Abstract
Methanosarcina acetivorans C2A encodes three putative hydrogenases, including one cofactor F420-linked (frh) and two methanophenazine-linked (vht) enzymes. Comparison of the amino acid sequences of these putative hydrogenases to those of Methanosarcina barkeri and Methanosarcina mazei shows that each predicted subunit contains all the known residues essential for hydrogenase function. The DNA sequences upstream of the genes in M. acetivorans were aligned with those in other Methanosarcina species to identify conserved transcription and translation signals. The M. acetivorans vht promoter region is well conserved among the sequenced Methanosarcina species, while the second vht-type homolog (here called vhx) and frh promoters have only limited similarity. To experimentally determine whether these promoters are functional in vivo, we constructed and characterized both M. acetivorans and M. barkeri strains carrying reporter gene fusions to each of the M. acetivorans and M. barkeri hydrogenase promoters. Generally, the M. acetivorans gene fusions are not expressed in either organism, suggesting that cis-acting mutations inactivated the M. acetivorans promoters. The M. barkeri hydrogenase gene fusions, on the other hand, are expressed in both organisms, indicating that M. acetivorans possesses the machinery to express hydrogenases, although it does not express its own hydrogenases. These data are consistent with specific inactivation of the M. acetivorans hydrogenase promoters and highlight the importance of testing hypotheses generated by using genomic data.
Methanogens are a phylogenetically diverse group of archaea, but the number of substrates that they use for growth and methanogenesis is limited. Using very similar central metabolic pathways, some methanogens utilize H2-CO2, while others use acetate or methylated compounds. Methanosarcina is the only genus that contains members capable of utilizing all these substrates, whereas most methanogens can use only one substrate. Nevertheless, not all Methanosarcina species are capable of using all methanogenic substrates. Instead, there is significant diversity within the genus Methanosarcina with respect to which substrates are utilized. To our knowledge, every Methanosarcina species isolated to date is capable of growth on methanol and other methylated compounds, and most species can use acetate; however, the ability to use H2-CO2 is less widespread. Interestingly, the ability to utilize H2 seems to correlate with the environments from which individual species were isolated. Accordingly, the majority of Methanosarcina species isolated from freshwater environments, such as Methanosarcina barkeri, are capable of utilizing H2-CO2 (18-20, 29, 32, 44). The majority of marine Methanosarcina isolates, such as isolates of Methanosarcina acetivorans, do not grow on H2-CO2 (9, 23, 24, 30, 38, 39, 45, 51).
The ability of methanogens to utilize H2 as a reductant requires a class of phylogenetically related enzymes called hydrogenases. In Methanosarcina, all hydrogenases are Ni-Fe enzymes that contain two core subunits, termed the large and small subunits (50). The large subunit contains two motifs that coordinate the Ni-Fe active site, RXCGXCXXXH and DPCXXCXXH. The small subunit contains 4 to 10 conserved Cys residues that coordinate Fe-S clusters.
The characterized Ni-Fe hydrogenases require posttranslational modification to become enzymatically active (3). While this modification has not been examined in Methanosarcina, it has been extensively studied in Escherichia coli, where the gene products of hypABCDE and hypF are responsible for insertion of Ni and Fe into the hydrogenase active site and coordination of —C☰O and —C☰N groups to the Fe. Homologs of these genes are found in each of the sequenced Methanosarcina genomes, suggesting that posttranslational activation occurs by similar mechanisms in these organisms.
The biochemical and physiological roles of hydrogenases have been studied in some detail in M. barkeri and Methanosarcina mazei. The Ech hydrogenase of M. barkeri has been purified and characterized. This enzyme is ferredoxin dependent and involved in coupling an electrochemical gradient to the reduction of CO2 to formyl-methanofuran (35, 47). Deletion of ech eliminates growth on acetate and H2-CO2 due to blocks in methanogenesis and prevents growth on methanol-H2-CO2 due to a biosynthetic block (36).
The Frh hydrogenase, which has also been purified from M. barkeri, couples oxidation of H2 to reduction of cofactor F420 (11). During growth on H2-CO2, Frh provides the reducing equivalents (via F420H2) for reduction of methenyl tetrahydromethanopterin (methenyl-H4MPT) to methylene-H4MPT and for reduction of methylene-H4MPT to methyl-H4MPT. M. barkeri frh is expressed during growth on H2-CO2, as expected. However, it is also expressed during growth on methanol (49), where its function is unknown. Molecular analysis revealed the presence of a second F420-reducing hydrogenase operon (encoding a protein with ca. 86 to 88% amino acid identity to M. barkeri Frh) in M. barkeri, designated fre, but the physiological role of this second operon is also unknown (49).
A third type of hydrogenase has been characterized from M. mazei. The so-called F420-nonreducing hydrogenase (also called viologen-reducing hydrogenase) transfers electrons from H2 to methanophenazine, which is the electron donor for reduction of the heterodisulfide of coenzyme M and coenzyme B to the free thiols of coenzyme M and coenzyme B (48). The M. mazei genome was found to contain operons encoding two copies of this hydrogenase, named vho and vht (whose products exhibited ca. 95% amino acid sequence identity). Transcription of vho occurs during growth on methanol, trimethylamine (TMA), H2-CO2, and acetate, while vht is transcribed during growth on methanol, TMA, and H2-CO2 but not during growth on acetate (6, 7). The relative role of each methanophenazine-reducing hydrogenase during methanogenesis has yet to be addressed.
Recent genome sequencing demonstrated that M. acetivorans has genes encoding three hydrogenases in its chromosome (frh and two vht-type hydrogenase genes), but the role of these hydrogenases in an organism incapable of growth on H2-CO2 is unclear (12). Furthermore, M. acetivorans does not produce detectable levels of hydrogenase during growth on methanol or methanol-H2-CO2 despite the fact that the operons are present in the chromosome (13). The reason for this discrepancy has not been addressed yet. Consistent with this observation, early proteomic studies failed to detect any of the hydrogenase subunits or maturation proteins, while nonquantitative microarray experiments revealed the presence of transcripts only for vhtG and hypA and not for other genes in these putative transcriptional units (26-28). A later study found a single peptide that could be attributed to VhtA but, again, no peptide from any other hydrogenase subunit or maturation protein (25). These data may explain previous studies showing that M. acetivorans produces barely detectable levels of hydrogenase activity in crude cell extract during growth on acetate (37). The reason that M. acetivorans displays hydrogenase activity during growth on acetate but not during growth in the presence of H2 is also unclear.
The availability of the M. acetivorans, M. mazei, and M. barkeri genome sequences (8, 12, 31) allows in silico analysis of the underlying reasons for the lack of H2 metabolism in M. acetivorans. Hypotheses generated from genomic data can then be tested in vivo using recently developed genetic techniques (43). In this study, M. acetivorans predicted hydrogenase amino acid sequences and promoters were compared to analogous regions in M. barkeri and M. mazei to gain insight into possible mechanisms of M. acetivorans hydrogenase inactivation. Reporter gene fusions to each hydrogenase promoter from M. barkeri and M. acetivorans were then examined to test the expression of the operons in vivo.
MATERIALS AND METHODS
Sequence analysis.
All sequence data used are data from previously published or publicly available genomes (8, 10, 12, 21, 31). DNA sequences were aligned using default ClustalW settings from Biology Workbench (http://workbench.sdsc.edu). Protein sequences were aligned using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/). Phylogeny was analyzed in PAUP* 4.0b10 using the maximum parsimony method with exhaustive sampling of tree topologies and 1,000 bootstrap replicates.
Growth of Methanosarcina strains.
Methanosarcina strains were grown as single cells (46) in anaerobic tubes sealed with butyl rubber stoppers at 37°C on HS media (34). In liquid media, 50 mM TMA, 120 mM acetate, or 125 mM methanol was provided as a growth substrate under an N2-CO2 (80:20) headspace. H2-CO2 (80:20) was provided for growth at an overpressure of 140 kPa when it was combined with methanol and at an overpressure of 220 kPa when it was used as the sole growth substrate. Growth on agar-solidified media has been described previously (4, 46). Puromycin was added to a final concentration of 2 μg ml−1 to select for the presence of pac (40). 8-Aza-2,6-diaminopurine was added to a final concentration of 20 μg ml−1 to select against the presence of hpt (41).
Plasmids and strains.
Standard techniques were used for plasmid manipulation and isolation in E. coli (1). Relevant plasmids and their functions are listed in Table 1. E. coli WM3118 was used as the host strain for plasmids containing oriV, so that copy number could be induced prior to plasmid purification (14). BW25141 was the host strain for Π-dependent plasmids (15). DH10B was the host strain for all other plasmids (Invitrogen, Carlsbad, CA). Promoter fusion plasmids were constructed by first PCR amplifying ca. 1 kb of DNA upstream of the translational start site of the first gene of each operon of interest. This PCR fragment was then cloned in frame into pAMG82, creating a translational fusion to uidA using ATG as the start site (14).
TABLE 1.
Plasmids used
| Plasmid | Purpose | Reference or source |
|---|---|---|
| pAMG82 | Parent plasmid for promoter fusions with ATG start site | 14 |
| pAMG84 | M. barkeri vht promoter fusion to uidA | This study |
| pAMG85 | M. acetivorans vht promoter fusion to uidA | This study |
| pAMG89 | M. acetivorans hyp promoter fusion to uidA | This study |
| pAMG90 | M. barkeri hyp promoter fusion to uidA | This study |
| pAMG91 | M. acetivorans vhx promoter fusion to uidA | This study |
| pAMG92 | M. acetivorans frh promoter fusion to uidA | This study |
| pAMG93 | M. barkeri frh promoter fusion to uidA | This study |
| pAMG94 | M. barkeri fre promoter fusion to uidA | This study |
| pAMG99 | M. barkeri ech promoter fusion to uidA | This study |
| pAMG101 | M. barkeri vhx promoter fusion to uidA | This study |
Construction of Methanosarcina strains.
Promoter fusion plasmids were transformed into M. acetivorans WWM82 (Δhpt::φC31int-attP) and M. barkeri WWM85 (Δhpt::φC31int-attP) (14) via liposome-mediated transformation (4, 33). These nonreplicating plasmids integrated into the M. acetivorans and M. barkeri chromosomes via φC31 Int-mediated site-specific recombination. Promoter fusions were verified to be integrated into each strain as single copies using the four-primer PCR screen as described previously (14).
During the course of this study, we determined that the M. acetivorans transformation efficiency was higher when organisms were grown on 50 mM TMA or 50 mM methanol than when they were grown on 125 mM methanol (A. Bose and W. W. Metcalf, unpublished data). Therefore, while M. barkeri strains were constructed using either 125 mM or 50 mM methanol as the growth substrate on agar plates, some M. acetivorans strains were constructed using methanol and some M. acetivorans strains were constructed using TMA. No phenotypic differences were observed between strains isolated on 125 mM methanol, strains isolated on 50 mM methanol, and strains isolated on 50 mM TMA.
β-Glucuronidase activity determination.
Strains were adapted to each growth substrate (without puromycin) for at least 15 generations prior to measurement. Triplicate cultures were measured on at least two separate days. β-Glucuronidase activity levels were determined as described previously (42). UV-visible absorbance spectra were recorded with a Hewlett Packard 8453 diode array spectrophotometer. β-Glucuronidase activity was calculated by following cleavage of p-nitrophenyl-β-d-glucuronide at 415 nm (12,402 mM−1 cm−1). The protein concentration was determined by the Bradford method (5), using bovine serum albumin as the standard. The limit of detection is 0.4 mU mg protein−1.
RESULTS
M. acetivorans lacks ech but contains frh, vht, and vhx hydrogenase genes.
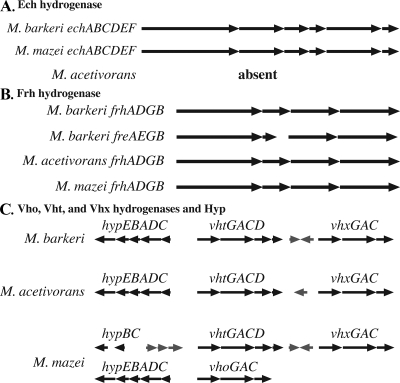
Three types of hydrogenases are thought to be required for growth of Methanosarcina on H2-CO2: a ferredoxin-reducing Ech hydrogenase, an F420-reducing hydrogenase, and a so-called F420-nonreducing (methanophenazine-reducing) hydrogenase. M. acetivorans was first hypothesized to not grow on H2-CO2 due to the lack of ech (12). However, more recent studies indicate that M. acetivorans does not produce any functional hydrogenase when it is grown on methanol or methanol-H2-CO2 (13). To address the mechanism of inactivation of the M. acetivorans hydrogenases, genomic regions containing putative hydrogenase genes were aligned to compare the M. acetivorans sequences to those of M. barkeri and M. mazei, both of which grow on H2-CO2 (Fig. 1).
FIG. 1.
Alignments of Methanosarcina genomic regions encoding hydrogenases. The black arrows represent hydrogenase and hydrogenase-related genes. All other genes are represented by gray arrows.
While M. acetivorans lacks genes encoding the Ech hydrogenase, which is present in both M. barkeri and M. mazei (Fig. 1A), each sequenced Methanosarcina isolate contains one full F420-reducing hydrogenase operon (frh) (Fig. 1B). M. barkeri has a second copy of the F420-reducing hydrogenase (fre) operon, which does not encode a homolog of the maturation peptidase FrhD (49). It seems possible, given the very high levels of amino acid identity, that the Fre hydrogenase could be processed by FrhD and therefore may represent a functional hydrogenase. However, the second copy may not be required for growth on H2-CO2, because M. mazei grows on H2-CO2 with only a single frh operon.
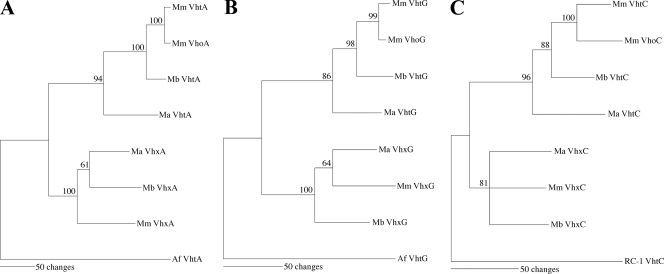
M. mazei was reported previously to contain two operons encoding the F420-nonreducing hydrogenase, designated vho and vht (viologen hydrogenase one and viologen hydrogenase two), which are ca. 95% identical to each other (7). However, Methanosarcina genome sequences reveal a more complex scenario (Fig. 1C). M. mazei also contains a third, more divergent operon homologous to vho and vht. Here, we refer to this third operon as vhx (for vho-like hydrogenase with unknown [x] function). M. barkeri and M. acetivorans each encode two F420-nonreducing hydrogenases: one hydrogenase most similar to the vho/vht-encoded hydrogenase and one hydrogenase most similar to the vhx-encoded hydrogenase. Phylogenetic analysis of the predicted protein sequences indicates that Vho and Vht group together to the exclusion of Vhx (Fig. 2) and that Vho likely arose from a recent duplication within the M. mazei lineage. Interestingly, the M. mazei vho and vhx operons each lack a homolog of vhtD, encoding the putative maturation peptidase, which should be required for posttranslational maturation of the hydrogenase enzyme.
FIG. 2.
Maximum parsimony phylogenetic trees for Methanosarcina Vht and Vhx subunits. Mm, M. mazei; Mb, M. barkeri; Ma, M. acetivorans; RC-I, rice cluster I methanogen; Af, Archaeoglobus fulgidus. Bootstrap scores greater than 50 are indicated at the nodes. (A) VhtA/VhxA subunits; (B) VhtC/VhxC subunits; (C) VhtG/VhxG subunits.
Just upstream of vht, M. barkeri contains a putative hyp operon, which is presumed to be required for posttranslational modification of the Ni-Fe hydrogenases (Fig. 1C). This location of hyp is conserved in M. acetivorans. In M. mazei, hyp is adjacent to vho instead of vht-vhx. An additional copy of hypB and hypC is located upstream of M. mazei vht.
Known active site residues are conserved in M. acetivorans hydrogenases.
M. acetivorans produces no detectable hydrogenase enzyme activity under most growth conditions (13). Numerous types of changes could account for this inactivation, including disruption of transcription or translation of hydrogenase genes, mutation of residues essential for enzyme function, lack of posttranslational modification of the enzymes, or a combination of the these factors. To address the mechanism of inactivation, the inferred amino acid sequences of hydrogenase subunits (encoded by ech, frh, vht, and vhx) from M. barkeri, M. acetivorans, and M. mazei were first aligned to determine if important amino acid residues are conserved.
While the M. acetivorans genome does not encode an Ech hydrogenase, Ech hydrogenase subunits from M. barkeri and M. mazei show 82% to 97% identity. All known essential residues are present in each strain (see Fig. S1 in the supplemental material). EchA and EchB are predicted integral membrane proteins thought to be involved in ion translocation. EchC is the hydrogenase small subunit, which contains four Cys residues conserved in all hydrogenase small subunits. EchE is the hydrogenase large subunit and contains the conserved RXCGXCXXXH and DPCXXCXXH motifs that coordinate the hydrogenase Ni-Fe center. EchF is a polyferredoxin and contains two prototypical [4Fe-4S] cluster-binding sites. Conserved acidic amino acids in EchA, EchB, EchC, EchE, and EchF are thought to be involved in coupling electron transfer to ion translocation (16, 22; R. Hedderich, personal communication).
In Frh, all known essential active site residues are conserved in M. acetivorans, as well as in M. barkeri and M. mazei (see Fig. S2 in the supplemental material). FrhA is the hydrogenase large subunit and contains the conserved RXCGXCXXXH and DPCXXCXXH Ni-coordinating residues. FrhD is a putative maturation peptidase and contains the proposed catalytic Asp residue. FrhG is the hydrogenase small subunit and includes two conserved [4Fe-4S] motifs and six other conserved Cys residues thought to coordinate metal centers. FrhB is the F420-binding subunit. Furthermore, pairwise comparisons of each subunit reveal that M. acetivorans Frh is not more divergent than the M. barkeri and M. mazei copies (see Table S1 in the supplemental material). Thus, there is no obvious reason that the M. acetivorans frh operon should not produce active hydrogenase. Nevertheless, it is still possible that other nonobvious mutations may inactivate the Frh hydrogenase.
Essential amino acid residues are also generally conserved in M. acetivorans Vht and Vhx. VhtG, the hydrogenase small subunit, contains 11 conserved Cys residues (see Fig. S3 in the supplemental material), 10 of which are presumed to coordinate Fe-S centers (7). The VhtG homologs of each organism contain most of the CXXCXnGXCXXXGXmGCPP motif commonly found in hydrogenase small subunits but lack the final Pro residue. Interestingly, the VhxG homologs from M. barkeri and M. mazei also lack the first Cys residue in the motif, while it is present in M. acetivorans VhxG. The ability of M. barkeri and M. mazei Vhx to function as a hydrogenase in the absence of this Cys residue is not known. Another interesting feature, conserved in each sequence of subunit G, is the Tat signal peptide (RRXFXKX18V/AXA), indicating that this hydrogenase is probably localized to the periplasmic side of the membrane (2, 7). VhtA/VhxA is the hydrogenase large subunit and contains the RXCGXCXXXH and DPCXXCXXH Ni-binding motifs. VhtC/VhxC is the cytochrome b subunit, which contains two His residues proposed to coordinate heme (7). However, the second His residue is not conserved in VhxC, while several other His residues are conserved. Therefore, one of the other His residues may coordinate heme instead. VhtD is predicted to be a maturation peptidase that contains a conserved, catalytic Asp residue. Like Frh/Fre discussed above, it seems possible that the VhtD peptidase may process each of the Vht, Vho, and Vhx hydrogenases.
The level of identity was calculated for each pairwise set of Vht and Vhx subunits (see Table S2 in the supplemental material). While the levels of identity between Frh subunits are approximately equal for all three organisms, M. acetivorans Vht is more divergent from M. barkeri and M. mazei Vht, and the levels of identity are 8 to 16% lower between M. acetivorans and either M. barkeri or M. mazei than between M. barkeri and M. mazei. Despite this divergence, M. acetivorans Vht and Vhx contain all of the proposed active site residues and vht and vhx may encode functional hydrogenases.
The vht and ech promoters are well conserved, while the frh and vhx promoters are not well conserved.
The lack of obvious inactivating mutations in the important catalytic and structural residues in the hydrogenase-encoding regions suggests that the M. acetivorans hydrogenase operons have the potential to encode functional proteins. Therefore, the putative promoter region of each operon (ca. 1 kb upstream of each translational start site) was examined for conserved sequences involved in transcription and translation initiation. Although we have not mapped the transcription start sites of these operons, we are not aware of any archaeal transcriptional units whose start sites are not within 1 kbp of the coding sequence.
The echA genes from M. barkeri and M. mazei contain a conserved ATG start site and a putative ribosome binding site (RBS) (see Fig. S4 in the supplemental material). While the ech promoters have not been characterized, a putative TATA box and BRE element are present.
The frh promoters show very little conservation between the organisms (see Fig. S5 in the supplemental material). The TTG translational start site is the only portion that is absolutely conserved. The sequence for the putative RBS is conserved, but the spacing is different for the two M. barkeri promoters compared to the M. mazei and M. acetivorans promoters. Neither characterized M. barkeri transcriptional start site (49) is conserved in either M. mazei or M. acetivorans.
The vht promoters, on the other hand, are highly conserved in all three Methanosarcina strains, from the putative BRE element to the start site (see Fig. S6 in the supplemental material). Using the characterized M. mazei promoters as a reference, all four promoters share a conserved ATG start site, RBS, transcriptional start site, TATA box, and BRE element.
Unlike the vht promoters, the vhx promoters share only limited identity near the putative translational start site (see Fig. S7 in the supplemental material). No putative RBS could be identified upstream of either potential translational start site, and the transcriptional start site has not been identified.
M. barkeri hydrogenase promoters are functional, while M. acetivorans hydrogenase promoters are nonfunctional.
The analysis described above shows that the lack of hydrogenase activity in M. acetivorans extracts is not due to obvious mutations in either the coding regions or the promoters. To experimentally test the functionality of the Methanosarcina hydrogenase promoters, translational fusions to uidA to the first genes in each operon from M. barkeri and M. acetivorans were constructed. For frh and fre the promoter fusions were constructed using an ATG start site rather than the native TTG translational start site. The 10 uidA fusions (M. acetivorans Pvht, Pvhx, Pfrh, and hyp and M. barkeri Pech, Pvht, Pvhx, Pfrh, Pfre, and Phyp) were inserted into both the M. barkeri chromosome and the M. acetivorans chromosome via φC31 Int-mediated site-specific integration (14).
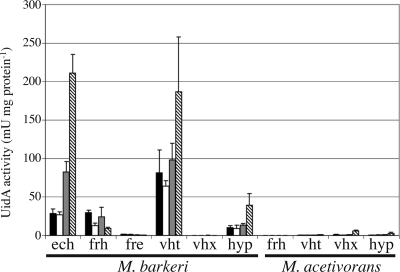
In M. barkeri, the M. barkeri Phyp, Pvht, Pech, and Pfrh promoters are all expressed on all substrates (Fig. 3). Phyp and Pvht are expressed at the same level on methanol or methanol-H2-CO2, but they are upregulated ca. twofold on acetate. Pech is also expressed at the same level on methanol and methanol-H2-CO2, but it is upregulated ca. 10-fold on acetate. Pfrh, on the other hand, is expressed at similar levels on H2-CO2 and methanol, but it is downregulated two- to threefold both on acetate and methanol-H2-CO2. M. barkeri Pfre is expressed at only a very low level and is downregulated on acetate, like Pfrh. No expression was detected for M. barkeri Pvhx. The M. acetivorans promoter fusions in M. barkeri were expressed at a level near or below the background level for nearly all fusions on each substrate. The exception is M. acetivorans Pvhx, which was upregulated ca. fivefold on acetate.
FIG. 3.
Hydrogenase promoter activity in M. barkeri. β-Glucuronidase activity (UidA activity) was measured for M. barkeri strains carrying single-copy reporter gene fusions to each M. barkeri and M. acetivorans hydrogenase promoter. The growth substrates used were methanol (black bars), methanol plus H2 (open bars), H2-CO2 (gray bars), and acetate (striped bars).
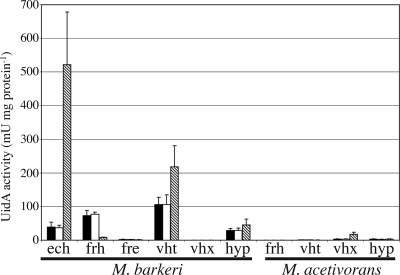
The same pattern was generally seen when these promoter fusions were inserted into the M. acetivorans chromosome (Fig. 4). The exception to this pattern is the expression of M. barkeri Pfrh on methanol-H2-CO2. In M. barkeri, this promoter is downregulated on methanol-H2-CO2 compared to the expression on methanol alone. In M. acetivorans, the expression levels were the same on methanol with and without H2-CO2. Additionally, the regulation is more drastic in M. acetivorans: M. barkeri frh expression is 10-fold lower on acetate than on methanol. These data are fully consistent with previous proteomic and microarray experiments showing that the levels of expression of the M. acetivorans hydrogenases and maturation proteins are at or below the limit of detection (25-28).
FIG. 4.
Hydrogenase promoter activity in M. acetivorans. β-Glucuronidase activity (UidA activity) was measured for M. acetivorans strains carrying single-copy reporter gene fusions to each M. barkeri and M. acetivorans hydrogenase promoter. The growth substrates used were methanol (black bars), methanol plus H2 (open bars), and acetate (striped bars).
DISCUSSION
The data presented here provide additional mechanistic evidence regarding the inability of M. acetivorans to metabolize hydrogen. The previously published genome sequence established that ech is not present, presumably due to deletion (12). However, genetic studies clearly showed that the lack of ech alone did not explain the dearth of hydrogen metabolism in this organism (13). Here we show that the remaining M. acetivorans hydrogenases have been inactivated at the level of transcription and/or translation.
Several mechanisms could be envisioned for this hydrogenase inactivation. For example, the M. acetivorans promoters could be inactivated by a trans-acting factor, such as a nonfunctional activator or a constitutive repressor. If this occurs, one would expect all 10 hydrogenase promoters to be inactive in M. acetivorans and active in M. barkeri (which clearly expresses its own hydrogenases). Alternatively, each M. acetivorans promoter could be inactivated via cis-acting mutations, resulting in genes that are not transcribed. If this model were correct, all the M. acetivorans promoters should be nonfunctional in both M. acetivorans and M. barkeri, while the M. barkeri promoters should be active in both M. acetivorans and M. barkeri. Because the M. acetivorans promoters showed similarly low levels of activity in both M. acetivorans and M. barkeri, while the M. barkeri promoters were expressed at similar levels in both organisms, it seems that the latter mechanism is present; i.e., the M. acetivorans genes are nonfunctional due to cis-acting mutations in either transcriptional or translational sequence elements. Nevertheless, the mutations resulting in this inactivation are not always obvious.
The M. acetivorans vht promoter shares the conserved BRE element, TATA box, +1 transcriptional start site, RBS, and ATG translational start site that have been characterized in M. mazei. One explanation for the lack of vht expression in M. acetivorans could be that this promoter has lost either the binding site for a transcriptional activator or a required secondary structure in the mRNA somewhere in the nonconserved region. The M. acetivorans frh promoter, on the other hand, has little, if any, similarity to the M. barkeri and M. mazei promoters, which are also dissimilar to each other. Thus, the sequence motifs needed for frh expression cannot be determined by such a comparative approach. Because the M. barkeri and M. mazei frh promoters are so dissimilar, M. mazei may use a different regulatory mechanism for frh than M. barkeri uses.
Based upon operon structure and proximity to vhx, we refer to the operon encoding the first M. barkeri methanophenazine-reducing hydrogenase as vht rather than vho, but interestingly, the expression of M. barkeri vht more closely resembles the reported expression pattern of M. mazei vho. In M. mazei, vho is expressed on methanol, acetate, and H2-CO2, while vht is not expressed on acetate (6). The fact that the M. mazei vho and vht promoters share so many features but are expressed differently indicates that another level of regulation has yet to be elucidated. This regulation may be absent from M. acetivorans. Because vhtD encodes the putative maturation peptidase, it is unclear whether M. mazei vho can produce a functional hydrogenase during growth on acetate in the absence of vhtD expression.
The function of vhx is still cryptic. The Vhx gene is present in the genomes of all three sequenced Methanosarcina species, indicating it was in the genome since before the organisms diverged from each other. All known essential Vht residues are present in Vhx with the exception of the first conserved Cys in VhxG in M. barkeri and M. mazei. Thus, whether Vhx can function as a hydrogenase also remains unclear. However, M. barkeri vhx does not seem to be expressed. Therefore, either very low levels are required or Vhx no longer has a function in M. barkeri under laboratory conditions.
In contrast, vhx seems to be the only M. acetivorans hydrogenase operon expressed (albeit at a low level), and it is upregulated on acetate. Furthermore, M. acetivorans VhxG is the only VhxG that contains the first conserved Cys present in VhtG. Taken together, these findings may explain the very low levels of hydrogenase detected in M. acetivorans during growth on acetate (37) but not during growth on methanol or methanol-H2-CO2 (13). Presuming that Vhx requires proteolytic processing like other hydrogenases, it is also unclear what protein would perform this cleavage in the absence of vht expression. Therefore, the physiological role of Vhx in M. acetivorans remains unclear as well.
The differential regulation of M. barkeri frh suggests that M. barkeri and M. acetivorans can both sense a flux of carbon between CO2 and methyl-H4MPT. Accordingly, we observed that the expression of M. barkeri Pfrh is decreased on acetate in both organisms but is decreased on the combination of methanol, H2, and CO2 only in M. barkeri. A commonality between M. barkeri growth on acetate and M. barkeri growth on methanol-H2-CO2 is a decrease in this flux of carbon between CO2 and methyl-H4MPT. Methanogenesis from acetate or methanol-H2-CO2 does not require this portion of the pathway, and it may be relegated to a strictly biosynthetic role (13). Indeed, the levels of many of the enzymes in this portion of the pathway are reduced during growth on acetate (17). During methanogenesis from methanol, on the other hand, 25% of the methyl groups are oxidized to CO2 via this pathway, while all carbon passes through this pathway during methanogenesis from H2-CO2. Therefore, a decrease in the concentration of one of these methanogenic intermediates during growth on acetate and on methanol-H2-CO2 may serve as the signal to downregulate expression of frh. In M. acetivorans, however, one would not expect to see this effect on methanol-H2-CO2 because the organism does not use H2. Thus, even in the presence of H2, methanol is still presumably oxidized via this pathway, and the level of M. barkeri Pfrh expression would be predicted to be high. Instead, flux through this pathway should decrease in M. acetivorans only during growth on acetate, resulting in decreased M. barkeri Pfrh expression.
Like the physiological role of vhx, the physiological role of fre is unknown. This work suggests that M. barkeri fre has minor importance in M. barkeri due to a low level of expression. Furthermore, M. mazei grows on H2-CO2 without fre, so fre must not be absolutely required. It is therefore not surprising that fre is expressed at a much lower level than frh, implying that it has decreased importance under laboratory conditions. Similar to Vhx, Fre lacks a homolog of the maturation peptidase encoded by frhD. Therefore, Fre functionality may be strictly dependent on the presence of the frh operon for posttranslational processing.
Finally, this study demonstrates the importance of testing the hypotheses derived from genome sequences. Based solely on a comparison of genomic data, M. acetivorans was predicted to produce a hydrogenase, implying that H2 could play a significant role in its physiology. While no deleterious mutations were found within the coding region of the M. acetivorans hydrogenase genes, biochemical, genetic, and now expression data suggest that these genes are inactive. In support of this idea, we have been unable to transform M. acetivorans with plasmids encoding the active Vht hydrogenase from M. barkeri, suggesting that M. acetivorans cannot tolerate Vht hydrogenase expression, at least under the conditions tested (data not shown). It should be noted, however, that it remains possible that we simply have not yet determined appropriate conditions needed to elicit expression of the M. acetivorans genes. If such conditions exist, then it is clear that the regulatory mechanism of the M. acetivorans genes is distinct from that of the orthologous M. barkeri genes. If they are never expressed, they should be considered pseudogenes despite the lack of obvious deleterious mutations in the coding regions.
Supplementary Material
Acknowledgments
This work was supported by grants to W.W.M. from The National Science Foundation (grant MCB0212466) and the Department of Energy (grant DE-FG02-02ER15296) and by NIH Cell and Molecular Biology Training Grant T32GMO7283) to A.M.G.
Footnotes
Published ahead of print on 6 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22393-404. [DOI] [PubMed] [Google Scholar]
- 3.Blokesch, M., A. Paschos, E. Theodoratou, A. Bauer, M. Hube, S. Huth, and A. Bock. 2002. Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30674-680. [DOI] [PubMed] [Google Scholar]
- 4.Boccazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J. Bacteriol. 1822611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Deppenmeier, U. 1995. Different structure and expression of the operons encoding the membrane-bound hydrogenases from Methanosarcina mazei Go1. Arch. Microbiol. 164370-376. [DOI] [PubMed] [Google Scholar]
- 7.Deppenmeier, U., M. Blaut, S. Lentes, C. Herzberg, and G. Gottschalk. 1995. Analysis of the vhoGAC and vhtGAC operons from Methanosarcina mazei strain Gö1, both encoding a membrane-bound hydrogenase and a cytochrome b. Eur. J. Biochem. 227261-269. [DOI] [PubMed] [Google Scholar]
- 8.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4453-461. [PubMed] [Google Scholar]
- 9.Elberson, M. A., and K. R. Sowers. 1997. Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae. Int. J. Syst. Bacteriol. 471258-1261. [DOI] [PubMed] [Google Scholar]
- 10.Erkel, C., M. Kube, R. Reinhardt, and W. Liesack. 2006. Genome of rice cluster I archaea—the key methane producers in the rice rhizosphere. Science 313370-372. [DOI] [PubMed] [Google Scholar]
- 11.Fiebig, K., and B. Friedrich. 1989. Purification of the F420-reducing hydrogenase from Methanosarcina barkeri (strain Fusaro). Eur. J. Biochem. 18479-88. [DOI] [PubMed] [Google Scholar]
- 12.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guss, A. M., B. Mukhopadhyay, J. K. Zhang, and W. W. Metcalf. 2005. Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H(2) metabolism between closely related species. Mol. Microbiol. 551671-1680. [DOI] [PubMed] [Google Scholar]
- 14.Guss, A. M., M. Rother, J. K. Zhang, G. Kulkarni, and W. W. Metcalf. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 1836384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedderich, R., and L. Forzi. 2005. Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 1092-104. [DOI] [PubMed] [Google Scholar]
- 17.Jablonski, P. E., A. A. DiMarco, T. A. Bobik, M. C. Cabell, and J. G. Ferry. 1990. Protein content and enzyme activities in methanol- and acetate-grown Methanosarcina thermophila. J. Bacteriol. 1721271-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis, G. N., C. Strompl, D. M. Burgess, L. C. Skillman, E. R. Moore, and K. N. Joblin. 2000. Isolation and identification of ruminal methanogens from grazing cattle. Curr. Microbiol. 40327-332. [DOI] [PubMed] [Google Scholar]
- 19.Jussofie, A., F. Mayer, and G. Gottschalk. 1986. Methane formation from methanol and molecular-hydrogen by protoplasts of new methanogenic isolates and inhibition by dicyclohexylcarbodiimide Arch. Microbiol. 146245-249. [Google Scholar]
- 20.Kandler, O., and H. Hippe. 1977. Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Arch. Microbiol. 11357-60. [DOI] [PubMed] [Google Scholar]
- 21.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390364-370. [DOI] [PubMed] [Google Scholar]
- 22.Künkel, A., J. A. Vorholt, R. K. Thauer, and R. Hedderich. 1998. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur. J. Biochem. 252467-476. [DOI] [PubMed] [Google Scholar]
- 23.Lai, M. C., and C. J. Shih. 2001. Characterization of Methanococcus voltaei strain P2F9701a: a new methanogen isolated from estuarine environment. Curr. Microbiol. 42432-437. [DOI] [PubMed] [Google Scholar]
- 24.Lai, M. C., C. M. Shu, M. S. Chiou, T. Y. Hong, M. J. Chuang, and J. J. Hua. 1999. Characterization of Methanosarcina mazei N2M9705 isolated from an aquaculture fishpond. Curr. Microbiol. 3979-84. [DOI] [PubMed] [Google Scholar]
- 25.Lessner, D. J., L. Li, Q. Li, T. Rejtar, V. P. Andreev, M. Reichlen, K. Hill, J. J. Moran, B. L. Karger, and J. G. Ferry. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. USA 10317921-17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., Q. Li, L. Rohlin, U. Kim, K. Salmon, T. Rejtar, R. P. Gunsalus, B. L. Karger, and J. G. Ferry. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q., L. Li, T. Rejtar, B. L. Karger, and J. G. Ferry. 2005. Proteome of Methanosarcina acetivorans. Part I. An expanded view of the biology of the cell. J. Proteome Res. 4112-128. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., L. Li, T. Rejtar, B. L. Karger, and J. G. Ferry. 2005. Proteome of Methanosarcina acetivorans. Part II. Comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4129-135. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., D. R. Boone, R. Sleat, and R. A. Mah. 1985. Methanosarcina mazei LYC, a new methanogenic isolate which produces a disaggregating enzyme. Appl. Environ. Microbiol. 49608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyimo, T. J., A. Pol, H. J. Op den Camp, H. R. Harhangi, and G. D. Vogels. 2000. Methanosarcina semesiae sp. nov., a dimethylsulfide-utilizing methanogen from mangrove sediment. Int. J. Syst. Evol. Microbiol. 50171-178. [DOI] [PubMed] [Google Scholar]
- 31.Maeder, D. L., I. Anderson, T. S. Brettin, D. C. Bruce, P. Gilna, C. S. Han, A. Lapidus, W. W. Metcalf, E. Saunders, R. Tapia, and K. R. Sowers. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 1887922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mah, R. A., M. R. Smith, and L. Baresi. 1978. Studies on an acetate-fermenting strain of Methanosarcina. Appl. Environ. Microbiol. 351174-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 942626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalf, W. W., J. K. Zhang, X. Shi, and R. S. Wolfe. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 1785797-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meuer, J., S. Bartoschek, J. Koch, A. Kunkel, and R. Hedderich. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265325-335. [DOI] [PubMed] [Google Scholar]
- 36.Meuer, J., H. C. Kuettner, J. K. Zhang, R. Hedderich, and W. W. Metcalf. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. USA 995632-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, M. J., and J. G. Ferry. 1984. Carbon monoxide-dependent methyl coenzyme M methylreductase in acetotrophic Methanosarcina spp. J. Bacteriol. 160526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni, S., C. R. Woese, H. C. Aldrich, and D. R. Boone. 1994. Transfer of Methanolobus siciliae to the genus Methanosarcina, naming it Methanosarcina siciliae, and emendation of the genus Methanosarcina. Int. J. Syst. Bacteriol. 44357-359. [DOI] [PubMed] [Google Scholar]
- 39.Ni, S. S., and D. R. Boone. 1991. Isolation and characterization of a dimethyl sulfide-degrading methanogen, Methanolobus siciliae HI350, from an oil well, characterization of M. siciliae T4/MT, and emendation of M. siciliae. Int. J. Syst. Bacteriol. 41410-416. [DOI] [PubMed] [Google Scholar]
- 40.Possot, O., P. Gernhardt, A. Klein, and L. Sibold. 1988. Analysis of drug resistance in the archaebacterium Methanococcus voltae with respect to potential use in genetic engineering. Appl. Environ. Microbiol. 54734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 701425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rother, M., P. Boccazzi, A. Bose, M. A. Pritchett, and W. W. Metcalf. 2005. Methanol-dependent gene expression demonstrates that methyl-coenzyme M reductase is essential in Methanosarcina acetivorans C2A and allows isolation of mutants with defects in regulation of the methanol utilization pathway. J. Bacteriol. 1875552-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rother, M., and W. W. Metcalf. 2005. Genetic technologies for Archaea. Curr. Opin. Microbiol. 8745-751. [DOI] [PubMed] [Google Scholar]
- 44.Simankova, M. V., S. N. Parshina, T. P. Tourova, T. V. Kolganova, A. J. Zehnder, and A. N. Nozhevnikova. 2001. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst. Appl. Microbiol. 24362-367. [DOI] [PubMed] [Google Scholar]
- 45.Singh, N., M. M. Kendall, Y. Liu, and D. R. Boone. 2005. Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica. Int. J. Syst. Evol. Microbiol. 552531-2538. [DOI] [PubMed] [Google Scholar]
- 46.Sowers, K. R., J. E. Boone, and R. P. Gunsalus. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 593832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stojanowic, A., and R. Hedderich. 2004. CO2 reduction to the level of formylmethanofuran in Methanosarcina barkeri is non-energy driven when CO is the electron donor. FEMS Microbiol. Lett. 235163-167. [DOI] [PubMed] [Google Scholar]
- 48.Tietze, M., A. Beuchle, I. Lamla, N. Orth, M. Dehler, G. Greiner, and U. Beifuss. 2003. Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chembiochem 4333-335. [DOI] [PubMed] [Google Scholar]
- 49.Vaupel, M., and R. K. Thauer. 1998. Two F420-reducing hydrogenases in Methanosarcina barkeri. Arch. Microbiol. 169201-205. [DOI] [PubMed] [Google Scholar]
- 50.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25455-501. [DOI] [PubMed] [Google Scholar]
- 51.von Klein, D., H. Arab, H. Volker, and M. Thomm. 2002. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6103-110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.