Abstract
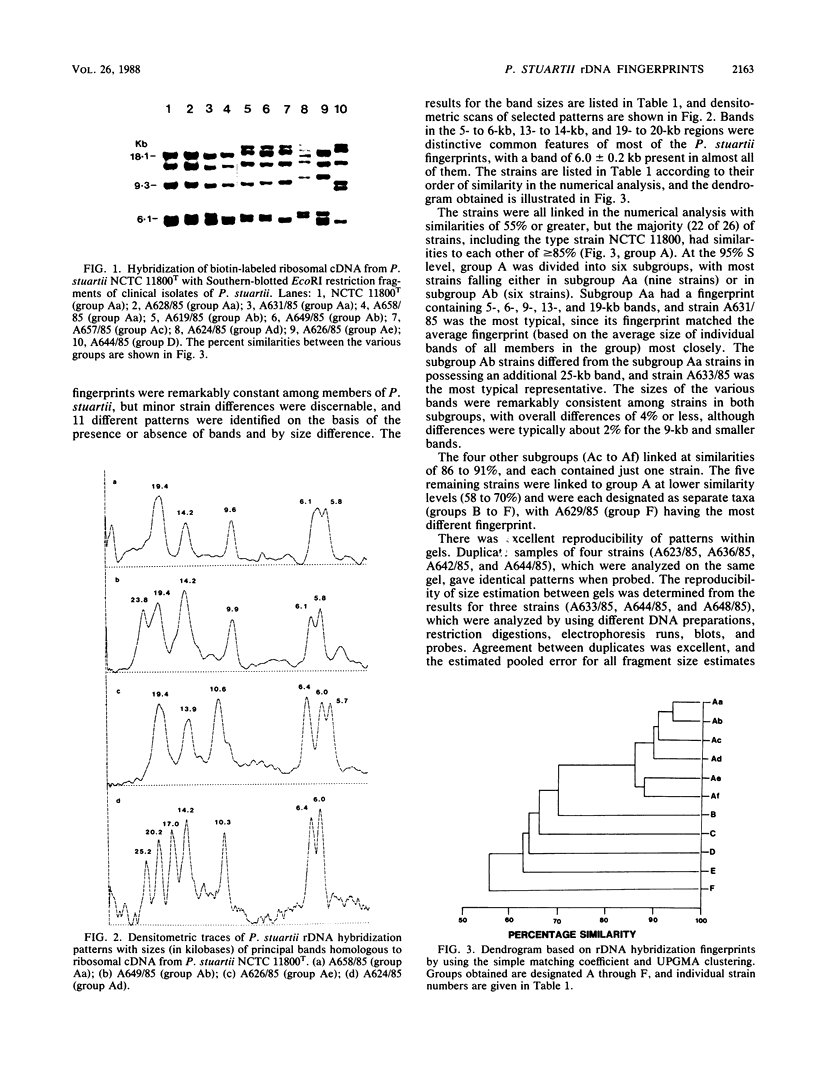
Chromosomal DNA from 26 strains of Providencia stuartii isolated mainly in hospitals in the United Kingdom and reference strains of P. stuartii, P. rustigianii, and Proteus vulgaris were digested with the restriction endonucleases EcoRI and HindIII. After electrophoresis in agarose gels, the fragments were subjected to Southern blot hybridization analysis with a biotin-labeled cDNA probe transcribed from a mixture of 16S and 23S rRNA from P. stuartii NCTC 11800T. The pattern of bands (the rDNA fingerprint), which depended on restriction fragment length polymorphisms containing rRNA genes, was used as a measure of minor genomic variation within and between species. The P. stuartii clinical isolates had similar total digest patterns, but the rDNA fingerprints revealed some heterogeneity between strains, with EcoRI digests providing better strain discrimination than HindIII. Such rDNA fingerprints comprised between five and seven bands with sizes in the range of 5 to 28 kilobases. The 11 different EcoRI patterns were compared by numerical analysis, and several groups or subgroups of strains were identified. Over half (15 of 26) of the urease-negative isolates (subgroups Aa and Ab) had patterns that differed only by the presence or absence of a 25-kilobase band. Urease-negative strains from other clinical material were more heterogeneous in their patterns. No correlation was apparent between strain pattern group and urease production or geographic location of isolate. The P. stuartii rDNA fingerprints were quite distinct from those of allied Providencia and Proteus species and provided a more sensitive measure of minor genomic differences than total DNA digests did.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bercovier H., Kafri O., Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986 May 14;136(3):1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Goldmann D. A. New microbiological techniques for hospital epidemiology. Eur J Clin Microbiol. 1987 Jun;6(3):344–347. doi: 10.1007/BF02017637. [DOI] [PubMed] [Google Scholar]
- Grant R. B., Penner J. L., Hennessy J. N., Jackowski B. J. Transferable urease activity in Providencia stuartii. J Clin Microbiol. 1981 Mar;13(3):561–565. doi: 10.1128/jcm.13.3.561-565.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Hawkey P. M., Bennett P. M., Hawkey C. A. Cryptic plasmids in hospital isolates of Providencia stuarti. J Med Microbiol. 1984 Oct;18(2):277–284. doi: 10.1099/00222615-18-2-277. [DOI] [PubMed] [Google Scholar]
- Hawkey P. M., Bennett P. M., Hawkey C. A. Evolution of an R plasmid from a cryptic plasmid by transposition of two copies of Tn1 in Providencia stuartii. J Gen Microbiol. 1985 Apr;131(4):927–933. doi: 10.1099/00221287-131-4-927. [DOI] [PubMed] [Google Scholar]
- Hawkey P. M., Penner J. L., Linton A. H., Hawkey C. A., Crisp L. J., Hinton M. Speciation, serotyping, antimicrobial sensitivity and plasmid content of Proteeae from the environment of calf-rearing units in South West England. J Hyg (Lond) 1986 Dec;97(3):405–417. doi: 10.1017/s0022172400063592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey P. M. Providencia stuartii: a review of a multiply antibiotic-resistant bacterium. J Antimicrob Chemother. 1984 Mar;13(3):209–226. doi: 10.1093/jac/13.3.209. [DOI] [PubMed] [Google Scholar]
- Küntzel H., Piechulla B., Hahn U. Consensus structure and evolution of 5S rRNA. Nucleic Acids Res. 1983 Feb 11;11(3):893–900. doi: 10.1093/nar/11.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., D'Souza T. M., Magee P. T. Strain and species identification by restriction fragment length polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol. 1987 Apr;169(4):1639–1643. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Owen R. J., Beck A., Borman P. Restriction endonuclease digest patterns of chromosomal DNA from nitrate-negative Campylobacter jejuni-like organisms. Eur J Epidemiol. 1985 Dec;1(4):281–287. doi: 10.1007/BF00237103. [DOI] [PubMed] [Google Scholar]
- Owen R. J., Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 1987 Apr 24;15(8):3631–3631. doi: 10.1093/nar/15.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. W., Sampson J. S., Plikaytis B. B. Evaluation of a commercial gene probe for identification of Legionella cultures. J Clin Microbiol. 1986 Feb;23(2):217–220. doi: 10.1128/jcm.23.2.217-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]