Abstract
The histone variant H2A.Z (Htz1p) has been implicated in transcriptional regulation in numerous organisms, including Saccharomyces cerevisiae. Genome-wide transcriptome profiling and chromatin immunoprecipitation studies identified a role for Htz1p in the rapid and robust activation of many oleate-responsive genes encoding peroxisomal proteins, in particular POT1, POX1, FOX2, and CTA1. The Swr1p-, Gcn5p-, and Chz1p-dependent association of Htz1p with these promoters in their repressed states appears to establish an epigenetic marker for the rapid and strong expression of these highly inducible promoters. Isw2p also plays a role in establishing the nucleosome state of these promoters and associates stably in the absence of Htz1p. An analysis of the nucleosome dynamics and Htz1p association with these promoters suggests a complex mechanism in which Htz1p-containing nucleosomes at fatty acid-responsive promoters are disassembled upon initial exposure to oleic acid leading to the loss of Htz1p from the promoter. These nucleosomes reassemble at later stages of gene expression. While these new nucleosomes do not incorporate Htz1p, the initial presence of Htz1p appears to mark the promoter for sustained gene expression and the recruitment of TATA-binding protein.
The organization of DNA into chromatin provides cells with a key regulatory mechanism for gene expression by limiting the access of the genome to the transcriptional machinery. The nucleosome represents a basic structural unit of chromatin and posttranslational modifications of histones serve as signals to define active, repressed, or inert chromatin states. In addition, chromatin states and gene expression can be influenced by the dynamics of histones and their nonallelic variants. Indeed, the exchange of canonical histones for histone variants appears to be a key mechanism by which the transcriptional machinery overcomes the restricted access imposed by nucleosome positioning (1). Of the many classes of histone variants discovered, the Z variant of H2A is perhaps the best characterized. H2A.Z differs from the canonical H2A histone in both the length and sequence of the C terminus (37) and is conserved from yeast to mammals (15). Early studies with H2A.Z in Tetrahymena spp. showed that H2A.Z incorporation is linked with transcriptionally active chromatin (35). The Saccharomyces cerevisiae orthologue of H2A.Z is called Htz1p and is encoded by HTZ1. Although HTZ1 is not an essential gene under standard laboratory growth conditions, Htz1p is implicated in transcriptional regulation. Global chromatin studies have revealed that Htz1p preferentially associates with the two nucleosomes flanking the nucleosome free region of promoters (12, 18, 26, 41), and this association is inversely proportional to transcription rates (3, 18).
Studies of the role of Htz1p in transcriptional regulation at specific promoters, such as those of GAL1 and PHO5 (1, 30) indicate that the presence of Htz1p at promoters is dynamic; Htz1p is bound in their repressed states but dissociates during the activation process. Accordingly, it is proposed that nucleosomes containing Htz1p are poised to undergo nucleosome displacement, allowing for rapid transcriptional responses (41).
The yeast S. cerevisiae is an excellent model for understanding the mechanisms of cellular responses to induced perturbations. Upon the exposure of yeast to fatty acids, such as oleate, cells respond by dramatically altering their gene expression patterns, inducing genes required for peroxisomal β-oxidation and peroxisome biogenesis (32). Genetic screens to identify proteins specifically required for efficient fatty acid metabolism in S. cerevisiae (34) identified metabolic enzymes, proteins required for the biogenesis of the organelle, signaling proteins and transcriptional regulators, and chromatin modifiers. Among this latter class of proteins, this approach identified genes encoding Htz1p, RNA polymerase II, mediator subunits, and components of chromatin remodeling complexes. We thus seek to understand the nature of how chromatin is regulated and remodeled in response to exposure to fatty acids and the specific role Htz1p plays in these regulation/remodeling processes.
In this report, transcriptomes of wild-type (WT) and htz1Δ strains were compared during exposure to oleic acid. While the loss of Htz1p reduced the expression of many genes, genes involved in the fatty acid response were particularly sensitive. A model is proposed in which Htz1p-containing nucleosomes at fatty acid-responsive promoters are disassembled upon initial exposure to oleic acid, leading to the loss of Htz1p from the promoter. These nucleosomes reassemble at later stages of gene expression. While these nucleosomes do not incorporate Htz1p, the initial presence of Htz1p appears to mark the promoter for sustained gene expression and the recruitment of TATA-binding protein (TBP).
MATERIALS AND METHODS
Strains and growth conditions.
All yeast strains used in this study are indicated in Table 1. Haploid strains with myc-tagged genes were made by genomically tagging target genes with the sequence encoding 13 copies of the c-myc epitope from pFA6a-13MYC (20) by homologous recombination into BY4742 (WT) using a previously described PCR-based procedure (2). The strains were verified by PCR analysis of the tagged gene loci and Western blot analysis of the fusion proteins. An examination of the growth characteristics of each strain suggests that the chimeras did not alter protein function. For all experiments, the control strains were otherwise isogenic to the test strains. The strains were cultured at 30°C in the following media: YPD (1% yeast extract, 2% peptone, 2% glucose) and SCIM (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% yeast extract, 0.5% peptone, 0.079% complete supplement mixture, 0.5% ammonium sulfate) containing 0.5% Tween 40 (wt/vol) and 0.2% (wt/vol) oleate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open |
| BY4742 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open |
| YWY004 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 | Open |
| YWY042 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 POT1-GFP::natMX | This study |
| YWY007 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 POT1-GFP::natMX pRS416 | This study |
| YWY005 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 POT1-GFP::natMX | This study |
| YWY009 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 POT1-GFP::natMX pRS416 | This study |
| YWY010 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 POT1-GFP::natMX pCM305 | This study |
| YWY011 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 POT1-GFP::natMX pCM330 | This study |
| YWY012 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 htz1::kanMX4 POT1-GFP::natMX pCM314 | This study |
| YWY013 | MATα leu2Δ0 ura3Δ0 HA-HTZ1 | 41 |
| YWY0165 | MATα leu2Δ0 ura3Δ0 HA-HTZ chz1::kanMX4 | This study |
| YWY0166 | MATα leu2Δ0 ura3Δ0 HA-HTZ gcn5::kanMX4 | This study |
| YWY0177 | MATα leu2Δ0 ura3Δ0 HA-HTZ swr1::kanMX4 | This study |
| YWY206 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 SPT15-13MYC::kanMX4 | This study |
| YWY207 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 SPT15-13MYC::kanMX4 htz1::hphMX | This study |
| Plasmids | ||
| pRS416 | CEN6-ARS4 URA3 | Open |
| pCM314 | CEN6-ARS4 URA3 HA-htz1K14A | 23 |
| pCM330 | CEN6-ARS4 URA3 HA-htz1K14R | 23 |
| pCM305 | CEN6-ARS4 URA3 HA-HTZ1 | 23 |
RNA preparation and microarray analysis.
Yeast cultures were grown at 30°C to a density of ∼1 × 107 cells/ml. The cells were collected and immediately frozen in liquid nitrogen. Total RNA was isolated by hot acid phenol extraction. The total RNA was treated with RNase-free DNase I and purified with a Qiagen RNeasy kit. Microarray labeling and hybridization reactions were performed as previously described (7). Two color microarrays, comparing RNA from the experimental conditions (WT and htz1Δ cells grown in oleate [SCIM] for 6 h) to RNA from the control WT cells grown in glucose-containing medium (YPD), were performed using Agilent whole-genome S. cerevisiae arrays. All experiments were performed with duplicate experimental and duplicate technical replicates of each condition, and the log10 of the average mRNA abundance ratios are reported. Differentially expressed genes were identified by maximum-likelihood analysis (λ ≥ 100) (14, 32) and significantly affected genes in the mutants were identified by a change in expression of twofold or more compared to the expression in the relevant WT strains.
For the quantitative reverse transcription-PCR (qRT-PCR), total RNA was directly reverse transcribed using the First Strand cDNA synthesis kit from Fermentas (catalog no. K1611). cDNAs were treated by RNase H and diluted 1/100 for the qPCR. The RT-PCR was done using a 7900HT fast real-time PCR system and a DyNAmo Flash SYBR green qPCR kit (F-415L; NEB) with gene-specific oligonucleotides. mRNA levels were normalized relative to ACT1 mRNA levels from three independent RT-PCR analyses. A list of primers used for the qRT-PCRs is available on request.
ChIP and real-time PCR.
For each chromatin immunoprecipitation (ChIP) experiment, yeast strains were first grown in glucose medium (YPD) to a density of ∼1 × 107 cells/ml and then transferred to oleate medium (SCIM) for the times indicated in each figure. The ChIP experiments were performed as described in reference 33 with the following modifications. For the hemagglutinin (HA)-Htz1p ChIP, cells were crossed-linked with 1% formaldehyde for 45 min at room temperature. Two micrograms of anti-HA antibody (12CA5) was prebound to 50 μl of pan-mouse immunoglobulin G Dynabeads (Dynal Biotech) and then incubated with 1 mg (protein) of supernatant from the sheared chromatin overnight at 4°C. The TBP (Spt15p-Myc) ChIP was performed as described in reference 33. Cells were cross-linked with 1% formaldehyde for 2 h at room temperature. Two microliters of anti-Myc antibody (9E11; Abcam) was prebound to 50 μl of pan-mouse immunoglobulin G Dynabeads and then incubated with 1 mg (protein) of supernatant from sheared chromatin overnight at 4°C.
All ChIP experiments were performed in triplicate. The purified ChIP samples were used in the qPCR analysis. Real-time qPCR was performed by using an iCycler instrument (ABI 7900) and a DyNAmo Flash SYBR green qPCR kit. The average of the results of three independent replicates is reported as the relative amplification of each target of interest compared to a normalization control amplicon, within the nonpromoter IGRi YMR325W. The primer sequences are available on request. The occupancy level was determined by dividing the relative abundance of an experimental target by the relative abundance of a control target. This ratio represents the enrichment of ChIP DNA over the input DNA for a specific target versus the control target.
FACS analysis.
The fluorescence-activated cell sorter (FACS) analysis procedures were performed as previously described (27). The fluorescence intensities of individual cells were measured using a FACSCalibur flow cytometer (BD Biosciences). The data analysis was performed using WinMDI 2.8 (available from http://FACS.scripps.edu/).
Nucleosome scanning assay (NuSA).
A total of 200 ml of cells at an optical density at 600 nm of 1.0 in glucose medium or after transfer to oleate-containing medium prior to treatment with 1% formaldehyde for 20 min, followed by a 5-min incubation in 125 mM glycine. The cell permeabilization, micrococcal nuclease digestion, protein degradation, and DNA purification steps were performed as described previously (38). The DNA samples were then treated with RNase A and analyzed in a 2% agarose gel to quantify the nucleosomal content. The bands corresponding to mononucleosomal DNA were extracted using a Qiagen gel extraction kit. A qPCR analysis of the digested DNA was performed. A list of the qPCR primers is available on request; they cover the promoter regions of POT1, CTA1, POX1, and FOX2 with overlapping amplicons averaging 100 bp in size. To define nucleosome occupancy, the protection value of each amplicon was normalized to the CEN3 values as described previously (6). The N+1 nucleosome refers to the first nucleosome downstream of the transcription start site, which is located at the open reading frame regions. The N−1 nucleosome refers to the first nucleosome upstream of the transcription start site, which is located at the promoter regions.
RESULTS
Htz1p is required for the transcriptional activation of a subset of oleic acid-responsive genes.
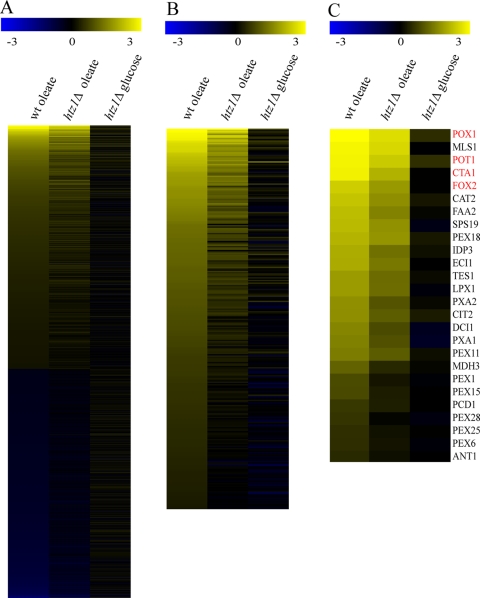
Transcriptome profiling was used to obtain a global understanding of how Htz1p contributes to gene expression in response to external stimuli. To do so, we focused on gene induction upon the shift from glucose to oleic acid growth conditions. This condition was chosen because we and others have previously shown that this transition leads to dramatic alterations in the gene expression patterns (16, 32, 33), because genes involved in fatty acid metabolism are significantly induced under these conditions, and because it has been shown that S. cerevisiae htz1Δ strains have a specific growth defect when grown on fatty acids (19, 34). In accordance with previous genome-wide analyses of oleate responses (16, 32, 33), a large portion of the genome responds to the transition (Fig. 1A, column 1). Reflecting the nonfermentative metabolism of oleate by the coordinated activities of peroxisomes and mitochondria, the most significantly enriched classes of induced genes include genes linked to mitochondrial respiration and peroxisomal lipid metabolism (hypergeometric distribution analysis of gene ontology terms oxidative phosphorylation, electron transport chain, and aerobic respiration, P < 10−10; components of the mitochondrial respiratory chain, P ≈ 10−13; fatty acid oxidation and peroxisome organization and biogenesis, P ≈ 10−6; and the peroxisomal compartment, P ≈ 10−12). By comparison, there were many genes that were relatively unresponsive in htz1Δ cells (Fig. 1A, column 2; Fig. 1B). This included genes that were both poorly induced and genes that were poorly repressed in htz1Δ cells compared to the WT (Fig. 1A). Among the genes induced upon oleate exposure, 292 were expressed at least twofold less in htz1Δ cells than in WT cells (Fig. 1B). Interestingly, these poorly induced genes were most enriched for those annotated with peroxisomal functions and components but were not enriched for annotations of mitochondrial components or aspects of mitochondrial respiration (fatty acid and lipid oxidation, P ≈ 4.0 × 10−12; peroxisomes, P ≈ 9 × 10−20) (Fig. 1C). Indeed, 26 genes (of 57 total) encoding peroxisomal proteins showed significantly reduced transcription in an htz1Δ background (Fig. 1C). These data suggest that Htz1p is required for the regulated expression of a large number of genes upon the transition from one state to another. In the case of the transition to oleate, genes linked to peroxisomal fatty acid oxidation are normally highly induced and their expression is the most significantly affected in the absence of Htz1p.
FIG. 1.
The robust expression of oleate-responsive genes expression is dependent on HTZ1. (A) Comparison of changes in the mRNA levels of all yeast genes in WT (left column) and htz1Δ cells (middle column) after induction in oleate medium for 6 h. Shown are the relative expression levels (log10) of genes that were determined to be significantly (λ ≥ 100) altered in cells on oleate (compared to WT cells on glucose). Relative expression levels are shown using the scale of the yellow-blue heat map (top). Genes are ordered top to bottom based on relative expression in WT cells on oleate. Approximately 1,000 genes were significantly induced, and 1,000 genes were repressed and changed in expression at least twofold. Genes that were reduced in expression were significantly enriched for functions related to ribosomal biogenesis (hypergeometric distribution analysis of gene ontology terms, P ≈ 10−50). Induced genes were enriched for oxidative phosphorylation, the electron transport chain, and aerobic respiration (P ≈ 10−10); components of the mitochondrial respiratory chain (P ≈ 10−13); fatty acid oxidation and peroxisome organization and biogenesis (P ≈ 10−6); and the peroxisomal compartment (P ≈ 10−12). For comparison, the relative expression of each gene in htz1Δ cells in oleate (middle column) and glucose (right column) is shown. (B) The same as described for panel A, but shown are the relative expression levels of 292 genes significantly (λ ≥ 100) altered in WT cells on oleate and expressed at least twofold less than their expression levels in WT cells. This list is enriched for genes linked to fatty acid and lipid oxidation (P ≈ 4.0 × 10−12) and peroxisomes (P ≈ 9 ×10−20). (C) The same as described for panel B, but shown are genes encoding peroxisomal proteins significantly (λ ≥ 100) altered in WT cells on oleate and expressed at least twofold less in htz1Δ cells.
HTZ1 is required for normal peroxisomal β-oxidation.
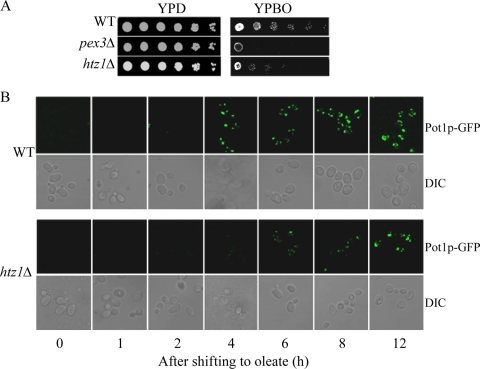
The finding that normally highly induced genes linked to fatty-acid oxidation are poorly expressed in htz1Δ cells is consistent with the finding that cells lacking HTZ1 show a specific impairment of fatty acid metabolism (19, 34). Like mutants defective in peroxisomal function (e.g., pex3Δ), htz1Δ cells exhibit a growth defect on fatty acid-containing medium (YPBO) but not on glucose-containing medium (YPD) (Fig. 2A) or on other nonfermentable carbon sources such as glycerol (YPG) or acetate (YPA) requiring mitochondrial function (34). As expected, the WT cells grew normally on different carbon sources.
FIG. 2.
The deletion of HTZ1 leads to delayed peroxisome biogenesis. (A) The deletion of HTZ1 impairs cell growth on oleate-containing medium. Strains were grown to mid-logarithmic phase in liquid YPD medium, and equal amounts of cells were serially diluted 10-fold onto YPD and incubated at 30°C for 3 days and onto oleate-containing YPBO and incubated at 30°C for 5 days. (B) The fluorescent images of WT and htz1Δ cells shown are expressing the peroxisomal matrix protein Pot1p fused with GFP (Pot1p-GFP) at different time points of oleate incubation and were captured on a TCS SP2 laser scanning spectral confocal microscope.
To examine the effect of Htz1p on the organelle itself, we examined peroxisomes by fluorescence microscopy. WT and htz1Δ cells expressing peroxisomal thiolase Pot1p, tagged by genomic integration with green fluorescent protein (GFP), were incubated in oleate medium and observed over a time course of induction by direct fluorescence confocal microscopy (Fig. 2B). In glucose-containing medium, peroxisomes were barely detectable. However, upon the shift to oleic acid, WT cells induced the expression and import of Pot1p-GFP as indicated by the accumulation of punctate fluorescent structures (29). However, there was a dramatic delay in the appearance of punctate GFP fluorescence in htz1Δ cells compared with that in the WT cells induced over the same time period. Together, these data suggest that peroxisome biogenesis per se is not defective in htz1Δ cells. Rather, the defect in the ability to metabolize oleate effectively is a result of the relatively poor expression of genes required for (peroxisomal) fatty acid metabolism.
Transcriptional response of POT1, POX1, FOX2, and CTA1.
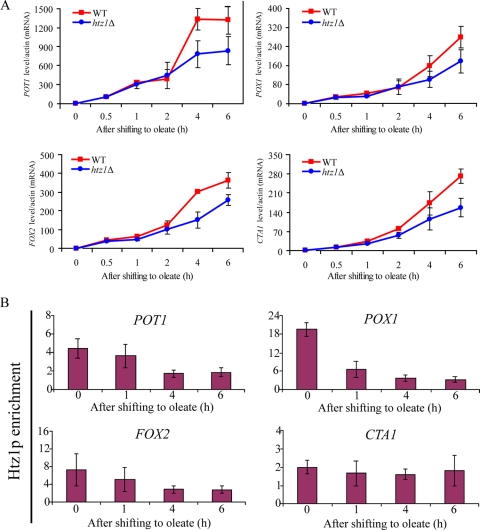
To further examine the molecular defects associated with the loss of Htz1p, we focused on four strongly induced peroxisomal matrix enzymes encoded by POT1, FOX2, POX1, and CTA1 (Fig. 3A, red). These genes are normally repressed on glucose and strongly induced on oleic acid (32). qRT-PCR of these mRNAs demonstrated that in the absence of Htz1p, each of these genes was repressed as in the WT cells, but their induction was impaired upon the transition to oleate medium (Fig. 3A). Interestingly, the expression of each of these genes appeared to be most significantly affected at the later time points after the transition to oleate (compare 4 and 6 h of induction to 0.5 and 1 h of induction). These data suggest that the loss of Htz1p did not dramatically alter the initial response but was important for the sustained expression of these four genes.
FIG. 3.
Htz1p dynamically dissociates from oleate-responsive promoters upon induction. (A) POT1, POX1, FOX2, and CTA1 mRNA levels were determined by RT-PCR in WT and htz1Δ strains over a time course of oleate induction. The signal obtained from ACT1 mRNA was used as a loading control for normalization. Error bars represent standard deviations from the means of three independent experimental values. (B) Htz1p enrichment at four promoters was determined by qPCR during oleate induction. Relative enrichment values (y axes) are the averages of the results from three independent ChIPs with qPCR determination performed twice per biological replicate. Nonpromoter IGRi YMR325W was used as an internal control to normalize signals of promoter enrichment. In response to oleate induction, Htz1p was lost from the POT1, POX1, FOX2, and CTA1 promoters.
Having demonstrated a role for Htz1p in the normal regulation of POT1, FOX2, POX1, and CTA1 expression in the presence of oleic acid, we next sought to determine if Htz1p binds the cognate promoters of these genes using the ChIP of a strain expressing an HA-tagged version of Htz1p. The cells were grown under either repressed (glucose) or activated (oleate) conditions. Htz1p-HA was immunoprecipitated with anti-HA antibody, and the isolated DNA was analyzed by PCR. This analysis revealed that Htz1p was bound to each of the four promoters (POT1, FOX2, POX1, and CTA1) in their repressed states (Fig. 3B). These data are consistent with the genome-wide characterization of the levels of Htz1p association with these promoters (41). The association of Htz1p with these promoters was dynamic; when cells were shifted to oleic acid-activating conditions, Htz1p levels on the POT1, POX1, and FOX2 promoters were dramatically reduced. Dissociation from the CTA1 promoter was not observed. These data suggest that the loss of Htz1p from promoters is coincident with gene activation, but that dissociation is not required for the induction of all genes.
Swr1p-, Chz1p-, and Gcn5p-dependent association of Htz1p to promoters.
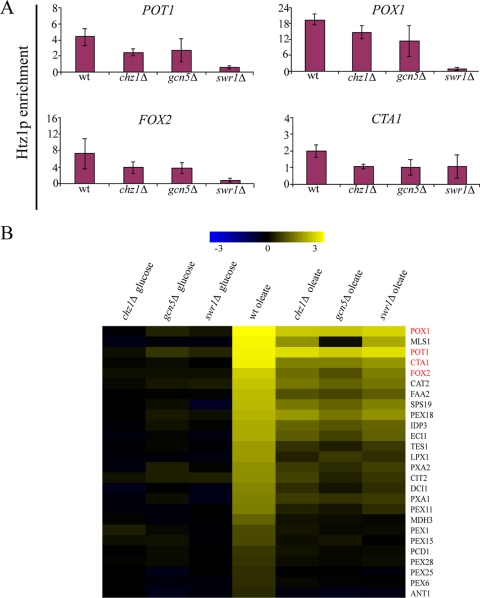
Swr1p, Chz1p, and Gcn5p have been implicated in modulating Htz1p association at promoter regions. Swr1p is part of the SWR1-C multisubunit protein complex, necessary for Htz1p deposition at repressed promoters (24). Chz1p was recently identified as a histone chaperone that preferentially interacts with Htz1p (21), and Gcn5p is the histone acetyltransferase subunit of the SAGA complex (36). To investigate whether these factors affect Htz1p binding to the oleate-responsive promoters and subsequent expression, the association of Htz1p with POT1, POX1, FOX2, and CTA1 promoters was investigated in cells lacking these proteins under conditions of repression (2% glucose), and the expression of these genes was monitored upon oleate induction (Fig. 4). Similar to its role at the well-studied GAL1 promoter, Swr1p is required for Htz1p binding to oleate responsive promoters, suggesting a common role for Swr1p at disparate, highly inducible promoters. Likewise, Gcn5p was required for efficient Htz1p binding. This suggests that Gcn5p, which plays a role as a coactivator of transcription through histone acetylation (11), controls the binding or stability of Htz1p at repressed promoters. This may also be via histone acetylation. In the absence of Chz1p, Htz1p occupancy at each of the four promoters was decreased. As expected, the amount of Htz1p on each of these promoters in mutant strains remained low upon the switch to oleate (data not shown).
FIG. 4.
Swr1p-, Chz1p-, and Gcn5p-dependent association of Htz1p with promoters. (A) In vivo association of Htz1p with the POT1, POX1, FOX2, and CTA1 promoters was measured by ChIP in the WT, chz1Δ, gcn5Δ, and swr1Δ strains in 2% glucose medium. ChIP was performed in glucose-containing medium. Error bars represent standard deviations from the means of three independent experimental values and two technical replicates of each. (B) Comparison of changes in the mRNA levels of all yeast genes in WT, chz1Δ, gcn5Δ, and swr1Δ strains after induction in oleate medium for 6 h. As described in the legend to Fig. 1C, genes encoding peroxisomal proteins significantly (λ ≥ 100) altered in WT cells on oleate and expressed at least twofold less in htz1Δ cells are shown.
Microarray analyses of the gcn5Δ, swr1Δ, and chz1Δ mutants support a model in which initial Htz1p association with the promoter is required for subsequent full induction. The expression levels of POT1, FOX2, POX1, and CTA1 were significantly reduced in the mutant strains compared to those in the WT upon oleate induction. Moreover, all 26 genes encoding peroxisomal proteins that showed transcriptional defects in htz1Δ cells (Fig. 1C) were similarly reduced in their expression at least twofold in the gcn5Δ, swr1Δ, and chz1Δ mutants compared to that of the WT (Fig. 4B). Together, these data suggest that factors functionally associated with Htz1p, such as the chromatin remodeling complex component Swr1p, histone acetyltransferase Gcn5p, and chaperone Chz1p, regulate the deposition or maintenance of Htz1p at repressed promoters, which, in turn, facilitates the rapid activation of transcription.
The acetylation of Htz1p is required for efficient transcriptional induction.
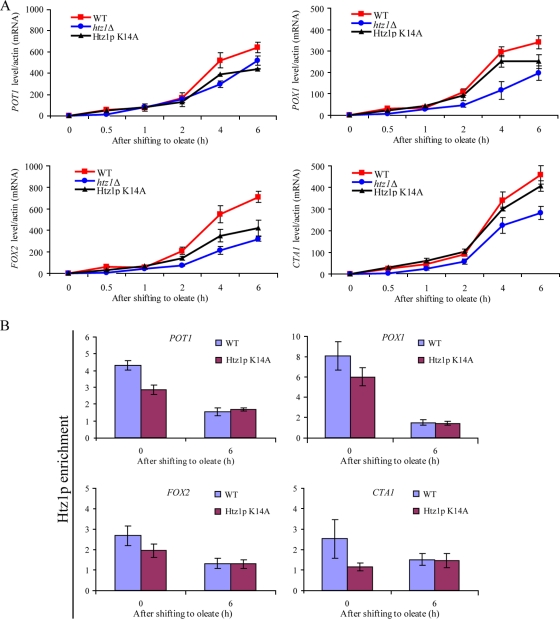
The acetylation of Htz1p is known to occur at sites of active transcription (23). The finding that Gcn5p is required for the expression of oleate-responsive genes suggests that acetylation on Htz1p is required for the oleate response. To address this question, plasmids expressing either one of two acetylation mutants of Htz1p (pCM314 [Htz1p K14A] or pCM330 [Htz1p K14R]) were introduced into htz1Δ cells and expression was monitored by FACS, confocal microscopy, and qRT-PCR. FACS and confocal microscopy demonstrated that Pot1p-GFP fluorescence in cells carrying pCM305 (WT HTZ1) was stronger than that in those cells carrying empty plasmid (pRS416) or acetylation mutants (pCM330 or pCM314) during a time course of oleate incubation, but the peroxisomes were morphologically normal (data not shown). mRNA levels of POT1, CTA1, FOX2, and POX1, determined by qRT-PCR, were consistent with the GFP reporter analysis (Fig. 5A). The K14A acetylation mutant of Htz1p showed a defect in the normal induction of POT1, CTA1, FOX2, and POX1. In addition, cells expressing Htz1p K14A also exhibited a growth defect on fatty-acid-containing medium but not on glucose-containing medium. This growth defect was less pronounced than that of the null mutant of HTZ1 (data not shown). The association of Htz1p K14A with these oleate responsive promoters at two time points (0 h and 6 h) also revealed that Htz1p K14A association was diminished under glucose conditions (Fig. 5B). Significant differences in the association of Htz1p K14A on these promoters were not observed during 6 h of oleate induction. These data indicate that the acetylation of Htz1p is required for association with oleate-responsive promoters under repressed conditions and that the acetylation of Htz1p is not required for the dissociation of Htz1p from oleate-responsive promoters during oleate induction (Fig. 5B). These data collectively indicate that the acetylation of Htz1p is important for the expression of fatty acid-responsive genes and normal peroxisomal matrix protein assembly.
FIG. 5.
The acetylation of Htz1p is required for efficient transcriptional induction. (A) POT1, POX1, FOX2, and CTA1 mRNA levels were determined by RT-PCR in the WT, htz1Δ, and Htz1p K14A mutant strains over a time course of oleate induction. The signal obtained from ACT1 mRNA was used as a loading control for normalization. Error bars represent standard deviations from the means of three independent experimental values. (B) Enrichment of WT Htz1p and the Htz1p K14A mutant at four promoters was determined by qPCR during glucose and oleate induction for 6 h. Relative enrichment values (y axes) are the averages of the results from three independent ChIP experiments with two technical replicates of each. Nonpromoter IGRi YMR325W was used as an internal control to normalize signals of promoter enrichment.
TBP is not efficiently recruited to oleate-inducible promoters in the absence of Htz1p.
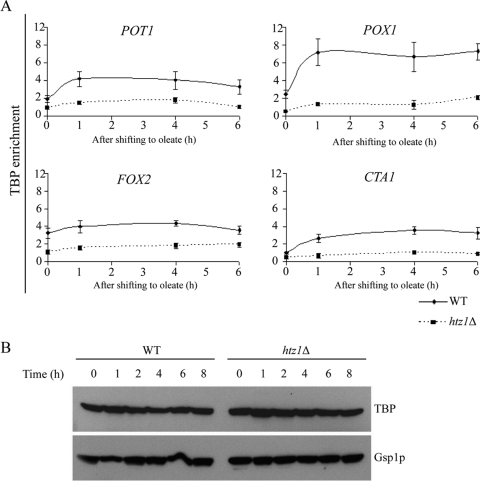
We next directly analyzed the in vivo binding of the transcriptional machinery to repressed and activated promoters in both the WT and htz1Δ strains (Fig. 6A). As expected, the binding of TBP to the POT1, POX1, and CTA1 promoters increased with gene expression in oleate in WT cells. The abundance of TBP did not significantly increase on the FOX2 promoter following oleic acid induction but was present at higher initial levels than the other three other promoters studied. Nonetheless, at all four promoters in htz1Δ cells, TBP binding was significantly reduced compared to that of the WT cells. The reduced levels of TBP binding were not attributable to decreased cellular levels of TBP. Western blot analysis of both WT and htz1Δ cells demonstrated that TBP levels were equivalent between the strains and did not significantly change during oleate induction (Fig. 5B). These results suggest a positive function for the Htz1p-containing nucleosomes in the recruitment of TBP to the repressed promoters during the process of transcriptional activation.
FIG. 6.
Recruitment of TBP during oleate induction requires Htz1p. (A) The association of TBP with the POT1, POX1, FOX2, and CTA1 promoters was determined by ChIP using anti-Myc antibodies, followed by gene-specific PCR. The relative enrichment ratio is plotted at four time points (0, 1, 4, and 6 h) of induction in oleate. ACT1 was used as an internal control to normalize signals of promoter enrichment. Error bars show the standard deviation from three independent experimental values with two technical replicates of each. (B) Deletion of HTZ1 did not affect TBP expression during oleate induction. The WT strain and htz1Δ strains expressing genomically integrated TBP were grown in 2% glucose overnight and then transferred to oleate-containing SCIM medium at the indicated time points. Samples containing equal amounts of protein were analyzed by Western blotting with anti-Myc antibody to visualize TBP expression. A polyclonal antibody directed against Gsp1p was used as a loading control.
Htz1 regulates nucleosome-promoter association during activation.
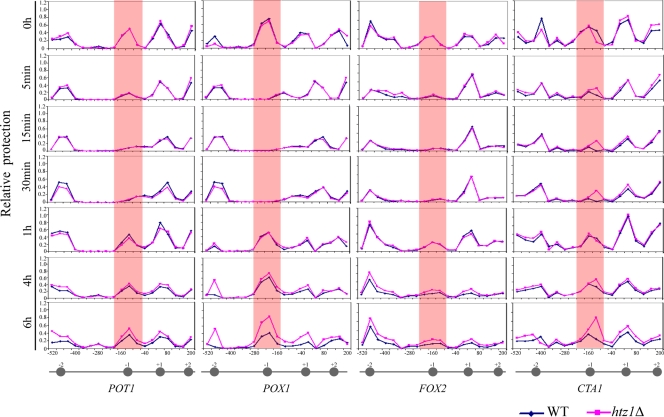
A NuSA was used to investigate the role of Htz1p in modulating chromatin structure by measuring nucleosome occupancy and location within oleate-responsive promoters during activation (POT1, POX1, FOX2, and CTA1) (Fig. 7). Mononucleosome-associated DNA was isolated from yeast cells before and after oleate induction, and real-time qPCR was used to measure dynamic nucleosome occupancy during activation in WT and htz1Δ cells. The precise positions of the nucleosomes were determined by qPCR corresponding to their known positions (17). Overall, the gross nucleosome position pattern at each of the four promoters under repressed conditions was the same in the WT and htz1Δ cells. The major nucleosome changes were observed at position N-1 in each promoter. These nucleosomes appeared to begin disassembly from each promoter at the earliest time point measured (5 min) and continued through to the 30-min time point. After this initial disassembly, nucleosomes were detected to have begun reassembly after 1 h of induction (Fig. 7). These reassembled nucleosomes likely do not contain Htz1p. As shown in Fig. 3B, Htz1p is progressively lost from these promoters during the 6-h period of induction. Notably, the nucleosomes of each promoter appeared to be more protected at the later time points (6 h) in htz1Δ cells than in the WT. This was most evident at the N-1 position of the promoters of POX1 and CTA1 (and at the N-2 position of POX1). These data suggest that upon oleate treatment, the nucleosome proximal to the initiation site in each promoter disassembles, leading to Htz1p loss and initial transcriptional activation. During prolonged expression, nucleosomes reassemble, but these reassembled nucleosomes do not contain Htz1p.
FIG. 7.
Htz1p regulates the occupancy of specific nucleosomes on the POT1, POX1, FOX2, and CTA1 promoters. The NuSA was used to determine the nucleosome positioning and density at the POT1, POX1, FOX2, and CTA1 promoters during oleate induction (the time of induction is indicated on the left) in the WT and HTZ1 deletion strains. Each point represents the relative protection of each PCR amplicon, quantified by real-time PCR and normalized to a centromeric control. The position of each amplicon (referenced to the middle of each amplicon) within the promoter is shown on the x axis. The approximate location of a nucleosome is represented by a gray circle with the nucleosome number referred to in the text shown above the circle.
Interplay between Htz1p and chromatin-remodeling factor Isw2p.
While Htz1p is proposed to contribute to nucleosome disassembly during induction, the results presented above indicate that the overall chromatin structure at the promoters was not extensively perturbed in htz1Δ cells. To gain insight into the potential additional mechanisms at play during the transcriptional induction, we considered additional chromatin-bound proteins. One such protein is Isw2p. Isw2p is an ATP-dependent chromatin-remodeling factor that has previously been shown to be required for the maintenance of chromatin structure at the POT1 promoter (8, 9, 39). We therefore investigated Isw2p function at the POT1, POX1, FOX2, and CTA1 promoters in WT and htz1Δ cells.
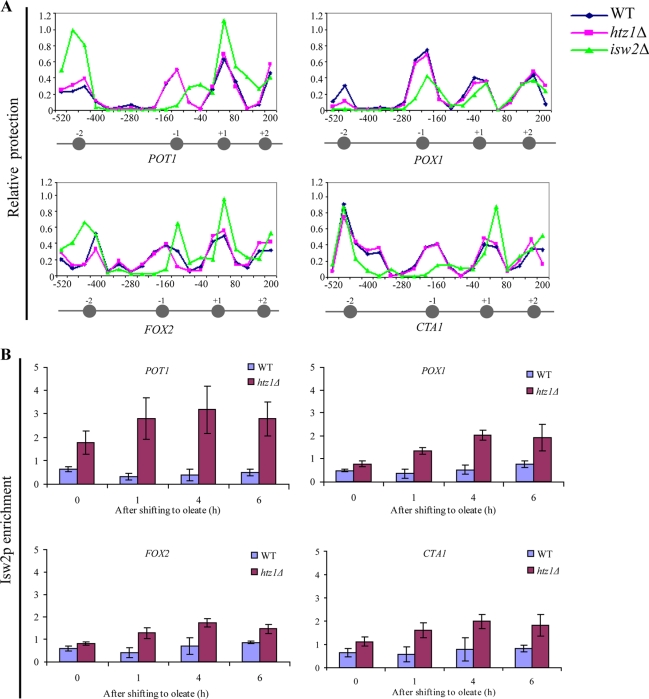
Nucleosome protection assays of isw2Δ cells led to significant changes in the nucleosome structure in all four promoters (Fig. 8A). These data indicate that Isw2p plays a role in the chromatin structure of these four promoters and are consistent with previous work on the POT1 promoter (8, 9, 39).
FIG. 8.
Isw2p can associate with oleate-responsive promoters in the absence of Htz1p. (A) The NuSA was used to determine the nucleosome positioning and density at the POT1, POX1, FOX2, and CTA1 promoters during repression (2% glucose) in WT, htz1Δ, and isw2Δ strains. Each point represents the relative protection of each PCR amplicon, quantified by real-time PCR and normalized to a centromeric control. The approximate location of a nucleosome is represented by a gray circle with the nucleosome number referred to in the text shown above the circle. (B) The association of Isw2p (as a C-terminal myc fusion) with the POT1, POX1, FOX2, and CTA1 promoters was determined by ChIP using anti-Myc antibodies, followed by gene-specific PCR. The relative enrichment ratio is plotted at four time points (0, 1, 4, and 6 h) of induction in oleate. ACT1 was used as an internal control to normalize the signals of promoter enrichment. Error bars show the standard deviations from three independent experimental values with two technical replicates of each.
Next, we used ChIP to assay the ability of Isw2p to associate with the four oleate-responsive promoters. Previous work has shown that in WT cells, Isw2p does not stably associate with these promoters, which suggests that under normal conditions, Isw2p contributes to the nucleosome structure at these Htz1p-containing POT1, POX1, FOX2, and CTA1 promoters through transient interactions (9). Similarly, we found very low levels of Isw2p association with these promoters in WT cells, in glucose and after oleate induction. However, substantial amounts of Isw2p were observed in association with each of these promoters in the htz1Δ cells. Isw2p remained associated with these promoters during their activation, suggesting a role in establishing chromatin dynamics in the absence of Htz1p (Fig. 8B).
DISCUSSION
The exposure of yeast cells to oleate results in a large-scale reorganization of gene expression regulatory networks and provides an excellent experimental system for understanding the mechanisms of gene expression at both the molecular and network levels (28, 33). The resulting changes in gene expression are widespread, representing the reorganization of regulatory networks governing numerous categories of gene function. For example, genes involved in protein translation and glycolysis are repressed, reflecting the shift in growth rates and metabolism (16, 32, 33). Likewise, genes linked to mitochondrial respiration and peroxisomal fatty acid metabolism are induced, reflecting the shift of the cells to nonfermentative β-oxidation as an energy source (16, 32, 33). The ability of yeast cells to adapt to this shift is dependent on the HTZ1 gene encoding the histone variant Htz1p/H2A.Z (19, 32). The data presented here demonstrate that Htz1p plays a critical role in this transition by contributing to the recruitment of TBP to oleate-responsive genes leading to the rapid and robust expression of highly inducible genes.
Transcriptome profiling studies presented here demonstrate that the expression of genes that are normally highly responsive to oleate is impaired in the absence of Htz1p. Because, under these conditions, many of the most strongly induced genes are required for peroxisomal β-oxidation and peroxisome proliferation, lack of Htz1p renders cells unable to respond efficiently to the transition and metabolize the fatty acids.
In order to elucidate the stepwise molecular function of Htz1p in the transcriptional regulation of these genes, we generated and compared various time course datasets to analyze chromatin states before and after the switch to oleic acid. We used ChIP to assay the dynamic association of Htz1p at the promoters of four model genes (POT1, POX1, FOX2, and CTA1) encoding peroxisomal matrix enzymes, the expression of which was perturbed by deletions of HTZ1. Htz1p has been proposed to preferentially bind repressed promoters, facilitating the rapid activation of the associated genes (41). Consistent with the current models, we demonstrate that Htz1p tends to be bound to these promoters in their repressed states (glucose) and disassociates from these promoters once the cells are exposed to oleate; however, this association and dissociation pattern occurs at levels that are promoter specific. The methods employed here did not reveal a significant dissociation of Htz1p from the CTA1 promoter. The data suggest that Htz1p levels on the CTA1 promoter are lower (approximately twofold lower than the control regions) than the other promoters examined. Thus, Htz1p does not appear to dissociate from the CTA1 promoter following oleic acid induction. The mechanisms underlying the promoter-specific effects of Htz1p and other epigenetic factors remain fertile ground for future study.
In addition, the data presented here support previous studies of both yeast and mammalian cells that demonstrate that Htz1p is deposited at promoters by the chromatin-remodeling protein Swr1p (24, 40). Similarly, as in other transcriptional responses (23), Gcn5p/Esa1p-mediated acetylation at Lys14 of Htz1p is required for efficient transcriptional activation. Moreover, Gcn5p/Esa1p-mediated acetylation at Lys14 of Htz1p is required for the efficient association of Htz1p at some oleate responsive promoters. The significantly decreased association of the Htz1p K14R mutant was observed under repressive conditions (i.e., glucose), and this decreased binding of Htz1p was also observed in cells lacking the enzyme (Gcn5p) responsible for Htz1p acetylation. Htz1p acetylation mutant cells also displayed defects in peroxisome proliferation and growth on oleic acid, similar to an HTZ1 null mutant. These data demonstrate that the acetylation of Htz1p, mediated by Gcn5p, is required for association with oleate-responsive promoters during repressed conditions and for normal transcriptional induction contributed by Htz1p.
Among the known effectors of Htz1p, Chz1p is relatively less well characterized. Luk et al. (21) identified a role for Chz1p as a nuclear chaperone for Htz1p, though the functional relationship between Chz1p and Htz1p with respect to transcriptional regulation remained uncharacterized. Here, we report that Chz1p, like Swr1p and Gcn5p, is also involved in the deposition of Htz1p at repressed promoters.
With respect to the role of Htz1p in TBP recruitment, studies of the GAL promoters have drawn different conclusions. In a recent study, TBP recruitment to the GAL1 promoter in htz1Δ strains was indistinguishable from that of the WT cells (10). However, earlier studies showed the Htz1p-dependent enrichment of TBP to GAL1 and GAL10 promoters during a time course of galactose induction (1). In the case of the fatty acid-inducible promoters tested here, the absence of Htz1p led to a significant reduction in the recruitment of TBP during oleate induction. We did not observe an increased enrichment of TBP at the FOX2 promoter during oleate induction. The dynamics of TBP binding appear to be promoter specific. The relative abundance of TBP at the FOX2 promoter prior to induction by oleic acid (compared to later time points) suggests that the activation of FOX2 does not require additional TBP binding and that the gene exists in a state poised for its activation upon receiving the correct signals (i.e., oleic acid). A comprehensive investigation of the dynamic and quantitative role of Htz1p in the recruitment of factors such as mediator and TBP to different promoters throughout the genome remains for future studies.
Our data suggest that the activation of repressed genes leads to a dynamic reorganization of chromatin structure. Specifically, upon oleate treatment, the nucleosome proximal to the initiation site in each promoter disassembles. This coincides with the ejection of Htz1p from the promoter. These data are in agreement with previous studies that indicate that nucleosome disassembly from promoters during activation provides access to the transcriptional machinery (25). While Htz1p is proposed to contribute to nucleosome disassembly during induction, surprisingly, the apparent rate of nucleosome disassembly at the oleate-responsive promoters was not dramatically different in htz1Δ cells. After initial disassembly, the nucleosomes reassemble (∼1 h after induction). Interestingly, these new nucleosomes do not appear to contain Htz1p, but the levels of transcription are nonetheless higher in WT cells than in cells lacking Htz1p. These data suggest that the initial presence of Htz1p ensures a normal transcriptional response and provides an epigenetic mark that persists after its loss, ensuring high levels of expression. A close examination of the data from nucleosome protection assays suggest that, in the absence of Htz1p, nucleosomes in the promoter regions of oleate-responsive genes are relatively more assembled, which may cause reduced expression levels at these later time points. The reassembly of nucleosomes during the coincident high levels of gene expression suggests that transcriptional activity is not simply related to an overall openness of chromatin at activated promoters and obstruction at repressed promoters. Rather, the precise dynamic placement and specific constituents of individual nucleosomes at promoters mechanistically regulate transcription by modulating the access of transacting factors to specific sites. Further, the characterization of the dynamics of the epigenetic marks, protein components of the nucleosomes, and chromatin-remodeling complexes at these promoters is required to delineate the mechanistic basis of the links between chromatin state and transcriptional activation.
The observed lower expression levels in cells lacking Htz1p may also be contributed by Isw2p. Isw2p is an energy-dependent chromatin-remodeling factor and negative regulator of gene expression (8, 9). When assayed for genome binding by ChIP-chip Isw2p association with the POT1, POX1, FOX2, and CTA1 promoters was not detected (9). Similarly, we found no enrichment of Isw2p at these promoters in WT cells. However, Isw2p bound to each promoter in the absence of Htz1p, and this association persisted through the 6 hours of induction. Therefore, the increased association of Isw2p to these four oleate-responsive promoters may account for the reduced expression levels in htz1Δ cells. It is also possible that in the WT cells Isw2p provides a complementary mechanism for chromatin structural changes independent of Htz1p. The loss of Htz1p provides an opportunity for Isw2p binding that is not normally functional in Htz1p-containing regions of chromatin. Further studies are required to understand the global relationship between Isw2p and Htz1p.
In mammalian cells histone variant H2A.Z can serve as a novel epigenetic marker of breast cancer progression as it is associated with lymph node metastasis and decreased breast cancer survival (13). In the plant Arabidopsis thaliana, histone H2A.Z is required for immune resistance to the phytopathogenic bacterium Pseudomonas syringae pv. tomato (22). In zebrafish, histone variant 2a z (H2afza) is essential for larval development through the generation of a lethal locus with a truncation of conserved carboxy-terminal residues in the protein (31). Taken together, these studies implicate histone H2A.Z in a number of diverse functions in different organisms. Because peroxisomes are highly dynamic and responsive eukaryotic organelles whose dysfunction is linked to a host of human conditions (4, 5), it is important to understand the roles of proteins like Htz1p that control aspects of chromatin structure and transcriptional responses preceding the proliferation of peroxisomes and fatty acid metabolism in S. cerevisiae.
Acknowledgments
We thank Bradley R. Cairns, Haiying Zhang, and Michael Grunstein for providing plasmids and strains and Jeff Ranish and members of the Aitchison laboratory for helpful comments and discussion during the course of this project.
This work was supported by grants GM075152, GMO76547, and RR022220 from the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 216270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel, and R. W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1311133-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446572-576. [DOI] [PubMed] [Google Scholar]
- 4.Bensinger, S. J., and P. Tontonoz. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454470-477. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J., and D. E. Moller. 2002. The mechanisms of action of PPARs. Annu. Rev. Med. 53409-435. [DOI] [PubMed] [Google Scholar]
- 6.Biddick, R. K., G. L. Law, and E. T. Young. 2008. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS ONE 3e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley, A. M., J. Aach, M. A. Steffen, and G. M. Church. 2002. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl. Acad. Sci. USA 997554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fazzio, T. G., M. E. Gelbart, and T. Tsukiyama. 2005. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 259165-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelbart, M. E., N. Bachman, J. Delrow, J. D. Boeke, and T. Tsukiyama. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19942-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gligoris, T., G. Thireos, and D. Tzamarias. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 274198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 2531-42. [DOI] [PubMed] [Google Scholar]
- 12.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua, S., C. B. Kallen, R. Dhar, M. T. Baquero, C. E. Mason, B. A. Russell, P. K. Shah, J. Liu, A. Khramtsov, M. S. Tretiakova, T. N. Krausz, O. I. Olopade, D. L. Rimm, and K. P. White. 2008. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ideker, T., V. Thorsson, A. F. Siegel, and L. E. Hood. 2000. Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J. Comput. Biol. 7805-817. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, J. D., V. T. Falciano, and M. A. Gorovsky. 1996. A likely histone H2A.F/Z variant in Saccharomyces cerevisiae. Trends Biochem. Sci. 21466-467. [DOI] [PubMed] [Google Scholar]
- 16.Koerkamp, M. G., M. Rep, H. J. Bussemaker, G. P. Hardy, A. Mul, K. Piekarska, C. A. Szigyarto, J. M. De Mattos, and H. F. Tabak. 2002. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell 132783-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 391235-1244. [DOI] [PubMed] [Google Scholar]
- 18.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 10218385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockshon, D., L. E. Surface, E. O. Kerr, M. Kaeberlein, and B. K. Kennedy. 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 17577-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 21.Luk, E., N. D. Vu, K. Patteson, G. Mizuguchi, W. H. Wu, A. Ranjan, J. Backus, S. Sen, M. Lewis, Y. Bai, and C. Wu. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25357-368. [DOI] [PubMed] [Google Scholar]
- 22.March-Diaz, R., M. Garcia-Dominguez, J. Lozano-Juste, J. Leon, F. J. Florencio, and J. C. Reyes. 2008. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53475-487. [DOI] [PubMed] [Google Scholar]
- 23.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303343-348. [DOI] [PubMed] [Google Scholar]
- 25.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 13474-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey, S. A., J. J. Smith, D. Orrell, M. Marelli, T. W. Petersen, P. de Atauri, H. Bolouri, and J. D. Aitchison. 2006. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat. Genet. 381082-1087. [DOI] [PubMed] [Google Scholar]
- 28.Ratushny, A. V., S. A. Ramsey, O. Roda, Y. Wan, J. J. Smith, and J. D. Aitchison. 2008. Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophys. J. 83715-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleem, R. A., B. Knoblach, F. D. Mast, J. J. Smith, J. Boyle, C. M. Dobson, R. Long-O'Donnell, R. A. Rachubinski, and J. D. Aitchison. 2008. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J. Cell Biol. 181281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103411-422. [DOI] [PubMed] [Google Scholar]
- 31.Sivasubbu, S., D. Balciunas, A. E. Davidson, M. A. Pickart, S. B. Hermanson, K. J. Wangensteen, D. C. Wolbrink, and S. C. Ekker. 2006. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 123513-529. [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, J. J., S. A. Ramsey, M. Marelli, B. Marzolf, D. Hwang, R. A. Saleem, R. A. Rachubinski, and J. D. Aitchison. 2007. Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol. Syst. Biol. 3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, J. J., Y. Sydorskyy, M. Marelli, D. Hwang, H. Bolouri, R. A. Rachubinski, and J. D. Aitchison. 2006. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol. Syst. Biol. 22006.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stargell, L. A., J. Bowen, C. A. Dadd, P. C. Dedon, M. Davis, R. G. Cook, C. D. Allis, and M. A. Gorovsky. 1993. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 72641-2651. [DOI] [PubMed] [Google Scholar]
- 36.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 71121-1124. [DOI] [PubMed] [Google Scholar]
- 38.Whitehouse, I., O. J. Rando, J. Delrow, and T. Tsukiyama. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 4501031-1035. [DOI] [PubMed] [Google Scholar]
- 39.Whitehouse, I., and T. Tsukiyama. 2006. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 13633-640. [DOI] [PubMed] [Google Scholar]
- 40.Wong, M. M., L. K. Cox, and J. C. Chrivia. 2007. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J. Biol. Chem. 28226132-26139. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]