Abstract
Myocardin, a coactivator of serum response factor (SRF), plays a critical role in the differentiation of vascular smooth muscle cells (SMCs). However, the molecular mechanisms regulating myocardin stability and activity are not well defined. Here we show that the E3 ligase C terminus of Hsc70-interacting protein (CHIP) represses myocardin-dependent SMC gene expression and transcriptional activity. CHIP interacts with and promotes myocardin ubiquitin-mediated degradation by the proteasome in vivo and in vitro. Furthermore, myocardin ubiquitination by CHIP requires its phosphorylation. Importantly, CHIP overexpression reduces the level of myocardin-dependent SMC contractile gene expression and diminishes arterial contractility ex vivo. These findings for the first time, to our knowledge, demonstrate that CHIP-promoted proteolysis of myocardin plays a key role in the physiological control of SMC phenotype and vessel tone, which may have an important implication for pathophysiological conditions such as atherosclerosis, hypertension, and Alzheimer's disease.
Phenotypic modulation of vascular smooth muscle (SM) cells (SMCs) plays a pivotal role in vascular development and remodeling during diseases. It is well established that the SMC phenotype is regulated by a wide range of extracellular cues. In response to vascular injury, vascular SMCs (VSMCs) dedifferentiate to a proliferative phenotype that is required for the various pathological states in atherosclerosis, neointimal hyperplasia, and hypertension (27, 28). VSMC differentiation is characterized by expressing the highest levels of SM contractile genes, whereas proliferating SMCs express reduced levels of these genes. Thus, changes in SM contractile gene expression are often used to mark SMC phenotypes. It has been known that most SM contractile genes such as SM α-actin, SM myosin heavy chain (SM-MHC), and SM22α are controlled by serum response factor (SRF), which binds to a sequence known as a CArG box (18, 28). The specific genes activated by SRF are determined by the intracellular signals, as well as the availability of positive and negative cofactors. One of these factors is the SRF cofactor myocardin, which activates SRF-dependent genes and functions as a “master regulator” of SMC differentiation during development (31, 32, 36).
Myocardin is expressed specifically in SM and cardiac muscle lineages and belongs to a SAP (SAF-A/B, acinus, PIAS) superfamily which has been implicated in cardiovascular development and adaptation of the cardiovascular system to hemodynamic stress (18, 28). Myocardin physically interacts with the MADS box transcription factor SRF, activating a subset of genes involved in cardiomyocyte and SMC differentiation in vivo (32). Recent reports from myocardin gain- and loss-of-function studies demonstrate that it is necessary and sufficient to initiate SMC differentiation (32). One study by Huang et al. shows that myocardin conditional mutant mice exhibit markedly diminished expression of SMC contractile proteins in the ductus arteriosus and a generalized defect in neural crest-derived SMC differentiation (14). Furthermore, several studies suggest that the expression and function of myocardin in SMC differentiation are tightly modulated, at least in part, via the RhoA/Rho kinase pathway (22, 24, 38). Recently, PIAS1 has been reported to regulate myocardin transactivity via its ligase-induced sumoylation (17). Phosphorylation of myocardin by GSK-3β also represses its transactivation and cardiomyocyte hypertrophy (2). However, the posttranslational mechanisms regulating myocardin protein stability and transcriptional activity in SMCs are not fully understood.
Increasing evidence suggests that chaperones and the ubiquitin (Ub)-proteasome system are responsible for maintaining the cell environment free from toxic misfolded proteins. One link that connects chaperones and the Ub-proteasome machinery is the chaperone-associated Ub ligase CHIP (C terminus of Hsc70-interacting protein). CHIP is most highly expressed in human striated muscle, aortic SMCs, endothelial cells, and other tissue cells (4). It has been shown that CHIP works in the protein quality control machinery at different levels and targets a number of Hsp70/90-associated substrates or aggregation-prone proteins for degradation by the proteasome (1, 7, 8, 15, 25, 39). Thus, these observations indicate that CHIP plays an important role in the regulation of cell growth, apoptosis, and neurodegeneration. It is still not clear, however, whether CHIP plays a physiological role in SMC differentiation and vessel tone. This prompted us to explore the possibility that CHIP is involved in the phenotypic modulation of VSMCs. Here we show that CHIP inhibits myocardin-dependent SMC gene expression and transcriptional activity. Furthermore, CHIP promotes phosphorylated myocardin ubiquitination and degradation by the proteasome, thereby inhibiting myocardin-dependent SMC differentiation and arterial tone.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents.
The expression plasmids containing wild-type (WT) and mutant CHIP and myocardin, His-Ub, hemagglutinin (HA)-SRF, the active form of GSK-3β S9A (Ac-GSK3β), the dominant-negative form of GSK-3β S9A (Dn-GSK-3β), CHIP small interfering RNA (siRNA), control siRNA, and SM22α- and ANF-luciferase reporters have been described previously (4, 6, 15, 31, 35). The antibodies used were anti-Flag antibody (M2; Sigma-Aldrich); anti-CHIP, anti-Myc (clone 9E10), and antimyocardin antibodies (Santa Cruz Biotechnology); anti-HA antibody (Roche); anti-Ub, anti-His, anti-GST, and anti-β-actin antibodies (Chemicon International Inc.); and anti-GSK-3β, anti-Hsp70, anti-Hsp90, and anti-mouse or anti-rabbit conjugated antibodies (Cell Signaling). MG132 and AR-A014418 were purchased from Calbiochem. Acetylcholine, phenylephrine, and sodium nitroprusside (SNP) were obtained from Sigma-Aldrich.
Cell culture and luciferase assays.
SMCs were isolated from the aortic media of 4-month-old male Sprague-Dawley rats and cultured as previously described (12). 293T and A10 cells were obtained from ATCC (Manassas, VA) and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, penicillin, and streptomycin at 37°C as described previously (20, 21). Transfection of cells was performed with Lipofectamine 2000 (Invitrogen) transfection reagent according to the protocol of the manufacturer.
Luciferase activity was assayed with the Luciferase Assay System (Promega) as described previously (20, 21). The data represent the means ± the standard errors of the means (SEM) of three independent experiments in triplicate and normalization to ß-galactosidase activity.
RNA analysis.
Total RNA was purified from cultured cells or arterial tissues with Trizol. SM gene analysis of SM22α, SM α-actin, SM-MHC, myocardin, SRF, smoothelin, and glyceraldehyde-3-phosphate dehydrogenase by reverse transcription (RT)-PCR was performed with primers designed to detect rat gene products as described previously (1, 23, 31). The relative quantities of mRNA were normalized against glyceraldehyde-3-phosphate dehydrogenase.
Immunoprecipitations, Western blotting, and GST pull-down assays.
Immunoprecipitation and immunoblotting were performed as described previously (20). Glutathione S-transferase (GST), GST-CHIP, and its mutant forms were prepared, and GST pull-down assays were performed as described previously (4). The FLAG-myocardin and corresponding deletion mutant proteins were expressed in 293 cells. The blots were developed with a chemiluminescence system, and the bands were scanned and densitometry analysis was performed with a Gel-pro 4.5 analyzer (Media Cybernetics).
Immunofluorescence.
Full-length myocardin cDNA was inserted into the green fluorescent protein (GFP) fusion vector pEGFP-C1 (Clontech) to create plasmid pEGFP-myocardin. Plasmids expressing Myc-CHIP and GFP-myocardin were cotransfected into A10 cells. After 24 h, cells were processed for immunofluorescence staining with the primary antibody (anti-Myc, 1:50 dilution) overnight at 4°C and an Alexa Fluor 488-conjugated secondary antibody (1:500 dilution). Samples were observed with a confocal microscope (20, 21).
Nickel-agarose chromatography and ubiquitination assays in vivo.
Cells were transfected with the indicated plasmids and lysed in Ni-agarose lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 5 mM imidazole, 0.05% Tween 20, 10 mM N-ethylmaleimide, complete protease inhibitor). Ub-conjugated proteins were purified by nickel chromatography (Ni-nitrilotriacetic acid-agarose; Qiagen) and detected by immunoblotting with anti-Flag or anti-His antibody (21). Ubiquitination in vivo was assayed as described previously (20).
Degradation assays.
To analyze the degradation kinetics of myocardin, 293T cells were seeded in six-well plates and transfected with myocardin and CHIP or the pcDNA3.1 vector as indicated. Protein lysates were prepared at the indicated time points after the addition of cycloheximide (50 μg/ml). The levels of myocardin were determined by immunoblotting, quantified at the indicated time points, and normalized to β-actin (20, 21).
In vitro ubiquitination reactions.
For CHIP-mediated ubiquitination, Flag-myocardin or mutant protein (amino acids [aa] 1 to 713) was precipitated from transfected 293 cells in the presence or absence of the GSK-3β inhibitor AR-A014418 (6 h) with 2 μg of anti-Flag antibodies. Purified Flag-myocardin and mutant protein (aa 1 to 713) were incubated in 20 μl of Ub reaction buffer-100 ng of E1-500 ng of UbcH5b-5 μg of Ub in the presence of bacterially purified GST only, GST-CHIP, or GST-CHIP ΔU-box (1 μg). After incubation at 30°C for 2 h, reaction products were analyzed by immunoblotting with the indicated antibodies as described previously (20).
Rat arterial contractility assays.
The thoracic aorta was isolated from anesthetized 2-month-old Sprague-Dawley rats by an approved institutional protocol according to Beijing Union Medical College guidelines. Aortic rings (4 mm in length) were mounted on 25-μm stainless steel wires in the chambers of a myograph (Danish Myo Technology A/S Inc.) in Krebs buffer (glucose at 5.5 mmol/liter, NaCl at 118.5 mmol/liter, KCl at 4.7 mmol/liter, MgSO4 at 1.2 mmol/liter, KH2PO4 at 1.2 mmol/liter, NaHCO3 at 25 mmol/liter, 37°C) gassed with 95% O2/5% CO2 at pH 7.4 and 37°C. The precontraction state of each aortic ring was determined by running a concentration-response curve for phenylephrine to establish maximum contraction. For relaxation studies with acetylcholine and SNP, rings were precontracted with phenylephrine at 80% maximum contraction as described previously (5, 26).
Transduction of aortic rings ex vivo.
Aortic segments were incubated in a 96-well plate at 37°C for 2 h with a viral suspension containing 3 × 108 PFU of adenovirus (Ad)-GFP or Ad-CHIP. Expression of GFP in the transduced carotid arteries was confirmed by observing fluorescence on fresh-frozen artery sections (10 μm) with a microscope (Nikon E600). For Western blot analysis, rings were rinsed twice with ice-cold phosphate-buffered saline and then lysed. Fifty micrograms of lysate was run on an 8% gel for the detection of myocardin and CHIP. β-Actin was used as a control for protein loading. The levels of SM22, SM-α-actin, and SM-MHC mRNAs in aortic rings were determined as described in the section on RNA analysis.
Statistical analysis.
Data are presented as means ± SEM. Differences between groups were evaluated with Student t tests. Concentration-response data were fitted to a logistic function by nonlinear regression and analyzed by a two-way analysis of variance. P values of less than 0.05 were regarded as significant.
RESULTS
CHIP represses the induction of myocardin-dependent SM contractile genes through downregulation of myocardin protein.
To investigate the role of CHIP in SMC differentiation, we first examined whether CHIP alters the expression of SM contractile genes. SMCs were transfected with CHIP plasmid, and the mRNA levels of SM contractile genes were quantified by RT-PCR. Transfection of cells with CHIP markedly decreased myocardin-induced SMC gene expression compared with that in control cells. Importantly, CHIP did not affect the mRNA levels of myocardin and myocardin-independent genes, including smoothelin and SRF (Fig. 1A and B). These results demonstrate that CHIP specifically inhibits myocardin-dependent SMC differentiation.
FIG. 1.
Overexpression of CHIP inhibits myocardin-induced SMC differentiation through downregulation of myocardin protein. (A) SMCs were transfected with CHIP or the empty vector for 24 h and then cultured in 2% horse serum for 48 h. The transcripts of SM genes, including those for SM22α, SM α-actin, SM-MHC, smoothelin, myocardin, and SRF, were examined by RT-PCR. (B) Quantitative analysis of the mRNA levels in panel A (n = 3, means ± SEM). *, P < 0.01 versus the vector. (C) SMCs were transfected with myocardin, WT CHIP, and U-box deletion mutant plasmids. Cells were cultured, and the mRNA levels of SM genes were analyzed as in panel A. (D) Quantitative analysis of mRNA transcripts in panel C (n = 3, means ± SEM). *, P < 0.01 versus myocardin alone. (E) SMCs were transfected with CHIP or vector plasmids and cultured in 2% horse serum for 0, 12, 24, 36, 48, or 60 h. The cell extracts were examined with antimyocardin, anti-CHIP, or β-actin antibody. (F) SMCs were transfected with increasing amounts of CHIP plasmid and cultured in 2% horse serum for 48 h. The cell extracts were examined as in panel E. (G, H) 293 cells were transfected with myocardin, SM22α, or ANF reporters and increasing amounts of WT CHIP or CHIP ΔU-box (0.25 and 0.5 μg). Luciferase (Luc) activity was measured (n = 3, means ± SEM). *, P < 0.01 versus myocardin alone. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Wt, wild type.
To examine whether Ub ligase activity is required for this effect, we coexpressed myocardin and WT CHIP or U-box deletion mutant CHIP (CHIP ΔU-box, loss of E3 ligase activity) and found that transfection with WT CHIP resulted in a marked inhibition of myocardin-dependent gene expression (SM22α, SM α-actin, and SM-MHC) compared with the control, whereas CHIP ΔU-box failed to page myocardin-induced gene expression (Fig. 1C and D), indicating that this effect is dependent on the E3 ligase activity of CHIP.
We next tested whether CHIP could regulate the stability of myocardin protein. SMCs were transfected with a CHIP plasmid for the indicated times of differentiation. The expression of myocardin sharply increased and reached its maximal level at 12 h but then gradually declined to a lower level until the completion of differentiation (Fig. 1E). Moreover, overexpression of CHIP decreased the level of endogenous myocardin in a dose-dependent manner (Fig. 1F), suggesting that CHIP downregulates the expression of myocardin at the protein level.
To further confirm that CHIP could directly inhibit myocardin-dependent transcriptional activity, luciferase assays were performed with SM22α and ANF promoters. As shown in Fig. 1G and H, overexpressing myocardin strongly activated these promoters. However, when WT CHIP was expressed at the same time, this induction was dramatically reduced in a dose-dependent manner, whereas CHIP ΔU-box was unable to repress myocardin-dependent gene transcriptional activity, indicating that the U-box domain is required for the inhibitory effect of CHIP on myocardin transactivation. Taken together, these data demonstrate that CHIP serves as a negative regulator of SMC differentiation through downregulation of myocardin protein and this effect is also dependent on its E3 ligase activity.
CHIP siRNA enhances the expression of SM contractile genes.
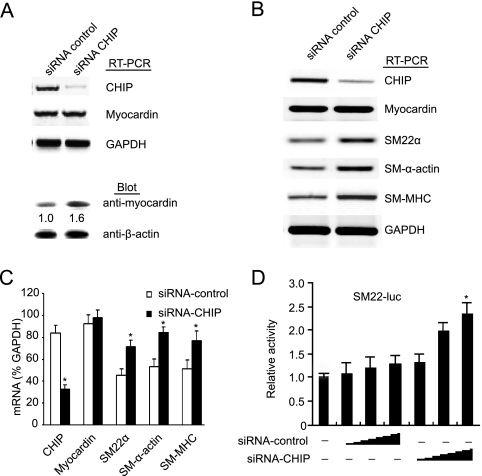
To further test the involvement of endogenous CHIP in the modulation of SMC differentiation, siRNA was used to inhibit endogenous expression of CHIP in SMCs. CHIP knockdown resulted in a moderate increase in the level of myocardin protein but no change in mRNA expression (Fig. 2A). Moreover, transfection of SMCs with CHIP siRNA also increased the expression of SM contractile genes, including SM22α, SM-α-actin, and SM-MHC, compared with transfection with control siRNA (Fig. 2B and C). Furthermore, transfection of SMCs with increasing amounts of CHIP siRNA resulted in a marked increase in SM22α promoter activity compared with that obtained with control siRNA (Fig. 2D). These findings suggest that endogenous CHIP plays a role in controlling the expression of myocardin-dependent SM contractile genes.
FIG. 2.
CHIP siRNA upregulates the expression of myocardin-induced SM contractile genes. (A) SMCs were transfected with control or CHIP siRNA and cultured in 2% horse serum for 48 h. CHIP and myocardin mRNA and protein levels were analyzed by RT-PCR and immunoblotting as indicated. (B) SMCs were transfected and cultured as in panel A. The transcripts of SMC genes, including those for SM22α, SM α-actin, SM-MHC, and myocardin, were examined by RT-PCR. (C) Quantitative analysis of the mRNA transcripts in panel B (n = 3, means ± SEM). *, P < 0.05 versus control siRNA. (D) SMCs were transfected with SM22α reporter, control siRNA, or CHIP siRNA for 48 h. Luciferase (luc) activity was measured (n = 3, means ± SEM). *, P < 0.01 versus control siRNA. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
CHIP interacts with myocardin.
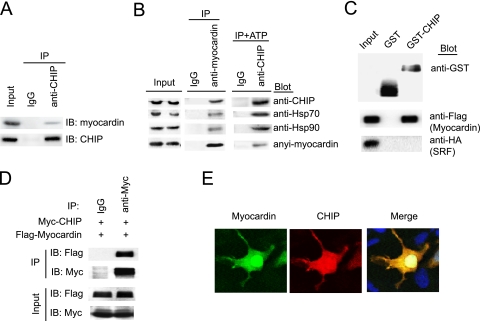
It has been known that one of the characteristics of an E3 kinase is the recognition of specific substrates. Next we examined whether endogenous CHIP interacts with myocardin in differentiated SMCs. The myocardin protein was detected in the immune complex precipitated with anti-CHIP antibody (Fig. 3A) and formed a complex with Hsp70/Hsp90 through CHIP in the absence or presence of ATP in SMCs (Fig. 3B), whereas no myocardin was found in the immune complex precipitated with a nonspecific immunoglobulin G (IgG) control (Fig. 3A and B). Also, Flag-myocardin expressed in 293 cells was pulled down by GST-CHIP fusion protein purified from bacteria but not between CHIP and SRF under this condition (Fig. 3C). Furthermore, immunoprecipitation analysis demonstrated that Flag-myocardin was detected in the Myc-CHIP immune complex, whereas no Flag-myocardin was found in control IgG (Fig. 3D). Finally, immunostaining indicated that CHIP was colocalized with myocardin in SMCs (Fig. 3E). Thus, myocardin forms a complex directly with CHIP in vivo and in vitro.
FIG. 3.
CHIP interacts with myocardin. (A) SMCs were cultured in 2% horse serum for 48 h, lysates were immunoprecipitated with anti-CHIP antibody or control IgG, and the immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted (IB) with antimyocardin (top) or anti-CHIP (bottom) antibody. (B) SMCs were cultured in 2% horse serum for 48 h. Cell lysates were subjected to coimmunoprecipitation (IP) with antimyocardin or anti-CHIP antibody in the absence or presence of ATP (10 mM) and rabbit IgG as a control. The presence of CHIP, Hsp70, Hsp90, or myocardin in the precipitated complexes was determined by immunoblotting as indicated. (C) CHIP interacts with myocardin in GST pull-down assays. GST fusion proteins were analyzed by immunoblotting with anti-GST antibody (top). The ability of Flag-myocardin (middle) or HA-SRF (bottom) expressed in 293 cells to be retained by GST or GST-CHIP fusion protein was analyzed by immunoblotting after binding reactions. (D) 293 cells were transfected with the indicated plasmids. Equal amounts of protein lysates were immunoprecipitated with the anti-Myc or control IgG serum and analyzed by immunoblotting with anti-Flag or anti-Myc antibody. (E) SMCs were cotransfected with Myc-CHIP and GFP-myocardin and immunostained with anti-Myc antibody. For quantitation, 150 cells per coverslip were counted and each treatment was performed in triplicate. A representative image is shown for each condition.
To identify the domain of CHIP that mediates the interaction with myocardin, a GST pull-down assay was performed with a series of GST-CHIP deletion mutant proteins as indicated. As shown in Fig. 4A and B, full-length CHIP (aa 1 to 303) bound strongly to myocardin and the U-box deletion mutant protein (aa 1 to 197) had less of an effect on binding to myocardin. However, the charged region deletion mutant protein (aa 1 to 142) failed to bind to myocardin in this assay, indicating that the charged domain (aa 143 to 197) of CHIP is required for binding to myocardin. Furthermore, the site of myocardin interaction with CHIP was determined with a series of truncated myocardin constructs and these assays demonstrate that residues of 714 to 935, containing the C-terminal transactivation domain (TAD) of myocardin, are necessary for the interaction with CHIP (Fig. 4C and D).
FIG. 4.
Mapping of the interactive domain of CHIP and myocardin. (A) The domain of CHIP involved in binding to myocardin. GST fusion proteins were analyzed by immunoblotting with anti-GST antibody (top). The ability of the truncated CHIP fusion proteins to bind to Flag-myocardin expressed in 293 cells was analyzed by immunoblotting with anti-Flag antibody (middle). Ten percent of the input is shown in the bottom panel. (B) Schematic representation of the CHIP mutant proteins used in panel A. (C) The domain of myocardin involved in binding to CHIP. GST-CHIP fusion proteins were analyzed by immunoblotting with GST antibody (top). The ability of the truncated myocardin proteins expressed in 293 cells to bind to GST-CHIP was analyzed by immunoblotting with anti-Flag antibody (middle). Ten percent of the input is shown in the bottom panel. (D) Schematic representation of deletion constructs of myocardin used in panel C. TPR, tetratricopeptide repeat; Q, a stretch of glutamine residues; SAP, SAF-A/B-acinus-PIAS domain; LZ, leucine zipper.
CHIP promotes myocardin ubiquitination for proteasome degradation.
Because the expression pattern of myocardin protein is inversely correlated with CHIP in SMCs (Fig. 1E and F and 2A) and CHIP formed a complex specifically with myocardin both in vivo and in vitro (Fig. 3 and 4), we sought to ascertain whether the effect of CHIP on myocardin protein is mediated through proteasome degradation. We first performed a pulse-chase assay and found that ectopic expression of CHIP resulted in a rapid decrease in myocardin protein compared with the vector control (Fig. 5A). Furthermore, transient expression of CHIP resulted in a marked reduction of myocardin protein in SMCs and this effect was abolished by the proteasome inhibitor MG132 (Fig. 5B).
FIG. 5.
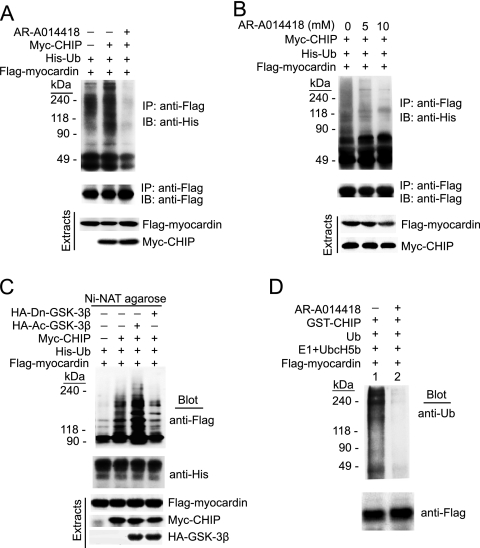
CHIP as an E3 ligase targets myocardin for proteasomal degradation. (A) 293 cells were transfected with a Flag-myocardin, Myc-CHIP, or vector plasmid and treated with cycloheximide (CHX, 10 μg/ml) at the indicated time points. The half-life of myocardin was determined by immunoblotting with anti-Flag antibody and quantified in the presence of a CHIP or vector plasmid (n = 3, means ± SEM). (B) The effect of CHIP on steady-state levels of myocardin was examined by transfection with a Myc-CHIP or vector plasmid in the presence of MG132 for 4 h. (C) The ubiquitination of endogenous myocardin in SMCs was tested by transfection with a Myc-CHIP or vector plasmid. Cell extracts were immunoprecipitated (IP) with antimyocardin antibody and immunoblotted (IB) with an anti-Ub (top) or antimyocardin (middle) antibody. The expression of myocardin and CHIP in the cell extracts was determined with antimyocardin or anti-Myc antibody (bottom). (D) 293 cells were transfected with a His-Ub, Myc-CHIP, or CHIP ΔU-box mutant plasmid in the presence of Flag-myocardin WT or mutant protein 1-713 and incubated with MG132 for 4 h before harvesting. Ub-conjugated proteins were purified by nickel chromatography and analyzed by immunoblotting with anti-Flag (top) or anti-His (middle) antibody. The expression of myocardin and CHIP in the cell extracts was determined with anti-Flag or anti-Myc antibody (bottom). (E) In vitro ubiquitylation reactions were performed with purified Ub, E1, E2-Ubc5b, GST alone, GST-CHIP, and GST-CHIP ΔU-box mutant protein in the presence of Flag-myocardin WT or 1-713 mutant protein. Reaction products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with anti-Ub (top) or anti-Flag antibody (bottom).
To determine whether CHIP promotes the ubiquitination of endogenous myocardin, a ubiquitination assay was performed with SMCs and myocardin-specific antibody. Coexpression of CHIP indeed induced the formation of high-molecular-weight species that can be recognized by anti-Ub antibody in the myocardin immunoprecipitates (Fig. 5C). Furthermore, CHIP E3 ligase activity was required for Ub conjugation to myocardin since myocardin ubiquitination was abrogated to a large degree by coexpression with a ligase-inactive mutant protein (CHIP ΔU-box) compared with WT CHIP (Fig. 5D, compared lane 4 with lane 5). As expected, myocardin lacking the TAD (mutant protein 1-713) was resistant to CHIP-mediated ubiquitination (Fig. 5D, lane 6).
To further confirm that CHIP can mediate myocardin ubiquitination in vitro, we reconstituted a reaction with the immunoprecipitated Flag-myocardin, GST-CHIP, GST-CHIP ΔU-box, and recombinant E1 and UbcH5b (29). Reactions were performed with Flag-myocardin expressed in 293T cells as a substrate. Consistent with in vivo results, ubiquitination of myocardin was detected in the presence of GST-CHIP (Fig. 5E, lane 6) whereas this effect was abolished by a U-box deletion in CHIP (Fig. 5E, compare lane 5 with lane 6). Furthermore, myocardin mutant protein 1-713, which lacks the CHIP-binding region, markedly attenuated the CHIP-induced ubiquitination of myocardin (Fig. 5E, compare lane 7 with lane 6). In addition, omitting E1, UbcH5b, Ub, or GST-CHIP abrogated the formation of poly-Ub chains (lanes 1 to 4). Collectively, these data indicate that CHIP functions as an E3 ligase and that the U-box domain of CHIP and the CHIP-binding region (TAD) of myocardin are both required for myocardin ubiquitination and degradation by the proteasome in vivo and in vitro.
CHIP ubiquitinates phosphorylated myocardin in vivo and in vitro.
It has been reported that myocardin protein is phosphorylated by GSK-3β (2). To test whether myocardin ubiquitination by CHIP requires its phosphorylation in vivo, 293 cells were transfected with His-Ub, Flag-myocardin, and Myc-CHIP and treated with AR-A014418, which is a known GSK-3β-specific inhibitor. Transfection with CHIP promoted myocardin ubiquitination in the absence of AR-A014418 but not in the presence of AR-A014418 treatment (Fig. 6A), and the ubiquitination of myocardin was reduced by AR-A014418 treatment in a dose-dependent manner (Fig. 6B). To further test whether overexpressing GSK-3β affects the ubiquitination of myocardin, 293 cells were transfected with Flag-myocardin, Myc-CHIP, Flag-Ub, Ac-GSK-3β, or Dn-GSK-3β and treated with the proteasome inhibitor MG132 for 4 h before harvesting. As shown in Fig. 6C, CHIP-mediated ubiquitination of myocardin was dramatically enhanced by Ac-GSK-3β whereas Dn-GSK-3β substantially paged the effect of CHIP. We then examined whether recombinant GST-CHIP ubiquitinates phosphorylated myocardin in vitro in the presence of E1, UbcH5b, and Flag-myocardin expressed in 293 cells pretreated without or with AR-A014418. Indeed, CHIP-mediated ubiquitination of myocardin was detected as high-molecular-weight bands by immunoblotting with anti-Ub in the absence of AR-A014418 whereas treatment with AR-A014418 abolished this effect (Fig. 6D, compare lane 2 with lane 1). Taken together, these results suggest that CHIP specifically ubiquitinates phosphorylated myocardin for degradation in vivo and in vitro.
FIG. 6.
CHIP promotes the ubiquitination of phosphorylated myocardin. (A) 293 cells were transfected with His-Ub, Flag-myocardin, and Myc-CHIP and left untreated or treated with AR-A014418 (10 mM) for 12 h in the presence of MG132 for 4 h before harvesting. Cell extracts were immunoprecipitated (IP) with the anti-Flag antibody and immunoblotted (IB) with the anti-His (top) or -Flag (middle) antibody. Cell extracts were subjected to immunoblotting with anti-Flag or anti-Myc antibody (bottom). (B) 293 cells were transfected with His-Ub, Flag-myocardin, and Myc-CHIP plasmids and treated with increasing amounts of AR-A014418 (0, 5, and 10 mM) for 12 h in the presence of MG132 for 4 h before harvesting. Cell extracts were immunoprecipitated and immunoblotted as in panel A. (C) 293 cells were cotransfected with vectors expressing His-Ub, Flag-myocardin, Myc-CHIP, GSK-3β S9A, or Dn-GSK in the presence of MG132 for 4 h before harvesting. Ub-conjugated proteins were purified by nickel chromatography and analyzed by immunoblotting with anti-Flag (top) and anti-His (middle) antibodies. The expression of myocardin, CHIP, and GSK-3β in the cell extracts was determined with anti-Flag, anti-Myc, or anti-HA antibody (bottom). (D) In vitro ubiquitination reactions were performed with purified Ub, E1, E2-UbcH5b, GST alone, or GST-CHIP. Flag-myocardin was precipitated from transfected 293 cells treated with AR-A014418 or left untreated for 12 h. Reaction products were analyzed as described in the legend to Fig. 5E.
CHIP gene transfer ex vivo reduces the expression of SMC contractile genes and contractility.
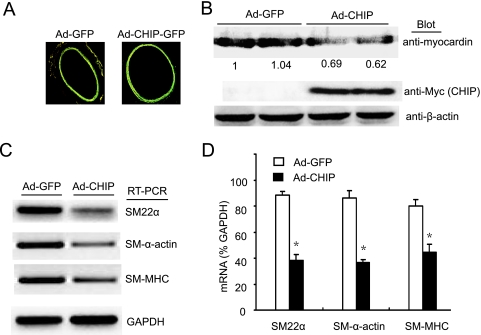
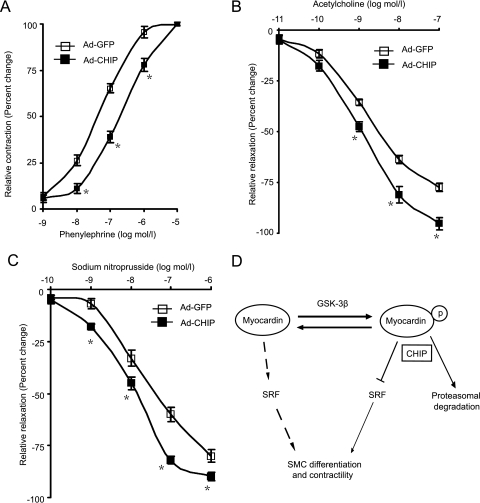
Because myocardin gene overexpression was associated with elevated contractile protein expression and contractility in cerebral VSMCs (5) and CHIP expression is inversely associated with myocardin protein level and its transactivation (Fig. 1 and 2), we hypothesized that overexpressing CHIP would reduce myocardin-dependent SMC contractile gene expression and contractility. Rat aortic rings were transduced ex vivo with Ad-CHIP or Ad-GFP, and the expression of contractile genes and the responses of transduced vessels to acetylcholine, phenylephrine, or SNP were examined. As shown in Fig. 7A, efficient gene transfer to the SMC layer of arteries, as indicated by GFP, was observed and Ad-CHIP transduction markedly decreased the expression of myocardin protein and SMC contractile genes compared with Ad-GFP controls in aortic rings (Fig. 7B to D). Consistent with these findings, in Ad-CHIP arteries, contractions in response to phenylephrine, a direct VSMC vasoconstrictor (3), were significantly reduced compared with those in Ad-GFP controls (Fig. 8A). In contrast, endothelium-dependent relaxations in response to acetylcholine were markedly increased (Fig. 8B). Furthermore, the effects of SNP, a nitric oxide donor that causes endothelium-independent VSMC relaxation (3), were also examined in vessels denuded of endothelium. As shown in Fig. 8C, SNP-mediated relaxations of Ad-CHIP-transduced rings in response to SNP were increased compared with those of Ad-GFP controls. These results suggest that CHIP is involved in the regulation of the SMC contractile phenotype in rat aortic rings.
FIG. 7.
CHIP gene transfer downregulates myocardin protein level and SM contractile gene expression in rat aortic rings. (A) Expression of GFP in aortic rings 2 days after transduction of Ad-GFP and Ad-CHIP was confirmed with a fluorescence microscope (magnification, ×40). (B) Myocardin and CHIP levels were determined by Western blot analysis. The blots are typical representatives of two different rings per group. (C) The expression of SM contractile gene mRNAs was detected by RT-PCR analysis. (D) Quantitative analysis of mRNA levels in panel C. Means ± SEM of four rings per group are shown. *, P < 0.01 versus Ad-GFP.
FIG. 8.
CHIP overexpression regulates the contractile phenotype in rat aortic rings. Shown are concentration-response curves for phenylephrine (A), acetylcholine (B), and SNP (C) in rat aortic rings transduced for 2 days with Ad-GFP (n = 4) or Ad-CHIP (n = 4). Means ± SEM from four rings per group are shown. *, P < 0.01 versus Ad-GFP. (D) Schematic model depicting the role of CHIP in SMC phenotype modulation via promoting myocardin degradation.
DISCUSSION
The cellular level of myocardin protein determines both its transcriptional activity and its responsiveness. The function of myocardin is regulated through a number of factors and signaling pathways (18, 28). However, the molecular mechanisms that regulate myocardin-mediated SMC differentiation and contractility are not fully elucidated. In the present study, we have investigated the relationship between CHIP and myocardin and its relevance for SMC differentiation and contractility. We clearly show that CHIP expression dramatically decreases myocardin stability and inhibits myocardin-dependent SMC differentiation through Ub-mediated degradation. The phosphorylation state is required for the ubiquitination of myocardin by CHIP. Importantly, CHIP gene transfer reduces the myocardin-mediated SMC contractile phenotype and arterial contractility.
CHIP was first identified as a protein that interacts with the C terminus of Hsp70 and shown to negatively regulate Hsp70 chaperone activity (4). Subsequently, CHIP was demonstrated to be a Ub ligase of the U-box family (4, 16). CHIP contains three major structural domains—a three-tetratricopeptide repeat domain that allows it to interact with chaperones such as Hsc70/Hsp70 and Hsp90, a charged domain, and a U-box domain that confers E3 Ub ligase activity on this cochaperone (4, 16). Recent studies have demonstrated that CHIP has a critical regulatory function in the protein quality control machinery of cells (4, 6, 9, 16) and is responsible for the misfolding-dependent ubiquitination and degradation of the glucocorticoid hormone receptor, ErbB2, cystic fibrosis transmembrane conductance regulator, androgen receptor, Ask1, p53, Tau, and ataxin 1 proteins (1, 7, 8, 10, 11, 15, 25, 39). Therefore, CHIP appears to play an important role in targeting chaperone-protein complexes to the proteasome. In this study, our data showed that CHIP expression dramatically decreased myocardin stability and inhibited myocardin-dependent SMC differentiation. Moreover, CHIP physically interacted with myocardin in vitro and in vivo and the charged region of CHIP was required for binding to myocardin. Furthermore, CHIP promoted myocardin ubiquitination and degradation by the proteasome and this process was dependent on its E3 ligase activity. These findings demonstrate that the cellular myocardin protein level is tightly controlled by CHIP-dependent proteasomal degradation. Importantly, CHIP-mediated degradation of myocardin represents a previously uncharacterized mechanism of inactivation of this SRF coactivator during SMC phenotype modulation.
In several cases, the target transcriptional factor proteins were first marked by phosphorylation for ubiquitination. For example, IκBα and β-catenin were phosphorylated by GSK-3β and then ubiquitinated by the Skp1/cullin 1/F-box complex (30, 37). Tau was also phosphorylated by GSK-3β and targeted to proteasomal degradation by CHIP (7, 19, 29). In the case of myocardin, its relevance for diverse physiological events in the posttranslational modifications, including phosphorylation and sumoylation, has been well investigated (2, 33, 34). Myocardin protein is known to be phosphorylated by GSK-3β. In contrast, GSK-3β pageade enhances myocardin-induced ANF transcription levels (2). In postphosphorylation, however, the fate of myocardin proteins remains unclear. In the present study, treatment with the GSK-3β-specific inhibitor AR-A014418 showed an inhibitory effect on the ubiquitination level of myocardin. Furthermore, cotransfection of Ac-GSK-3β resulted in a significant increase in CHIP-induced ubiquitination of myocardin whereas dominant-negative GSK-3β had an inhibitory effect. These results indicate that CHIP selectively recognizes and ubiquitinates phosphorylated myocardin in vivo and in vitro. Thus, it is not surprising that increased levels of CHIP, activated GSK3β, or both can accelerate the ubiquitination of myocardin.
Abnormal expression of SMC contractile genes and proliferation of VSMCs contribute to the various pathological states in atherosclerosis, hypertension, Alzheimer's disease, and the response to vascular injury (18). The expression of SM marker genes is regulated by the CArG box sequence and activities of SRF/myocardin (13). Although myocardin functions as a transcriptional coactivator that plays a critical role in SMC differentiation (32, 35), it is not fully understood whether myocardin can promote VSMC contractility. A recent study reported that elevated SRF/myocardin activity exhibits an Alzheimer's disease-like hypercontractile phenotype in human cerebral VSMCs and alters arterial vasodilation and contractility (5). In contrast, myocardin conditional mutant mice show a dramatic decrease in SMC contractile protein expression and a generalized defect in VSMC differentiation (14). These data place myocardin at a critical nodal point required for maintenance of the contractile SMC phenotype and contractility. However, it remains unclear what the physiological role of CHIP is in regulating the myocardin-mediated SMC phenotype and arterial contractility. Our results show that overexpression of CHIP in rat aortic rings markedly decreased the myocardin protein level and inhibited the expression of SMC contractile genes (Fig. 7B to D). Importantly, CHIP overexpression markedly reduced phenylephrine-induced contractility whereas endothelium-dependent and -independent relaxations in response to acetylcholine and SNP were increased in isolated aortic rings transduced with Ad-CHIP compared with those of Ad-GFP controls (Fig. 8A to C). These results suggest that CHIP plays a critical role in controlling the SMC phenotype and arterial tone.
In conclusion, here we provide comprehensive evidence that CHIP promotes the ubiquitination and degradation of phosphorylated myocardin by the proteasome. Through this mechanism, the effects of myocardin on SMC differentiation and vessel contractility are abolished by CHIP. The proposed mechanism is presented in Fig. 8D. Thus, the present study has identified a novel mechanism of posttranslational regulation of myocardin activity by CHIP that may open a new therapeutic approach to atherosclerosis and other cardiovascular disorders.
Acknowledgments
We are grateful to Da-zhi Wang for reagents and helpful discussion of the results of this study.
This study was supported by grants from the China Natural Science Foundation to H.L. (2006CB910306, 2006CB503805, 30670860, and 20060023051) and by grants from the National Institutes of Health to C.P. (R01 HL65619, R01 GM61728, and P01 AG024282).
We have no competing financial interests to declare.
Footnotes
Published ahead of print on 23 February 2009.
REFERENCES
- 1.Alberti, S., K. Bohse, V. Arndt, A. Schmitz, and J. Hohfeld. 2004. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 154003-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badorff, C., F. H. Seeger, A. M. Zeiher, and S. Dimmeler. 2005. Glycogen synthase kinase 3β inhibits myocardin-dependent transcription and hypertrophy induction through site-specific phosphorylation. Circ. Res. 97645-654. [DOI] [PubMed] [Google Scholar]
- 3.Bai, N., F. Moien-Afshari, H. Washio, A. Min, and I. Laher. 2004. Pharmacology of the mouse-isolated cerebral artery. Vasc. Pharmacol. 4197-106. [DOI] [PubMed] [Google Scholar]
- 4.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 194535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, N., R. D. Bell, R. Deane, J. W. Streb, J. Chen, A. Brooks, W. Van Nostrand, J. M. Miano, and B. V. Zlokovic. 2007. Serum response factor and myocardin mediate arterial hypercontractility and cerebral blood flow dysregulation in Alzheimer's phenotype. Proc. Natl. Acad. Sci. USA 104823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 393-96. [DOI] [PubMed] [Google Scholar]
- 7.Dickey, C. A., A. Kamal, K. Lundgren, N. Klosak, R. M. Bailey, J. Dunmore, P. Ash, S. Shoraka, J. Zlatkovic, C. B. Eckman, C. Patterson, D. W. Dickson, N. S. Nahman, Jr., M. Hutton, F. Burrows, and L. Petrucelli. 2007. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Investig. 117648-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickey, C. A., J. Koren, Y. J. Zhang, Y. F. Xu, U. K. Jinwal, M. J. Birnbaum, B. Monks, M. Sun, J. Q. Cheng, C. Patterson, R. M. Bailey, J. Dunmore, S. Soresh, C. Leon, D. Morgan, and L. Petrucelli. 2008. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc. Natl. Acad. Sci. USA 1053622-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esser, C., S. Alberti, and J. Hohfeld. 2004. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 1695171-188. [DOI] [PubMed] [Google Scholar]
- 10.Esser, C., M. Scheffner, and J. Hohfeld. 2005. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 28027443-27448. [DOI] [PubMed] [Google Scholar]
- 11.Galigniana, M. D., J. M. Harrell, P. R. Housley, C. Patterson, S. K. Fisher, and W. B. Pratt. 2004. Retrograde transport of the glucocorticoid receptor in neurites requires dynamic assembly of complexes with the protein chaperone hsp90 and is linked to the CHIP component of the machinery for proteasomal degradation. Brain Res. Mol. Brain Res. 12327-36. [DOI] [PubMed] [Google Scholar]
- 12.Hedin, U., J. Thyberg, J. Roy, A. Dumitrescu, and P. K. Tran. 1997. Role of tyrosine kinases in extracellular matrix-mediated modulation of arterial smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 171977-1984. [DOI] [PubMed] [Google Scholar]
- 13.Hendrix, J. A., B. R. Wamhoff, O. G. McDonald, S. Sinha, T. Yoshida, and G. K. Owens. 2005. 5′ CArG degeneracy in smooth muscle α-actin is required for injury-induced gene suppression in vivo. J. Clin. Investig. 115418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, J., L. Cheng, J. Li, M. Chen, D. Zhou, M. M. Lu, A. Proweller, J. A. Epstein, and M. S. Parmacek. 2008. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Investig. 118515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, J. R., C. Zhang, and C. Patterson. 2005. C-terminus of heat shock protein 70-interacting protein facilitates degradation of apoptosis signal-regulating kinase 1 and inhibits apoptosis signal-regulating kinase 1-dependent apoptosis. Cell Stress Chaperones 10147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, J., C. A. Ballinger, Y. Wu, Q. Dai, D. M. Cyr, J. Hohfeld, and C. Patterson. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 27642938-42944. [DOI] [PubMed] [Google Scholar]
- 17.Kawai-Kowase, K., M. S. Kumar, M. H. Hoofnagle, T. Yoshida, and G. K. Owens. 2005. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol. Cell. Biol. 258009-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai-Kowase, K., and G. K. Owens. 2007. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 292C59-C69. [DOI] [PubMed] [Google Scholar]
- 19.Kosik, K. S., and H. Shimura. 2005. Phosphorylated tau and the neurodegenerative foldopathies. Biochim. Biophys. Acta 1739298-310. [DOI] [PubMed] [Google Scholar]
- 20.Li, H. H., V. Kedar, C. Zhang, H. McDonough, R. Arya, D. Z. Wang, and C. Patterson. 2004. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 1141058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H. H., M. S. Willis, P. Lockyer, N. Miller, H. McDonough, D. J. Glass, and C. Patterson. 2007. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Investig. 1173211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, H. W., A. J. Halayko, D. J. Fernandes, G. S. Harmon, J. A. McCauley, P. Kocieniewski, J. McConville, Y. Fu, S. M. Forsythe, P. Kogut, S. Bellam, M. Dowell, J. Churchill, H. Lesso, K. Kassiri, R. W. Mitchell, M. B. Hershenson, B. Camoretti-Mercado, and J. Solway. 2003. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am. J. Respir. Cell Mol. Biol. 2939-47. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Z. P., Z. Wang, H. Yanagisawa, and E. N. Olson. 2005. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 9261-270. [DOI] [PubMed] [Google Scholar]
- 24.Mack, C. P., A. V. Somlyo, M. Hautmann, A. P. Somlyo, and G. K. Owens. 2001. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J. Biol. Chem. 276341-347. [DOI] [PubMed] [Google Scholar]
- 25.Muller, P., R. Hrstka, D. Coomber, D. P. Lane, and B. Vojtesek. 2008. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene 273371-3383. [DOI] [PubMed] [Google Scholar]
- 26.Mulvany, M. J., and W. Halpern. 1977. Contractile properties of small arterial resistance vessels in spontaneously hypertensive and normotensive rats. Circ. Res. 4119-26. [DOI] [PubMed] [Google Scholar]
- 27.Olson, E. S., and W. Dong. 2006. Nonlinearity in intracochlear pressure. ORL J. Otorhinolaryngol. Relat. Spec. 68359-364. [DOI] [PubMed] [Google Scholar]
- 28.Owens, G. K., M. S. Kumar, and B. R. Wamhoff. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84767-801. [DOI] [PubMed] [Google Scholar]
- 29.Shimura, H., D. Schwartz, S. P. Gygi, and K. S. Kosik. 2004. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2794869-4876. [DOI] [PubMed] [Google Scholar]
- 30.Strack, P., M. Caligiuri, M. Pelletier, M. Boisclair, A. Theodoras, P. Beer-Romero, S. Glass, T. Parsons, R. A. Copeland, K. R. Auger, P. Benfield, L. Brizuela, and M. Rolfe. 2000. SCF(β-TRCP) and phosphorylation dependent ubiquitination of IκBα catalyzed by Ubc3 and Ubc4. Oncogene 193529-3536. [DOI] [PubMed] [Google Scholar]
- 31.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105851-862. [DOI] [PubMed] [Google Scholar]
- 32.Wang, D. Z., and E. N. Olson. 2004. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 14558-566. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J., X. H. Feng, and R. J. Schwartz. 2004. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 27949091-49098. [DOI] [PubMed] [Google Scholar]
- 34.Wang, J., A. Li, Z. Wang, X. Feng, E. N. Olson, and R. J. Schwartz. 2007. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 27622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Z., D. Z. Wang, D. Hockemeyer, J. McAnally, A. Nordheim, and E. N. Olson. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428185-189. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z., D. Z. Wang, G. C. Pipes, and E. N. Olson. 2003. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 1007129-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida, T., M. H. Hoofnagle, and G. K. Owens. 2004. Myocardin and Prx1 contribute to angiotensin II-induced expression of smooth muscle α-actin. Circ. Res. 941075-1082. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, P., N. Fernandes, I. L. Dodge, A. L. Reddi, N. Rao, H. Safran, T. A. DiPetrillo, D. E. Wazer, V. Band, and H. Band. 2003. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 27813829-13837. [DOI] [PubMed] [Google Scholar]