Abstract
VP3 is a T7-like phage and was used as one of the typing phages in a phage-biotyping scheme that has been used for the typing of Vibrio cholerae O1 biotype El Tor. Here, we studied the receptor and other host genes of V. cholerae necessary for the lytic propagation of VP3. Six mutants resistant to VP3 infection were obtained from the random transposon insertion mutant bank of the sensitive strain N16961. The genes VC0229 and VC0231, which belong to the wav gene cluster encoding the core oligosaccharide (OS) region of lipopolysaccharide, were found to be interrupted by the transposon in five mutants, and the sixth mutant had the transposon inserted between the genes rhlB and trxA, which encode the ATP-dependent RNA helicase RhlB and thioredoxin, respectively. Gene complementation, transcription analysis, and the loss of VP3 sensitivity by the gene deletion mutants confirmed the relationship between VP3 resistance and VC0229, VC0231, and trxA mutation. The product of VP3 gene 44 (gp44) was predicted to be a tail fiber protein. gp44 could bind to the sensitive wild-type strain and the trxA mutant, but not to VC0229 and VC0231 mutants. The results showed that OS is a VP3 receptor on the surface of N16961, thioredoxin of the host strain is involved in the propagation of the phage, and gp44 is the tail fiber protein of VP3. This revealed the first step in the infection mechanism of the T7-like phage VP3 in V. cholerae.
Vibrio cholerae is the pathogenic agent of cholera and has caused seven pandemics. Only the O1 and O139 serogroups of V. cholerae have been associated with cholera epidemics, and strains of the O1 serogroup are subdivided into two biotypes, classical and El Tor (12). V. cholerae is the host for many bacteriophages, such as the tailed phage CP-T1 (18), K139 (39), and many filamentous phages (3, 24, 49). Phages infect and destroy host strains with high specificity, so phage-typing schemes were developed to differentiate strains of the same serogroups or genera in many bacteria, such as Salmonella enterica serovar Typhi (28) and Staphylococcus aureus (22). In V. cholerae, phage-typing schemes were also developed for typing of the O1 biotype El Tor (7) and O139 (5). A phage-biotyping scheme has been used for nearly 40 years in China for the typing of O1 El Tor strains (14). Based on the lytic patterns of the five typing phages, El Tor strains can be clustered into 32 phage types.
The first step of phage infection is the specific binding of phages to receptors on the host cell surface, triggering the ejection of phage DNA into the cell and initiating the propagation process (1). Some outer membrane proteins serve as receptors for phage adsorption, such as LamB (19), OmpA (8), and OmpC (52) of Escherichia coli and OmpK of Vibrio parahaemolyticus (23). Lipopolysaccharide (LPS) is a common phage receptor. Cytotoxin-converting ΦCTX from Pseudomonas aeruginosa (51) and FC3-10 from Klebsiella spp. (4) bind to the core region of LPS, while vibriophage CP-T1 (17) and ΦYeO3-12, specific for Yersinia enterocolitica serotype O:3 (35), bind to the O side chain. Other structures that can be used as receptors include flagella (32) and pili (49). Besides the bacterial receptors, there are some other host components necessary for the propagation of virulent phages, such as the host RNA polymerase involved in the transcription of phage genes and the components participating in the assembly of phage particles.
VP3 is one of the five typing phages in the phage-biotyping scheme (14). The complete genome sequence comprises 39,481 bp with an overall G+C content of 42.62%, and 52 open reading frames (ORFs) were predicted (our unpublished data). It is a T7-like phage by morphology and genome sequence comparison. Here, we have identified the receptor and other host genes of V. cholerae that are necessary for the lytic propagation of VP3. In addition, VP3 tail fiber protein was also predicted and proved to function as a receptor-binding protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. Phage VP3 was propagated on host strain 2477c. The El Tor strain N16961, whose genome was sequenced (20), is sensitive to VP3. Streptomycin (Sm)-resistant N16961-Smr was selected on Luria broth (LB) agar with Sm (100 μg/ml) and was also sensitive to VP3. All strains were grown in liquid or solid (15 g/liter agar) LB medium, which could be supplemented with kanamycin (Kan) (50 μg/ml), Sm (100 μg/ml), chloramphenicol (Cm) (15 μg/ml), or ampicillin (100 μg/ml). During the deletion mutant construction, solid LB medium with 20% sucrose and without NaCl was used to select plasmid excision from the chromosome.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| V. cholerae | ||
| 2477c | O1 El Tor, Ogawa | Laboratory stock |
| N16961 | O1 El Tor, Inaba | Laboratory stock |
| N16961-Smr | Spontaneous mutant of N16961; Smr | This study |
| N44G1 | N16961-Smr VC0229::mini-Tn5Km2 Smr Kanr VP3r | This study |
| N67B1 | N16961-Smr VC0231::mini-Tn5Km2 Smr Kanr VP3r | This study |
| N69H3 | N16961-Smr VC0229::mini-Tn5Km2 Smr Kanr VP3r | This study |
| N77C6 | N16961-Smr VC0229::mini-Tn5Km2 Smr Kanr VP3r | This study |
| N93C3 | N16961-Smr VC0231::mini-Tn5Km2 Smr Kanr VP3r | This study |
| N60C6 | N16961-Smr mutant with mini-Tn5Km2 inserted between rhlB and trxA; Smr Kanr VP3r | This study |
| C29 | N16961ΔVC0231 | This study |
| N16961-d0306 | N16961ΔtrxA | This study |
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK−) relA1 supE44 °(lac-proAB) [F′ traD36 proAB lacIqZ°M15] | TaKaRa |
| SM10(λpir) | thi thr leu tonA lacY supE recA::RP4-2-TC::Mu Km λpir | 44 |
| SM10-km2 | SM10(λpir) carrying plasmid pUTkm2; Kanr Ampr | This study |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Tiangen |
| Plasmids | ||
| pUTkm2 | Suicide plasmid carrying transposon mini-Tn5Km2; Kanr Ampr TnpA+ | 10 |
| pET30-a(+) | Expression vector; fl ori T7lac promoter Kanr | Novagen |
| pUC19 | Cloning vector; ori lacZ Ampr | TaKaRa |
| p0231UL | pUC19 carrying the upstream and downstream fragments flanking VC0231 | This study |
| pDS132 | Suicide plasmid; R6K ori mob RP4 cat sacB | 37 |
| pDS-0231 | pDS132 carrying the upstream and downstream fragments flanking VC0231 | This study |
| pTcat | pMD18-T::cat | 29 |
| p0306U-cat-L | pTcat carrying the upstream and downstream fragments flanking trxA | This study |
| pCVD442 | Suicide plasmid; R6K ori mob RP4 bla sacB | 11 |
| pCVD-0306 | pCVD442 with cat inserted between the upstream and downstream fragments flanking trxA | This study |
| pBR322 | Cloning vector; ColEI ori Ampr Tetr | TaKaRa |
| pBR0229c | pBR322 carrying gene VC0229 | This study |
| pBR0231c | pBR322 carrying gene VC0231 | This study |
| pBR0305c | pBR322 carrying gene rhlB | This study |
| pBR0306c | pBR322 carrying gene trxA | This study |
| pBR0305-6c | pBR322 carrying both gene rhlB and trxA | This study |
Propagation of phage VP3 and DNA extraction.
Twenty milliliters of 2477c culture (1 × 108 CFU/ml) and VP3 (1 × 108 PFU) were added to 80 ml of LB, incubated at 37°C, and shaken until the lysate was clear. The mixture was incubated at 56°C for 30 min and then centrifuged to remove cell debris. With a titer of 109 to 1010 PFU/ml, the supernatant was stored at 4°C as a stock solution. When phage DNA was extracted (40), the supernatant was treated with DNase I and RNase A, and then solid NaCl and polyethylene glycol 8000 were added, and it was left overnight at 4°C. Precipitated phage particles were recovered by centrifugation and were resuspended in 2 ml of SM buffer (40). The polyethylene glycol 8000 and cell debris were extracted from the phage suspension by adding an equal volume of chloroform, and it was centrifuged to recover the aqueous phase. Proteinase K and sodium dodecyl sulfate (SDS) were added to the solution, and it was incubated at 56°C for 1 h. An equal volume of phenol was added to the chilled phage suspension, mixed, and centrifuged at 3,000 × g for 5 min at room temperature. The aqueous phase was extracted with a 1:1 mixture of equilibrated phenol and chloroform and an equal volume of chloroform. DNA was precipitated with ethanol and dissolved in deionized water.
Transposon mutagenesis and the selection of VP3-resistant mutants.
Plasmid pUTkm2 (10) was transformed into E. coli SM10(λpir) (44) to obtain SM10-km2 (Kanr). Conjugation was performed between N16961-Smr as the recipient strain and SM10-km2 as the donor strain. The donor and recipient strain (optical density at 600 nm [OD600] ≈ 0.8) cultures were mixed 1:9 in a 1.5-ml microcentrifuge tube, washed twice with 1 ml of LB, resuspended in 100 μl of LB, and transferred onto 0.45-μm-pore-size membranes overlaid on LB plates. After being incubated at 37°C for 4 to 6 h, the membranes were washed with LB and the collected bacteria were diluted and plated onto the LB agar plates (Kan, 50 μg/ml, and Sm, 100 μg/ml) to select mutants.
Separated transconjugants were picked and inoculated into 150 μl of LB (Kan, 50 μg/ml, and Sm, 100 μg/ml) contained in 96-well plates (Corning Costar 3599) and incubated for 3 to 5 h until the OD was 0.5 to 0.8. This was monitored with a Tecan Genios Basic Microplate Reader at 395 nm. Then, 10 μl of the cultures was inoculated into 140 μl of LB (Kan, 50 μg/ml, and Sm, 100 μg/ml) with phage VP3 (1 × 108 PFU/ml) contained in new 96-well plates and incubated for 3 h. The wells with an OD395 of 0.5 or more were selected as the candidates for VP3-resistant mutants, which were confirmed subsequently by double-layer plaque assay. Briefly, 4 ml of melted 0.7% LB agar was mixed with 100 μl of cell cultures and poured onto an LB agar plate, and 10 μl of VP3 10-fold serial dilutions was dropped onto the plate when the upper layer solidified. After overnight incubation, mutants with no plaque formation were confirmed as VP3 resistant. The Kan resistance gene (kan) was detected by PCR with the primers pkan-low/pkan-up, and the transposase gene tnpA was detected with the primers ptnpA-low/ptnpA-up (Table 2). Strains with kan and without tnpA had the transposon inserted into chromosomes.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Restriction site |
|---|---|---|
| Detection of kan and tnpA | ||
| pkan-low | TGGCAAGATCCTGGTATCGGT | No |
| pkan-up | CAATCAGGTGCGACAATCTAT | No |
| ptnpA-low | AGGACAAACTGGCGCATAAC | No |
| ptnpA-up | TAACAGCCTGACCGCAACAA | No |
| Used in mutagenesis of VC0231 of N16961 | ||
| m0231U-1 | CTCGGTACCTTCTCGGCTGTCTGAATC | KpnI |
| m0231U-2 | CGGGGATCCGATGATCGTCGCCGCCAA | BamHI |
| m0231U-3 | TCCTCTAGATTCTCGGCTGTCTGAATC | XbaI |
| m0231L-1 | CGGGGATCCAGCGACTTGATGCTGAAA | BamHI |
| m0231L-2 | CAGGCATGCTGGTTTTCTGATGCTCAA | SphI |
| inner0231-1 | TATGATGGCTCTGTTCGG | No |
| inner0231-2 | ATTGACCTCCAGTGGCTT | No |
| Used in mutagenesis of trxA of N16961 | ||
| m0306U-1 | TTCGAGCTCCTTGAATGTGATTGAGGTTG | SacI |
| m0306U-2 | CGGGGATCCCTTTCACTCCAATGTGAT | BamHI |
| m0306L-1 | AGAGTCGACTGAGTCAGACCGCCAAG | SalI |
| m0306L-2 | CTTGCATGCAGTGTTCACTTTGAGCAATG | SphI |
| inner0306-1 | CGATGACGGTTTCGAGAA | No |
| inner0306-2 | CTACCACACTGCCATCTT | No |
| Used in construction of complementary plasmids | ||
| VC0229cU | GGGGATCCTTTTGGTATTTGTCACTT | BamHI |
| VC0229cL | GAGTCGACAGAATGGGAGCGAGGCGA | SalI |
| VC0231cU | GGGGATCCAGCTGTTGACCGAGTGTT | BamHI |
| VC0231cL | GAGTCGACTCCTATCATTACTGAGTG | SalI |
| VC0305cU | GGGGATCCTTGTCTGCACACTGCCAT | BamHI |
| VC0305cL | GAGTCGACTCGTTCTCGAAACCGTCA | SalI |
| VC0306cU | GGGGATCCGTCGGCGAACTTATGCTC | BamHI |
| VC0306cL | GAGTCGACATTAACGTGCGATAAACT | SalI |
Restriction sites are underlined.
Southern blot analysis and determination of transposon insertion sites.
Southern blot analysis was performed by a standard procedure (40). Total DNA was extracted from mutants and digested with restriction enzymes, such as EcoRI, PstI, KpnI, and SacI, and separated by electrophoresis. The fragment amplified from pUTKm2 with the primers pkan-low/pkan-up was labeled with digoxigenin using a DIG DNA Labeling and Detection Kit (Roche) and served as the DNA probe for hybridization. The blots were prehybridized for 2 h and hybridized for 12 h at 42°C in 50% formamide. If the restricted fragments containing the insert were of suitable size (2.5 kb to 4.4 kb in this study), they were ligated into pUC19 and sequenced to determine the transposon insertion sites.
Construction of deletion mutants and complementation experiments.
An in-frame VC0231 deletion mutant of V. cholerae N16961 was constructed by homologous recombination using the suicide plasmid pDS132 (37). The flanking sequences on the 5′ and 3′ sides of the fragment that was to be deleted were PCR amplified from N16961 chromosomal DNA using the primers m0231U-1/m0231U-2 and m0231L-1/m0231L-2 (Table 2), respectively. Then, the fragments were cloned into pUC19, generating the plasmid p0231UL. The fragment containing the in-frame deletion pattern of VC0231 was obtained by PCR from a p0231UL template using primers m0231U-3/m0231L-2 (Table 2) and cloned into pDS132 to construct the plasmid pDS-0231, which was conjugally transferred into N16961 from the donor strain E. coli SM10(λpir). Transconjugants were selected on thiosulfate-citrate-bile-sucrose (TCBS) agar with 15 μg/ml Cm and streaked onto LB agar plates with 20% sucrose and without NaCl. Colonies from sucrose selection medium were screened by PCR using the primers inner0231-1/inner0231-2 (Table 2), which were specific for the deleted fragment. Sequencing confirmed that the negatively PCR-amplified colonies were mutants.
A trxA deletion mutant of N16961 was constructed using the suicide plasmid pCVD442 (11) in a similar way. The flanking sequences on the 5′ and 3′ sides of trxA were PCR amplified using the primers m0306U-1/m0306U-2 and m0306L-1/m0306L-2 (Table 2), respectively, and were subsequently cloned into pTcat (29), generating the plasmid p0306U-cat-L. The plasmid pTcat was constructed previously in our laboratory by inserting a cat gene, which confers Cm resistance, on pMD18-T (TaKaRa). The plasmid p0306U-cat-L was restricted with SacI and SphI, and the 2-kb band containing the in-frame deletion pattern of trxA was purified and cloned into pCVD442 to construct the plasmid pCVD-0306. The following procedures were similar to those for the construction of the VC0231 deletion mutant, except that the sucrose selection medium contained Cm (15 μg/ml). Mutants were screened using the primers inner0306-1/inner0306-2 (Table 2) and were confirmed by sequencing.
When the complementary plasmid pBR0231c was constructed, the fragment containing VC0231 was PCR amplified from chromosomal DNA of N16961 with the primers VC0231cU/VC0231cL and inserted into pBR322. In the same way, the plasmids pBR0229c, pBR0305c, pBR0306c, and pBR0305-6c were constructed with the primers VC0229cU/VC0229cL, VC0305cU/VC0305cL, VC0306cU/VC0306cL, and VC0305cU/VC0306cL (Table 2), respectively. The fragment inserted into pBR0305-6c covered the genes rhlB and trxA. The complementary plasmids were transformed into different mutants, as mentioned in Results. The plasmid pBR322 was transformed into the mutants as a control. The sensitivity of the complemented mutants was examined by double-layer plaque assay.
Detection of gene transcription.
The transcription of rhlB and trxA was detected by reverse transcription (RT)-PCR. Total RNA was extracted with a Qiagen RNeasy Mini Kit from N16961, N60C6, and N60C6 complemented with different plasmids and reverse transcribed with SuperScript III Reverse Transcriptase (Invitrogen). The resulting cDNAs were used as PCR templates amplified with gene-specific primers.
Expression, purification, and FITC labeling of the predicted VP3 tail fiber protein.
A DNA fragment containing gene 44, which was predicted to encode VP3 tail fiber protein, was PCR amplified from VP3 DNA with the primers gp44NB1 (5′-TATACATATGCACCATCATCATCATCATTCAGGCACTCGTGCTCCT-3′) and gp44NB2 (5′-CGCACTCGAGTTAATTTAAAGGGATAGT-3′). The former contained the coding sequence of a hexahistidine tag and the restriction site for NdeI, and the latter contained the restriction site for XhoI (both underlined). The PCR product was digested with NdeI and XhoI and inserted into NdeI/XhoI-digested pET30-a (+). The resulting plasmid was transformed into the host strain E. coli BL21(DE3). Expression of His6-gp44 was induced with 0.4 mmol/liter isopropyl-β-d-thiogalactopyranoside (IPTG) at 28°C for 2 to 3 h. The total protein of the induced strain was separated by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by Western blotting using mouse anti-His antibody at a concentration of 1:1,000 and horseradish peroxidase-conjugated goat anti-mouse secondary antibody at a concentration of 1:500. His6-gp44 was purified with a His·Bind Purification Kit (Novagen) and labeled with a fluorescein isothiocyanate (FITC) labeling kit (Guangzhou Chang Rui Biotech, China) according to the manufacturer's instructions. His6-gp44 labeled with FITC is referred to as FITC-gp44 below.
Adsorption of FITC-gp44 to strains.
Strains were cultured in LB at 37°C until the OD600 was 0.3 to 0.4. Cells in 1 ml of the culture were collected by centrifugation at 4,000 × g for 4 min, washed once with phosphate-buffered saline (PBS), incubated for 1 h in FITC-gp44 solution, washed twice with PBS to remove the FITC-gp44 not adsorbed by the cells, and finally suspended in PBS. Two negative controls were made; one was E. coli BL21(DE3) treated with FITC-gp44 in parallel to the test strains, and the other was N16961 treated with PBS instead of the FITC-gp44 solution. To detect the dose dependence of agglutination of N16961 by FITC-gp44, cells of N16961 were incubated with twofold serial dilutions (1:2 to 1:32) of FITC-gp44 in PBS. All procedures were carried out at 4°C. After the FITC-gp44 solution was added, the samples were kept in the dark.
Each sample was mixed with an equal volume of 0.5% agarose preheated to 50°C, and 4 μl of the mixture was immediately dropped onto a slide, covered with a coverslip, and then observed with a fluorescence microscope (Olympus; BX51) and by confocal laser scanning microscopy (CLSM) (Olympus; FV500).
Nucleotide sequence accession number.
The GenBank accession number for VP3 gene 44 is FJ217960.
RESULTS
Selection of VP3-resistant transposon insertion mutants and determination of the insertion sites.
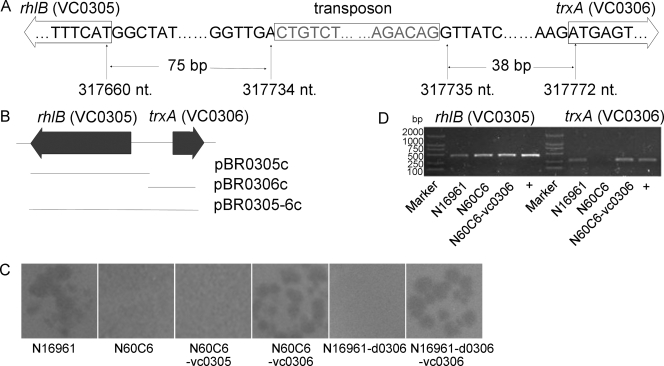
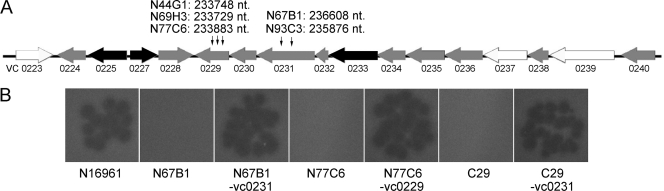
Nine thousand one hundred insertion mutants of strain N16961-Smr were generated by transposon mutagenesis with plasmid pUTKm2. By monitoring the growth rates of the mutants with the microplate reader when VP3 was coincubated and testing VP3 resistance by double-layer plaque assay, we finally obtained six VP3-resistant mutants (N44G1, N60C6, N67B1, N69H3, N77C6, and N93C3). These mutants were positive for kan and negative for tnpA by PCR amplification, suggesting the transposon had been inserted into chromosomes. Southern blot analysis showed a single band, implying each mutant had one copy of the transposon inserted. To clone the transposon insertion fragments in the mutants, the restriction fragments containing the transposon insert, which were 2.5 kb to 4.4 kb in size in the Southern blots, were inserted into vector pUC19 and sequenced. The mutation sites of three mutants (N77C6, N67B1, and N60C6) were determined by this method. In mutants N77C6 and N67B1, the transposon was inserted into the genes VC0229 and VC0231, respectively, both belonging to the wav gene cluster responsible for the synthesis of the core oligosaccharide (OS) region of LPS (34). In mutant N60C6, the transposon was inserted into the interval region between rhlB (locus tag VC0305), encoding the ATP-dependent RNA helicase RhlB, and trxA (locus tag VC0306), encoding thioredoxin (see Fig. 2A). The two genes are transcribed in opposite directions, and the insertion site lies in the 5′ noncoding regions of both genes. Primers were then designed flanking the genes mentioned above and were used to detect the other three mutants (N44G1, N69H3, and N93C3) by PCR. We found that the transposon was inserted into VC0229 in mutants N44G1 and N69H3 and into VC0231 in mutant N93C3. The transposon was inserted at different positions within the ORFs of VC0229 and VC0231 in these mutants (Fig. 1A). By random transposon mutagenesis, four genes that are probably necessary for VP3 infection were screened out, i.e., rhlB, trxA, and two genes in the wav gene cluster. We then studied the effects of the genes on VP3 infection.
FIG. 2.
trxA and VP3 infection. (A) In mutant N60C6, a transposon was inserted into the region between rhlB (VC0305) and trxA (VC0306) in the 5′ noncoding regions of both genes. nt., nucleotides. (B) Three complementary plasmids were constructed, each carrying a fragment containing rhlB, trxA, or both. The lines show the regions covered by the cloned fragments. (C) Response to VP3 infection by double-layer plaque assay. N60C6 became VP3 sensitive when it was transformed by pBR0306c (generating N60C6-vc0306), but not when the transforming plasmid was pBR0305c (generating N60C6-vc0305). (D) The transcription of rhlB and trxA was detected by RT-PCR. The lanes marked with a plus are positive controls.
FIG. 1.
Transposon insertion in the wav gene cluster and VP3 infection of the mutants. (A) Organization of the wav gene cluster of N16961 and the transposon insertion sites of the five VP3-resistant mutants. The large black arrows represent wav genes that were assumed to be essential. The white arrows represent genes that interfere with O-antigen attachment (34, 41). The grey arrows represent genes in the wav gene cluster. The transposon insertion positions are marked with small black arrows. The numbers after the mutant names stand for the nucleotide (nt) site just following the transposon in chromosome I of V. cholerae N16961. (B) Double-layer plaque assay of mutants (N67B1, N77C6, and C29) and complemented mutants (N67B1-vc0231, N77C6-vc0229, and C29-vc0231).
The wav gene cluster of V. cholerae N16961 is related to VP3 infection.
To further determine the relationship between VP3 resistance and the functions of genes in the wav cluster, a deletion mutant of N16961 in VC0231 (C29) was constructed by homologous recombination. Strain C29 showed resistance to VP3 when examined by double-layer plaque assay. The mutants N67B1, N93C3, and C29 regained VP3 sensitivity when they were complemented with the plasmid pBR0231c, and so did N44G1, N69H3, and N77C6 when complemented with pBR0229c (Fig. 1B).
V. cholerae trxA encoding thioredoxin is relevant for VP3 infection.
In mutant N60C6, the transposon lies in the 5′ noncoding regions of rhlB and trxA. To determine which gene (or both) is responsible for resistance to VP3 infection, three complementary plasmids were constructed and used to transform N60C6: pBR0305c, carrying rhlB; pBR0306c, carrying trxA; and pBR0305-6c, carrying both genes (Fig. 2B). N60C6 regained VP3 sensitivity when it was transformed with pBR0306c or pBR0305-6c, but not pBR0305c (Fig. 2C). We then tested whether both genes were transcribed in N60C6. When tested by RT-PCR, rhlB was still transcribed while trxA was not. The transcription of trxA was detected when N60C6 was complemented with pBR0306c (Fig. 2D). These results indicate that trxA was inactivated in N60C6, which led to the VP3 resistance. The trxA deletion mutant (N16961-d0306) resisted VP3 infection, and it became VP3 sensitive when transformed with pBR0306c (Fig. 2C), further confirming that trxA is indispensable for VP3 propagation.
gp44 of VP3 can adsorb to VP3-sensitive strains.
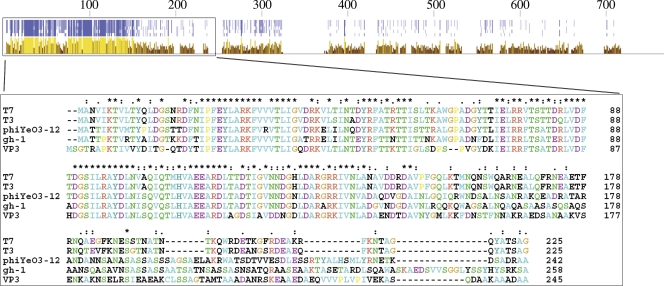
In order to find out whether the mutations influenced the adsorption of VP3 or the subsequent processes, the gene for VP3 tail fiber protein was predicted and expressed in vitro. The predicted gene 44 of VP3 encodes a protein (designated gp44) of 753 amino acid residues (aa), the N-terminal portion (aa 8 to 244) of which showed an identity of 48% with the N-terminal part of T7 gp17 (tail fiber protein; 553 aa) and showed high similarity to tail fiber proteins of other T7 family phages (Fig. 3). By searching in the Pfam database (13), VP3 gp44 was found to contain two Pfam-A (Phage_T7_tail and Collar) and one Pfam-B (Pfam-B_57397) domains. These domains are all common in phage tail fiber proteins, and domain Phage_T7_tail (corresponding to aa 15 to 244 of VP3 gp44) attaches the fibers to phage particles in phage T7 (45). Therefore, gp44 was predicted to be a tail fiber protein of VP3 and probably capable of binding to receptors with its C terminus.
FIG. 3.
Comparison of tail fiber protein sequences of phage T7, T3, ΦYeO3-12, gh-1, and VP3. T7, T3, ΦYeO3-12, and gh-1 are all T7-like phages. At the top are shown the identities and conservation of the full-length proteins. Darker blue dots mean higher identities. Lighter and higher yellow columns mean more conservation. The box shows multiple alignment of the N termini of the tail fiber proteins, where the similarity is particularly high.
VP3 gp44 was expressed in E. coli BL21(DE3) with a six-His tag added to the N terminus. The purified His6-gp44 formed two bands when separated on an SDS-PAGE gel; one was the predicted size, while the other was about double the size, and both contained the six-His tag as revealed by Western blotting. If the His6-gp44 sample was mixed with an equal volume of 7 M urea before SDS-PAGE, the larger band grew weaker and the smaller band grew stronger, suggesting the larger band was the dimer of His6-gp44.
His6-gp44 was labeled with FITC and used to treat VP3-sensitive N16961. Upon observation with a fluorescence microscope, many green masses were found in the sample, and most bacteria were agglutinated. Green-fluorescent masses were not found for the two negative controls, N16961 treated with PBS and E. coli BL21(DE3) treated with FITC-gp44 (data not shown). A similar phenomenon was also found under CLSM. The agglutination of N16961 changed with the amount of FITC-gp44 added (Fig. 4). The less FITC-gp44 added, the weaker the fluorescence and the smaller the bacterial masses formed, which showed dose dependence of gp44 for agglutination of N16961. These results suggest that gp44 adheres to VP3-sensitive strains by binding to receptors on the cell surface, and the possible oligomerization of gp44 causes the cells to agglutinate. If so, instead of VP3 particles, gp44 can be used to determine whether a strain can adsorb VP3.
FIG. 4.
Dose dependence analysis of FITC-gp44 for agglutination of N16961. Cells of N16961 were treated with twofold serial dilutions of prepared FITC-gp44 (1:2 to 1:32) and observed by CLSM. The dilution factors of FITC-gp44 are shown below.
Mutants with inactivated genes in the wav gene cluster lost the ability to adsorb phage VP3.
The adsorption of phage VP3 to the mutants was also examined by observing the binding of FITC-gp44 with a fluorescence microscope and by CLSM (Fig. 5). Mutants with inactivated genes in the wav gene cluster, including the five transposon mutants with the inactivated gene VC0229 or VC0231 and the deletion mutant C29, had no FITC-gp44 binding on the cell surface. The binding of FITC-gp44 was restored when the mutants were complemented with corresponding genes. Thus, mutations in the wav gene cluster probably altered the binding sites of VP3. Mutant N60C6 with inactivated trxA could adsorb FITC-gp44, suggesting that thioredoxin encoded by trxA is not involved in VP3 adsorption.
FIG. 5.
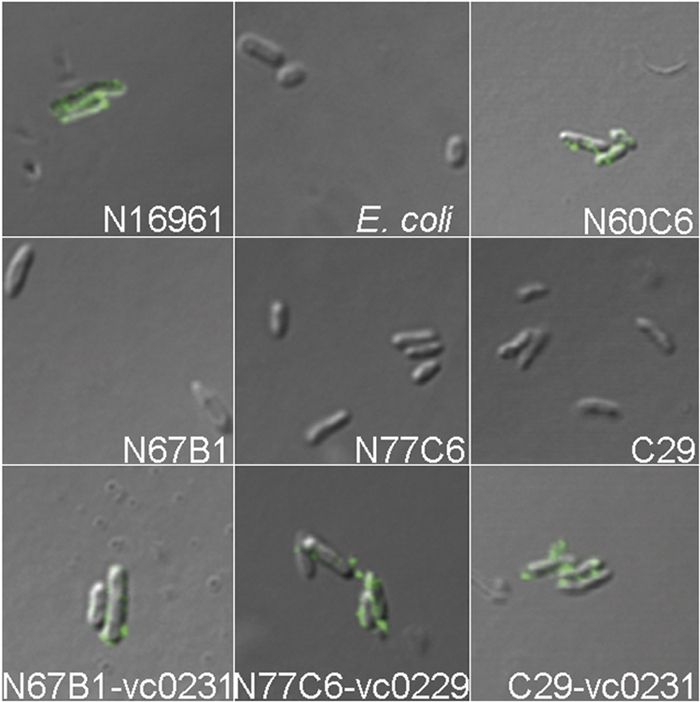
Binding of FITC-gp44 on the surfaces of different strains observed by CLSM. E. coli BL21(DE3) treated with FITC-gp44 was used as a negative control. N60C6 is an N16961 mutant with a transposon inserted into the intergenic region between rhlB and trxA. N67B1 and N77C6 are N16961 mutants with a transposon inserted into VC0231 and VC0229, respectively. C29 is an N16961 mutant with an in-frame deletion in VC0231. N67B1-vc0231, N77C6-vc0229, and C29-vc0231 are mutants with complementary plasmids.
DISCUSSION
A phage-biotyping scheme is used in China for subtyping of the V. cholerae O1 El Tor strains. VP3 is one of the five typing phages included in the scheme, and based on its genome sequence and morphology (our unpublished data), it belongs to the T7 group. In order to gain more knowledge about the infection mechanism of VP3, this study investigated VP3 receptors on the El Tor strain and other host genes necessary for VP3 propagation.
Many members of the T7 group use LPS as a receptor. The receptor of Y. enterocolitica phage ΦYeO3-12 is the O chain of serotype O:3, which consists of the rare sugar 6-deoxy-l-altropyranose (35); the receptor of lytic Pseudomonas putida bacteriophage gh-1 is proposed to be LPS, as well (27); and the LPS core is the binding site for T7 (38). The wav gene cluster of N16961, responsible for the synthesis of the OS region of LPS, lies in the 17.2-kb region between ORFs VC0223 and VC0240, including a group of glycosyl transferases and O-antigen ligase (Fig. 1A). It has been reported that waaA (VC0233), wavC (VC0227), and waaC (VC0225) are highly conserved and putatively essential core OS genes in V. cholerae (34). Among all the wav genes, only mutations in waaL (VC0237), wavA (VC0223), and wavL (VC0239) affect O antigen attachment (34, 41). Here, VC0229 and VC0231 were found to be relevant for N16961 sensitivity to phage VP3 in the process of phage attachment. Since genes related to O-antigen synthesis in these VC0229 and VC0231 insertion mutants are integral, O antigen is not proposed to function as the VP3 receptor. The exact functions of VC0229 and VC0231 are still unknown. VC0231 encodes a protein predicted to contain the catalytic domain of serine/threonine protein kinases. Genes encoding similar proteins exist in the V. parahaemolyticus O- and K-antigen biosynthesis gene cluster (GI193787939) and the Actinobacillus suis serotype K1 capsular gene cluster (GI29469153) ORF 7 (2). We predicted that the product of VC0231 possibly affects the activities of some enzymes needed in the process of OS synthesis. The OS was concluded to be the VP3 receptor on strain N16961.
Our results also implicate the gene trxA, encoding thioredoxin, in VP3 infection. In phage T7, E. coli thioredoxin is the processivity factor of T7 DNA polymerase (the gene 5 product of T7), increasing the processivity of nucleotide polymerization (31, 46), and influences the binding of T7 helicase to a DNA polymerase-thioredoxin complex (15). The situation may be similar for DNA polymerase of phage T3 (9). E. coli mutants with inactivated trxA become resistant to T7 (21, 38) but adsorb T7 normally (6). Similarly, thioredoxin is probably involved in the life process of VP3 as a subunit of VP3 DNA polymerase but is not involved in the binding of VP3. This supposition was also supported by the binding of FITC-gp44 on the surface of N60C6, which suggested that the inactivation of trxA did not alter the adsorption sites of phage VP3.
The tail fiber protein of a tailed phage has the receptor-binding function and decides the host specificity of the phage (16, 47, 50). VP3 gp44 is a homologue of tail fiber protein gp17 of phage T7, especially the proximal part. Three domains are predicted in gp44, and the proximal domain, Phage_T7_tail, is common in T7 family members, such as bacteriophage T7, T3 (36), K1F (43), and Pseudomonas phage gh-1 (27), and it links tail fibers to phage particles (45). The other two domains are also common in phage tail proteins. Domain Collar exists in the short tail fiber protein gp12 and the long tail fiber protein gp37 responsible for the receptor recognition of bacteriophage T4 (33, 48), while PfamB_PB057397 exists in ORF35 (tail fiber protein) of Vibrio phage K139 and some K139-like phages, such as Ch457 and E8498 (25). Therefore, gp44 of phage VP3 was predicted to be the tail fiber protein and to have a receptor-binding function; this was validated by the binding of gp44 to the surface of wild-type N16961. N16961 mutants with mutations in the wav gene cluster did not bind gp44, further proving the interaction of gp44 with the OS region of LPS.
We found that the expressed His6-gp44 tended to form oligomers, and strong denaturing conditions promoted the dissociation of the oligomer. In addition, fluorescence microscopy and CLSM showed that when FITC-gp44 was added, cells of VP3-sensitive strains agglutinated while cells of VP3-resistant wav gene mutants were dispersed. When N16961 and N60C6 were blended with a His6-gp44 solution dropped onto a slide, the cells were agglutinated, just like the reaction with V. cholerae antiserum, while E. coli BL21(DE3) and the wav gene mutants were not agglutinated (data not shown). These results implied that His6-gp44 is oligomeric in solution, which is the common form for phage tail fiber proteins. For example, T4 short tail fibers are trimeric gp12 (30), and tail fibers of T3 (26) and T7 (45) are trimers of the gp17 monomer. The agglutination of VP3-sensitive strains could be caused by the oligomerization of His6-gp44.
Conclusion.
Some bacteria cannot be lysed by a phage because they do not adsorb the phage particles, owing either to a lack of receptors on the cell surface or to the fact that the receptors are not accessible due to other cell surface components (42). Some bacteria are able to adsorb phages but fail to serve as host cells for phage multiplication if they lack any components necessary for phage propagation (1). We confirmed that mutants with an altered wav gene cluster resisted the virulent infection of VP3 because they could not adsorb the phage particles through the interaction between VP3 gp44 and the OS of V. cholerae. Thioredoxin, encoded by trxA, is necessary for VP3 proliferation and may be a subunit of VP3 DNA polymerase. These results can help us to understand the mechanism of T7-like phage infection of V. cholerae.
Acknowledgments
This research was supported by the National Basic Research Priorities Program (grant G1999054102) and partially by the High Tech Research and Development Program (grant 2006AA02Z425) of the Ministry of Science and Technology, Peoples Republic of China.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Adams, M. H. (ed.). 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY.
- 2.Boyce, J. D., J. Y. Chung, and B. Adler. 2000. Genetic organisation of the capsule biosynthetic locus of Pasteurella multocida M1404 (B:2). Vet. Microbiol. 72121-134. [DOI] [PubMed] [Google Scholar]
- 3.Campos, J., E. Martinez, E. Suzarte, B. L. Rodriguez, K. Marrero, Y. Silva, T. Ledon, R. del Sol, and R. Fando. 2003. VGJΦ, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTXΦ. J. Bacteriol. 1855685-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camprubi, S., S. Merino, V. J. Benedi, and J. M. Tomas. 1991. Isolation and characterization of bacteriophage FC3-10 from Klebsiella spp. FEMS Microbiol. Lett. 67291-297. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, A. K., A. N. Ghosh, G. B. Nair, S. K. Niyogi, S. K. Bhattacharya, and B. L. Sarkar. 2000. Development and evaluation of a phage typing scheme for Vibrio cholerae O139. J. Clin. Microbiol. 3844-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlin, M. 1974. Isolation and characterization of prototrophic mutants of Escherichia coli unable to support the intracellular growth of T7. J. Virol. 14509-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay, D. J., B. L. Sarkar, M. Q. Ansari, B. K. Chakrabarti, M. K. Roy, A. N. Ghosh, and S. C. Pal. 1993. New phage typing scheme for Vibrio cholerae O1 biotype El Tor strains. J. Clin. Microbiol. 311579-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., U. Chen-Schmeisser, I. Hindennach, and U. Henning. 1983. Apparent bacteriophage-binding region of an Escherichia coli K-12 outer membrane protein. J. Bacteriol. 153581-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson, J. F., R. Fox, D. D. Harris, S. Lyons-Abbott, and L. A. Loeb. 2003. Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res. 314702-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 594310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feeley, J. C. 1965. Classification of Vibrio cholerae (Vibrio comma), including El Tor vibrios, by infrasubspecific characteristics. J. Bacteriol. 89665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, S., S. Wu, and B. Liu. 1984. Characteristics of typing phages of Vibrio cholerae biotype El Tor. Fu Huo Luan Zi Liao Hui Bian 4:237-245.
- 15.Ghosh, S., S. M. Hamdan, T. E. Cook, and C. C. Richardson. 2008. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J. Biol. Chem. 28332077-32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundy, F. J., and M. M. Howe. 1984. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology 134296-317. [DOI] [PubMed] [Google Scholar]
- 17.Guidolin, A., and P. A. Manning. 1985. Bacteriophage CP-T1 of Vibrio cholerae. Identification of the cell surface receptor. Eur. J. Biochem. 15389-94. [DOI] [PubMed] [Google Scholar]
- 18.Guidolin, A., G. Morelli, M. Kamke, and P. A. Manning. 1984. Vibrio cholerae bacteriophage CP-T1: characterization of bacteriophage DNA and restriction analysis. J. Virol. 51163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazelbauer, G. L. 1975. Role of the receptor for bacteriophage lambda in the functioning of the maltose chemoreceptor of Escherichia coli. J. Bacteriol. 124119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgren, A., G. B. Kallis, and B. Nordstrom. 1981. A mutant thioredoxin from Escherichia coli tsnC 7007 that is nonfunctional as subunit of phage T7 DNA polymerase. J. Biol. Chem. 2563118-3124. [PubMed] [Google Scholar]
- 22.Hood, A. M. 1953. Phage typing of Staphylococcus aureus. J. Hyg. (London) 511-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue, T., S. Matsuzaki, and S. Tanaka. 1995. A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 125101-105. [DOI] [PubMed] [Google Scholar]
- 24.Jouravleva, E. A., G. A. McDonald, J. W. Marsh, R. K. Taylor, M. Boesman-Finkelstein, and R. A. Finkelstein. 1998. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect. Immun. 662535-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapfhammer, D., J. Blass, S. Evers, and J. Reidl. 2002. Vibrio cholerae phage K139: complete genome sequence and comparative genomics of related phages. J. Bacteriol. 1846592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, H., H. Fujisawa, and T. Minagawa. 1985. Purification and characterization of gene 17 product of bacteriophage T3. Virology 14622-26. [DOI] [PubMed] [Google Scholar]
- 27.Kovalyova, I. V., and A. M. Kropinski. 2003. The complete genomic sequence of lytic bacteriophage gh-1 infecting Pseudomonas putida—evidence for close relationship to the T7 group. Virology 311305-315. [DOI] [PubMed] [Google Scholar]
- 28.Lalko, J., and A. Gunnel. 1967. A new Vi-phage-type of Salmonella typhi and preparation of the typing phage. Bull. W. H. O. 36227-230. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, G., M. Yan, G. Qi, Y. Liu, S. Gao, and B. Kan. 2004. Chloramphenicol resistance marker exchange of lysogenic phage CTXΦ of Vibrio cholerae and its induction. Zhonghua Wei Sheng Wu Xue He Mian Yi Xue Za Zhi 24253-257. [Google Scholar]
- 30.Makhov, A. M., B. L. Trus, J. F. Conway, M. N. Simon, T. G. Zurabishvili, V. V. Mesyanzhinov, and A. C. Steven. 1993. The short tail-fiber of bacteriophage T4: molecular structure and a mechanism for its conformational transition. Virology 194117-127. [DOI] [PubMed] [Google Scholar]
- 31.Mark, D. F., and C. C. Richardson. 1976. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 73780-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merino, S., S. Camprubi, and J. M. Tomas. 1990. Isolation and characterization of bacteriophage PM3 from Aeromonas hydrophila the bacterial receptor for which is the monopolar flagellum. FEMS Microbiol. Lett. 57277-282. [DOI] [PubMed] [Google Scholar]
- 33.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 6786-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesper, J., A. Kraiss, S. Schild, J. Blass, K. E. Klose, J. Bockemuhl, and J. Reidl. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect. Immun. 702419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage ΦYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 1825114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 3191115-1132. [DOI] [PubMed] [Google Scholar]
- 37.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51246-255. [DOI] [PubMed] [Google Scholar]
- 38.Qimron, U., B. Marintcheva, S. Tabor, and C. C. Richardson. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. USA 10319039-19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reidl, J., and J. J. Mekalanos. 1995. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini-transposon to identify a phage-encoded virulence factor. Mol. Microbiol. 18685-701. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schild, S., A. K. Lamprecht, and J. Reidl. 2005. Molecular and functional characterization of O antigen transfer in Vibrio cholerae. J. Biol. Chem. 28025936-25947. [DOI] [PubMed] [Google Scholar]
- 42.Scholl, D., S. Adhya, and C. Merril. 2005. Escherichia coli K1's capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 714872-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholl, D., and C. Merril. 2005. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 1878499-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1784-791. [Google Scholar]
- 45.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200351-365. [DOI] [PubMed] [Google Scholar]
- 46.Tabor, S., H. E. Huber, and C. C. Richardson. 1987. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 26216212-16223. [PubMed] [Google Scholar]
- 47.Tetart, F., C. Desplats, and H. M. Krisch. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J. Mol. Biol. 282543-556. [DOI] [PubMed] [Google Scholar]
- 48.Thomassen, E., G. Gielen, M. Schutz, G. Schoehn, J. P. Abrahams, S. Miller, and M. J. van Raaij. 2003. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 331361-373. [DOI] [PubMed] [Google Scholar]
- 49.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 2721910-1914. [DOI] [PubMed] [Google Scholar]
- 50.Werts, C., V. Michel, M. Hofnung, and A. Charbit. 1994. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J. Bacteriol. 176941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokota, S., T. Hayashi, and H. Matsumoto. 1994. Identification of the lipopolysaccharide core region as the receptor site for a cytotoxin-converting phage, ΦCTX, of Pseudomonas aeruginosa. J. Bacteriol. 1765262-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, S. L., K. L. Ko, C. S. Chen, Y. C. Chang, and W. J. Syu. 2000. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J. Bacteriol. 1825962-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]