantification with the Cobas Amplicor Monitor assay revealed accurate results in four of the five cases with CTM failure, although the position of the critical mismatch in the downstream CTM primer is presumably identical. This finding suggests that not changes in the primer design but rather differences in PCR conditions (which are also not disclosed by the manufacturer) may be responsible for the CTM assay failures. A search for mismatches in the HIV-1 gag gene in the Los Alamos HIV database showed that about 2% (24 of 1,206) of all HIV-1 group M sequences had nucleotides other than the wild-type A at position 1488. However, 15 of these 24 variant sequences contained a G at this position, which did not show up in any of the viruses that were underquantified by the CTM assay. Therefore, it is unclear if a G at position 1488 can also lead to a high degree of underestimation of the viral load in the CTM assay. Nucleotides C and T, which were present in the underquantified samples, were observed in 2 (0.2%) and 7 (0.6%) of the 1,206 sequences, respectively, suggesting that a significant number of HIV-1-infected patients would have underquantified or false-negative results from the CTM assay. This assumption is supported by previous reports on underquantifications by the CTM assay compared to the results from a corresponding competitive PCR assay (2) or from another real-time PCR assay (1) for the evaluation of HIV-1 RNA. However, the reason for these underquantifications had not yet been unveiled. In consequence, viral loads in almost 1% of HIV-1-infected patients may be substantially underestimated. This may lead to inappropriate decisions about when to start therapy and may cause delayed detection of treatment failure and the development of drug resistance. Even more deleterious consequences can occur when this assay is used in blood donor screening, as seen in the case of patient 1. This patient was infected by transfusion with an erythrocyte concentrate from a blood donor whose infectious preseroconversion donation escaped routine detection by minipool PCR testing for HIV-1 RNA by the CTM assay. The cases described herein demonstrate that there is an urgent need for the modification of the CTM assay. Furthermore, for viruses with high degrees of genetic variability, it may be advantageous to base viral load assays on multiple target regions.
TABLE 1.
Comparison of results from HIV-1 assays
| Patient no. | Resulta from:
|
Degrees of underestimation (n-fold) by CTM assayb | HIV-1 subtypec | |||
|---|---|---|---|---|---|---|
| CTM | Cobas Amplicor Monitor HIV-1 v1.5 | Abbott RealTime HIV-1 | VERSANT HIV-1 branched-DNA 3.0 | |||
| 1 | 90 | 70,000 | 52,000 | 24,000 | 780, 580, 250 | B |
| 2 | Positive/<40 | NA | NA | 2,400 | NA, NA, >60 | B |
| 3 | Positive/<40 | 29,000 | 13,000 | 1,200 | >725, >325, >30 | CRF02_AG |
| 4 | 210 | 10,800 | 19,300 | NA | 50, 90, NA | A1 |
| 5 | ND/<40 | ND/<50 | 4.000 | NA | NA, >100, NA | F1 |
| 6 (control) | 3,100 | 3,600 | 2,600 | 8,200 | 1, 1, 3 | B |
| 7 (control) | 3,200 | NA | 1,500 | 1,000 | NA, 0.5, 0.3 | B |
All numbers are genome equivalents of HIV-1 RNA per milliliter of plasma. NA, not available; ND, RNA not detected.
Degrees of underestimation by the CTM assay compared to results from the Cobas Amplicor Monitor HIV-1 v1.5, Abbott RealTime HIV-1, and VERSANT HIV-1 branched-DNA 3.0 assays, respectively, are shown.
Subtype was determined from long terminal repeat, gag, and pol sequences in samples from patients 1, 2, and 3 and the control patients 6 and 7. For patients 4 and 5, sequences of the complete HIV-1 genome were used for subtyping. CRF, circulating recombinant form.
FIG. 1.
The 157-nucleotide sequence of the CTM HIV-1 amplicon shown corresponds to nucleotides 1359 to 1515 of the HXB2 reference sequence (GenBank accession no. K03455). The localization of the primers and probe of the Cobas Amplicor Monitor HIV-1 v1.5 assay is indicated by the lines above the consensus sequence. The position highlighted in bold in the alignment corresponds to nucleotide 1488.
Nucleotide sequence accession number.
The sequence of the CTM amplicon determined in this study has been deposited in GenBank under accession no. AM922506.
Footnotes
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Colson, P., C. Solas, J. Moreau, A. Motte, M. Henry, and C. Tamalet. 2007. Impaired quantification of plasma HIV-1 RNA with a commercialized real-time PCR assay in a couple of HIV-1-infected individuals. J. Clin. Virol. 39226-229. [DOI] [PubMed] [Google Scholar]
- 2.Damond, F., B. Roquebert, A. Bénard, G. Collin, M. Miceli, P. Yéni, F. Brun-Vezinet, and D. Descamps. 2007. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche COBAS AMPLICOR HIV-1 MONITOR version 1.5 and the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 assays. J. Clin. Microbiol. 453436-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler, J. F., L. K. de Souza Luna, C. Pedroso, B. D. Pedral-Sampaio, A. T. Queiroz, C. Brites, E. M. Netto, and C. Drosten. 2007. Rates of and reasons for failure of commercial human immunodeficiency virus type 1 viral load assays in Brazil. J. Clin. Microbiol. 452061-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]