Abstract
Activation of σ54-dependent gene expression essential for formation of flagella in Campylobacter jejuni requires the components of the inner membrane-localized flagellar export apparatus and the FlgSR two-component regulatory system. In this study, we characterized the FlgS sensor kinase and how activation of the protein is linked to the flagellar export apparatus. We found that FlgS is localized to the C. jejuni cytoplasm and that His141 of FlgS is essential for autophosphorylation, phosphorelay to the cognate FlgR response regulator, motility, and expression of σ54-dependent flagellar genes. Mutants with incomplete flagellar export apparatuses produced wild-type levels of FlgS and FlgR, but they were defective for signaling through the FlgSR system. By using genetic approaches, we found that FlgSR activity is linked to and downstream of the flagellar export apparatus in a regulatory cascade that terminates in expression of σ54-dependent flagellar genes. By analyzing defined flhB and fliI mutants of C. jejuni that form flagellar export apparatuses that are secretion incompetent, we determined that formation of the apparatus is required to contribute to the signal sensed by FlgS to terminate in activation of expression of σ54-dependent flagellar genes. Considering that the flagellar export apparatuses of Escherichia coli and Salmonella species influence σ28-dependent flagellar gene expression, our work expands the signaling activity of the apparatuses to include σ54-dependent pathways of C. jejuni and possibly other motile bacteria. This study indicates that these apparatuses have broader functions beyond flagellar protein secretion, including activation of essential two-component regulatory systems required for expression of σ54-dependent flagellar genes.
Responding to changing environmental and intracellular conditions in cells requires efficient communication networks that can rapidly receive and integrate signals. Two-component regulatory systems, which are distributed almost ubiquitously among prokaryotic organisms, allow bacteria to monitor their intracellular and extracellular environments and react by altering the expression of appropriate genes. These systems are typically comprised of a sensor histidine kinase and a response regulator protein (reviewed in references 46 and 65). The sensor protein contains a domain usually in the N-terminal portion that detects a specific signal, commonly through an interaction with another protein or a small effector molecule. Activation includes autophosphorylation of the sensor kinase and a conformational change that allows the transmitter domain, usually in the C-terminal portion, to activate a cognate response regulator via phosphotransfer. Some histidine kinases also have the ability to function as a phosphatase to remove a phosphate group from either themselves or their cognate response regulators when activity of the regulatory system is not favored.
The largest group of sensor histidine kinases includes those that are anchored to the cytoplasmic membrane and receive signals from the extracellular environment, allowing a cell to respond to external factors such as pH, temperature, or the presence of specific compounds (reviewed in reference 46). Since the monitoring of intracellular conditions is also vital to basic cellular processes, sensor kinases that are activated by alterations within bacteria have also evolved. These kinases include a relatively small group of kinases that are membrane anchored but respond to signals in the cytoplasm or periplasm and a larger group of soluble, cytoplasmic sensor kinases. Several members of the latter group have been characterized, such as NtrB, a kinase involved in nitrogen metabolism, whose activity is controlled by the PII protein (33). Nitrogen starvation results in uridylylation of PII, which blocks interaction with NtrB and causes the sensor protein to function as a kinase to initiate phosphorelay, culminating in phosphorylation of its cognate response regulator, NtrC. Under nitrogen-replete conditions, PII is deuridylylated and interacts with NtrB, allowing it to function as a phosphatase instead of as a kinase. Another example of a cytoplasmic histidine kinase that responds to intracellular conditions is KinA of Bacillus subtilis. Through interactions with two different proteins that inhibit the function of the kinase, KinA is responsive to the energy state of the bacterium or the ability of the cell to initiate replication (9, 58, 63). Activation of KinA begins a complex regulatory cascade leading to expression of genes essential for sporulation.
Flagellar assembly and chemotaxis systems also rely on two-component signaling systems to properly regulate bacterial motility (6, 62). The CheA kinase receives signals from a number of membrane-bound methyl-accepting chemotaxis protein (MCP) receptors (reviewed in references 3, 17, and 18). Motile bacteria respond via chemotaxis to small molecules that are attractants or repellants, and many of these effectors are bound by the periplasmic domains of MCPs. Through interactions of the cytoplasmic domains of MCPs with the CheA kinase, CheA is able to integrate and transmit these signals via phosphorelay to CheY, ultimately influencing the decision to continue swimming in a single direction or tumble and change direction.
Campylobacter jejuni is a gram-negative, microaerophilic bacterium commonly associated with a number of animals of agricultural significance, especially fowl. While the relationship between C. jejuni and avian species develops into commensalism, infection of humans causes gastroenteritis that can range from very mild enteritis to severe, bloody diarrheal episodes (7, 8). In both the developed and developing regions of the world, C. jejuni is responsible for a substantial percentage of cases of bacterial gastroenteritis (13, 53). In the United States, this bacterium is believed to be the leading single-species cause of diarrheal disease, resulting in significant loss in economic productivity (11).
C. jejuni is a highly motile organism owing to the presence of a single flagellum elaborated from one or both poles of the bacterium. Motility is critical for promoting optimal interactions between C. jejuni and avian or human hosts. Nonmotile variants of C. jejuni colonize the gastrointestinal tract of chicks at levels significantly lower than wild-type motile strains (25, 29, 50, 64, 66), and only motile strains can be recovered after coinfection of human volunteers with motile and nonmotile strains (5). In this organism, motility is a highly organized, regulated, and complex process, relying on the coordination of over 40 proteins to assemble a complete organelle (26). Although there are some similarities with the well-characterized regulatory cascades described for Escherichia coli and Salmonella species (12), genetic screens and in silico analyses indicate that there are several differences that distinguish the flagellar gene transcription and assembly processes in C. jejuni. While C. jejuni utilizes σ28 to activate transcription of the major flagellin (encoded by flaA) and other minor flagellum-associated proteins (10, 22, 23, 28, 30, 66), σ54 has been shown or proposed to be involved in transcription of the bulk of flagellar genes, including those encoding the hook, basal body, and minor flagellin (26, 28, 30, 66). The use of both σ28 and σ54 in these pathways indicates that flagellar gene transcription in C. jejuni is more similar to the regulatory cascades of species of Vibrio, Pseudomonas, and Helicobacter than to those of E. coli or Salmonella species (2, 16, 34, 39, 40, 47, 51, 56, 60).
In a transposon mutagenesis screen, a number of gene products were found to be required for σ54-dependent flagellar gene transcription in C. jejuni (30). These proteins include members of the flagellar export apparatus (FEA), FlhF (a putative GTPase), and the FlgSR two-component system, comprised of the FlgS sensor kinase and the FlgR response regulator (30). It was hypothesized that these proteins may act separately or in concert to integrate signals required to initiate transcription of σ54-dependent flagellar genes. Previous work characterized the unusual NtrC-like response regulator FlgR to understand the means by which this protein functions (35). Our group has also found that FlgR and FlgS are targets of phase variation, making FlgSR the only known two-component regulatory system in which both proteins are subject to this form of control (25, 27). However, the mechanism by which FlgS is activated and functions as a sensor kinase remains to be characterized. Sequence analyses indicate that this protein appears to contain domains common to many sensor histidine kinases, such as the ATP-binding catalytic domain and the histidine-containing phosphotransfer domain (61, 65). Although the homology is somewhat weaker in the N-terminal region of the protein, FlgS is similar to the flagellum-associated histidine kinases that are required for σ54-dependent flagellar gene expression and motility in species of Vibrio, Pseudomonas, and Helicobacter (14, 40, 51, 57). However, the signals that activate any of these kinases for positively influencing flagellar gene expression are uncharacterized.
In this work, we characterized FlgS and the activating signals that influence its ability to positively regulate flagellar gene expression and motility in C. jejuni. We first identified the histidine in the phosphotransfer domain that is autophosphorylated upon activation by FlgS. Through extensive experimentation, we characterized the origin of the signal that influences FlgS activation. Our research has led us to believe that (i) activation of FlgSR is dependent on the FEA and (ii) the signal for FlgS autophosphorylation may lie within the FEA, as formation of this apparatus appears to be necessary to promote expression of σ54-dependent flagellar genes. Our work expands previous models of Campylobacter flagellar gene regulation and motility by characterizing the FlgS sensor kinase and introducing potential mechanisms for activating this protein. Furthermore, our work suggests that the FEAs of a subset of motile bacteria that use σ54 to control expression of flagellar genes have broader functions than flagellar protein secretion, including influencing signaling pathways through two-component regulatory systems to activate flagellar gene expression.
MATERIALS AND METHODS
Bacterial strains.
C. jejuni strain 81-176 is a clinical isolate from a patient presenting with gastroenteritis and has been shown to promote commensal colonization of the chick gastrointestinal tract and to cause disease in human volunteers (5, 29). C. jejuni was routinely grown on Mueller-Hinton (MH) agar containing 10 μg/ml trimethoprim (TMP) under microaerobic conditions (85% N2, 10% CO2, and 5% O2) at 37°C. When necessary, strains were grown on MH agar containing 50 μg/ml kanamycin, 15 μg/ml chloramphenicol, or 0.5, 1, 2, or 5 mg/ml streptomycin. All C. jejuni strains were stored at −80°C in a solution of 85% MH broth and 15% glycerol. E. coli strains DH5α, XL1-Blue, and BL21(DE3)/pLysE were cultured with Luria-Bertani (LB) agar or broth containing 100 μg/ml ampicillin or 15 μg/ml chloramphenicol when required. All E. coli strains were stored at −80°C in a solution of 80% LB broth and 20% glycerol.
Construction of mutants.
All strains were constructed by using previously described protocols (28). To construct flgS(H141) mutants, pDRH310 (30) was subjected to PCR-mediated mutagenesis (45) to mutate the histidine codon at position 141 to a codon for alanine and then was verified by DNA sequence analysis. One plasmid, pDRH1276, was recovered and introduced into 81-176 Smr flgS::cat-rpsL (DRH441 [30]) and 81-176 Smr ΔastA flgS::cat-rpsL (DRH460 [30]) by electroporation. Mutants were recovered on MH agar containing streptomycin and verified by PCR analysis and DNA sequencing. Mutants used for further analysis were designated DRH1323 [81-176 Smr flgS(H141A)] and SNJ947 [81-176 Smr ΔastA flgS(H141A)].
We replaced native flgR with the flgRΔreceiver and flgRΔCTD alleles (where receiver indicates the N-terminal receiver domain and CTD indicates the C-terminal domain) in FEA mutants. For ΔfliP, ΔflhA, and ΔflhB mutants, flgR::kan-rpsL (pDRH443) was electroporated into strains 81-176 Smr ΔastA ΔfliP (DRH1016), 81-176 Smr ΔastA ΔflhA (DRH979), and 81-176 Smr ΔastA ΔflhB (DRH1734) (30). The resultant strains, 81-176 Smr ΔastA ΔfliP flgR::kan-rpsL (SNJ158), 81-176 Smr ΔastA ΔflhA flgR::kan-rpsL (DRH1765), and 81-176 Smr ΔastA ΔflhB flgR::kan-rpsL (DRH1830), were electroporated with pDRH1855 containing the flgRΔreceiver allele and pDRH1856 containing the flgRΔCTD allele (35). All transformants were selected on MH agar with streptomycin and verified by PCR and DNA sequencing.
C. jejuni ΔfliI mutants were constructed by first cloning the fliI locus into pUC19 (to generate pDRH1453) and then cloning an SmaI-digested kan-rpsL cassette (from pDRH427 [30]) into a PmeI site within the fliI coding sequence to generate pDRH1506. pDRH1506 was introduced into 81-176 Smr ΔastA (DRH461 [30]) by electroporation, generating 81-176 Smr ΔastA fliI::kan-rpsL (DRH2246), which was recovered on MH agar with kanamycin. pDRH1453 was then used in PCR-mediated mutagenesis (45) to delete a large portion of the coding sequence of the gene by fusing codon 4 to codon 453, creating pDRH1843. DRH2246 was then electroporated with pDRH1843 to replace fliI::kan-rpsL with the ΔfliI allele to create 81-176 Smr ΔastA ΔfliI (DRH2257).
Generation of flhB mutants first involved PCR-mediated mutagenesis (45) to create a point mutation, generating an StuI site in the coding sequence of flhB in pDRH666 (30) to create pSNJ355. This plasmid was then digested with StuI so that a cat-rpsL cassette generated by digestion of pDRH265 (28) with SmaI could be inserted with flhB. The resulting plasmid, pSNJ360, was then introduced into DRH461 (81-176 Smr ΔastA [(30]) by electroporation, replacing flhB with flhB::cat-rpsL to generate SNJ404 (81-176 Smr ΔastA flhB::cat-rpsL). PCR-mediated mutagenesis (45) with pDRH666 was used to generate point mutations and in-frame deletions within flhB. These mutations and the resulting plasmids included flhB(N267A) (pSNJ238), flhBΔ214-218 (pSNJ243), flhBΔ224-228 (pSNJ236), and flhBΔ244-253 (pSNJ237). These plasmids were introduced into SNJ404 by electroporation to replace the flhB::cat-rpsL allele with the different flhB alleles. Mutants were recovered on MH agar with streptomycin. The resulting strains included SNJ438 [81-176 Smr ΔastA flhB(N267A)], SNJ464 (81-176 Smr ΔastA flhBΔ214-218), SNJ428 (81-176 Smr ΔastA flhBΔ224-228), and SNJ475 (81-176 Smr ΔastA flhBΔ244-253). Mutants were verified by PCR and DNA sequencing.
To construct strains containing transcriptional reporters, plasmids pDRH532 (containing flgDE2::nemo), pDRH608 (containing flaA::astA), pDRH610 (containing flaB::astA), and pDRH669 (containing flgD::astA) were electroporated into C. jejuni to replace the native flgDE2, flaA, flaB, and flgD loci on the chromosome as previously described (30, 59). All mutants were recovered on MH agar containing kanamycin and were verified by PCR analysis.
Generation of polyclonal antiserum against C. jejuni proteins.
Generation of polyclonal murine antiserum against the RNA polymerase subunit A (RpoA) protein of C. jejuni involved first constructing primers with 5′ BamHI sites to amplify the coding sequence from codon 2 through the stop codon of rpoA from C. jejuni strain 81-176 (31). Ligation of this DNA fragment into the BamHI site of pQE30 (Qiagen) and transformation into E. coli XL1-Blue allowed recovery of pDRH2907, which encodes a His6-RpoA fusion protein. To purify the protein, a 1-liter culture in LB broth was grown to an optical density at 600 nm (OD600) of 0.5, and then the culture was induced for 4 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The bacteria were disrupted by two passages through an EmulsiFlex-C5 cell disrupter (Avesin) at 15,000 to 20,000 lb/in2. The protein was purified under native conditions from the soluble fraction with Ni-nitrilotriacetic acid agarose according to the manufacturer's instructions. Polyclonal murine antiserum was generated in mice by standard procedures using a commercial vendor (Cocalico Biologicals).
Detection of FlhB in C. jejuni required generation of rabbit polyclonal antiserum against the cytoplasmic domain of the protein. Because this portion of FlhB in Salmonella enterica serovar Typhimurium undergoes autoproteolytic processing between the asparagine and proline residues at positions 269 and 270 (19, 21, 48), we attempted to create a soluble, more stable protein to immunize rabbits for antiserum generation. We first used PCR-mediated mutagenesis (45) with pDRH666 (containing the wild-type flhB allele [30]) to change the codons for asparagine and proline at positions 267 and 268, respectively, to codons for alanines to generate pDRH2339. After construction of pDRH2339, we amplified a portion of the flhB(N267A P268A) sequence encoding amino acids 209 through 369 that encompasses the predicted entire unprocessed cytoplasmic domain of the protein. Primers were used in PCR so that in-frame BamHI sites were added to the 5′ end of the amplified product. The DNA was then cloned into BamHI-digested pGEX-4T-2 (GE Healthcare) in the correct orientation to produce a glutathione S-transferase (GST)-FlhBcyto(N267A P268A) fusion protein. The resulting plasmid was designated pDRH2367 and used to transform BL21(DE3). The resulting strain was grown in 3 liters of LB broth to mid-log phase and then induced with 25 mM IPTG for 3 h at 37°C. The bacteria were harvested and disrupted with an EmulsiFlex-C5 cell disrupter (Avesin) at 15,000 to 20,000 lb/in2. The soluble fraction was obtained by removing the insoluble material by centrifugation at 13,000 rpm for 2 h at 4°C. The soluble material was rocked with 2.4 ml of glutathione Sepharose 4B (GE Healthcare) for 30 min at room temperature. The protein was then purified according to the manufacturer's instructions. Despite our attempt to create a more stable, unprocessed version of the cytoplasmic domain of FlhB fused to GST, about one-third of the recovered purified protein had a molecular mass of approximately 29-kDa after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, which correlates with the size of GST rather than 46 kDa, the predicted size of the full-length fusion protein. Therefore, this protein was only partially stable. The purified products were used to immunize rabbits by standard procedures for antiserum generation using a commercial vendor (Cocalico Biologicals).
Immunoblot analyses of FlgS, FlgR, FlhB, and FlaA proteins.
C. jejuni strains were grown from frozen stocks on MH agar containing appropriate antibiotics at 37°C for 48 h and restreaked 16 h prior to use. SDS-PAGE and immunoblotting of FlgS and FlgR proteins were performed as previously described with anti-FlgS Rab11 and anti-FlgR Rab13 rabbit polyclonal antisera, respectively (25). Briefly, cells were resuspended from 16-h growth plates in MH broth and diluted to an OD600 of 0.8. One-milliliter samples were harvested by centrifugation and washed once with phosphate-buffered saline. For whole-cell lysates (WCLs), the pellet was resuspended in 50 μl 1× Laemmli buffer, and 4 μl (for FlgR analysis) or 7 μl (for FlgS analysis) of each resuspended pellet was loaded onto 10% SDS-PAGE gels.
For FlgS localization studies, 5-ml portions of cultures of wild-type and mutant strains at an OD600 of 0.8 were prepared as described above, resuspended in 10 mM HEPES (pH 7.4), and broken by sonication. Unbroken cells were removed by centrifugation at 13,000 × g for 5 min at 4°C, and the supernatant was transferred to a new tube and centrifuged at 13,000 × g for 30 min at 4°C to pellet the total membrane fraction (outer and inner membrane proteins). The supernatant contained soluble proteins (cytoplasmic and periplasmic proteins). Volumes representing equivalent cell numbers for the membrane and soluble proteins were analyzed by 10% SDS-PAGE after resuspension and boiling in 1× Laemmli buffer. For detection of FlgS, anti-FlgS Rab11 antiserum was used at a dilution of 1:10,000 (25). To detect proteins representative of the cytoplasmic fraction or inner membrane fraction, we analyzed the location of the RpoA cytoplasmic protein and the AtpF inner membrane protein by using anti-RpoA M59 antiserum at a dilution of 1:2,000 and anti-AtpF M3 antiserum at a dilution of 1:1,000 (4), followed by a goat anti-mouse secondary antibody.
To monitor the stability and location of FlhB proteins, bacteria were grown and 5-ml samples of cultures of wild-type and mutant strains were prepared and sonicated as described above. The total membrane fraction, containing inner and outer membrane proteins, was recovered by centrifugation at 13,000 rpm for 30 min at 4°C. The recovered pellet was suspended in 50 μl of 1× Laemmli buffer and loaded onto a 10% SDS-PAGE gel for immunoblot analysis. Primary anti-FlhB Rab476 antiserum was used at a dilution of 1:1,000 and was rocked with the membrane overnight at 4°C. The blot was then washed and incubated with a 1:10,000 dilution of goat anti-rabbit secondary antibody for 4 h at room temperature.
For analysis of FlaA secretion in C. jejuni strains, bacteria were grown and resuspended from plates as described above. WCLs from 1-ml portions of cultures of wild-type and mutant strains were prepared as described above. For recovery of outer membrane proteins, 5-ml cultures of each bacterial strain were prepared and sonicated, and the unbroken cells were removed by centrifugation at 13,000 rpm for 5 min at 4°C. Each supernatant was recovered and spun at 13,000 rpm for 30 min at 4°C. The pellet containing insoluble material representing the total membrane proteins (inner and outer membrane proteins) was resuspended in 1% N-laurylsarcosine (sodium salt) and incubated for 30 min at room temperature to solublize the inner membrane proteins. The outer membrane proteins were recovered as the insoluble pellet after centrifugation at 13,000 rpm for 30 min at 4°C. Volumes of sample corresponding to 200 μl and 700 μl of bacteria were loaded for analysis of WCLs and outer membrane proteins, respectively. Immunoblot analysis was performed with a 1:10,000 dilution of anti-FlaA LLI antiserum (42) and a 1:10,000 dilution of goat anti-rabbit secondary antibody.
Motility assays and transmission electron microscopy.
To analyze relative levels of motility, strains were grown on MH agar with TMP from freezer stocks for 48 h at 37°C under microaerobic conditions and then restreaked and grown for 16 h prior to use. Cells were resuspended in MH broth to an OD600 of 0.8, and a sterile needle was used to inoculate semisolid MH motility agar as described previously (28). Plates were incubated under microaerobic conditions for 24 to 36 h at 37°C and photographed. For transmission electron microscopy, 1-ml samples of bacteria in MH broth at an OD600 of 1.0 were centrifuged at 13,000 rpm for 3 min and then resuspended in 2% glutaraldehyde. After incubation for 1 h on ice, samples were stained with 1% uranyl acetate and visualized with an FEI Technai G2 Spirit BioTWIN transmission electron microscope.
Arylsulfatase reporter assays.
Strains were grown from frozen stocks for 48 h at 37°C under microaerobic conditions on MH agar with TMP or kanamycin and restreaked and grown for 16 h prior to the assay. Strains were analyzed for arylsulfatase activity by a previously described method (30), which was based on previously established methods (24, 67). Briefly, all strains were resuspended in phosphate-buffered saline to an OD600 of 0.8 to 1.0, washed in arylsulfatase assay buffer, and incubated with 10 mM nitrophenylsulfate and 1 mM tyramine for 1 h at 37°C. NaOH was added to terminate the assays, and the amount of nitrophenol present in each sample was determined spectrophotometrically at OD410. The number of arylsulfatase units produced by each strain was calculated by comparing the OD410 value of each sample to a standard curve obtained using known nitrophenol concentrations. One arylsulfatase unit is defined as the amount of enzyme catalyzing the release of 1 nmol of nitrophenol per h per OD600 unit. Each strain was tested in triplicate, and each assay was performed three times.
Purification of FlgS and FlgR proteins.
Wild-type His6-FlgR protein was purified as previously described (35). flgS(H141A) from pDRH1276 was amplified from codon 2 to the stop codon by PCR using primers that added in-frame 5′ and BamHI restriction sites to facilitate cloning into BamHI-digested pQE30 to generate pSNJ960. This plasmid was then transformed into XL1-Blue for induction and purification of the protein. Wild-type His6-FlgS and His6-FlgS(H141A) were purified by using previously described protocols (25).
Autophosphorylation of FlgS.
FlgS autophosphorylation assays were performed as described previously using purified His6-FlgS or His6-FlgS(H141A) in the presence of [γ-32P]ATP (35, 66). Briefly, 6 pmol His6-FlgS or His6-FlgS(H141A) was added to a buffer containing 50 mM Tris-HCl (pH 8.0), 75 mM KCl, 2 mM MgCl2, and 1 mM dithiothreitol. Ten microcuries of [γ-32P]ATP was then added. At each time point, a sample was removed and the reaction was stopped by addition of an equal amount of 2× SDS-PAGE loading buffer. Proteins were resolved by 10% SDS-PAGE, and the gels were dried and exposed to a phosphorimager screen. The screen was read with a Storm 820 phosphorimager (Amersham Biosciences), and the data were analyzed using the manufacturer's software.
FlgR phosphorylation.
In vitro phosphotransfer from His6-FlgS proteins to His6-FlgR was monitored as previously described (35, 66). For each reaction, 6 pmol of His6-FlgR was added to 6 pmol of His6-FlgS or His6-FlgS(H141A) that had been allowed to autophosphorylate for 2 min as described above. Reactions were stopped by addition of an equal volume of 2× SDS-PAGE loading buffer, and the samples were analyzed by SDS-PAGE. After drying, polyacrylamide gels were analyzed with a phosphorimager.
Real-time RT-PCR.
C. jejuni strains 81-176 Smr (DRH212), 81-176 ΔflhA (DRH946), 81-176 ΔflhB (SNJ471), and 81-176 ΔfliP (DRH1065) were grown from frozen stocks on MH agar containing appropriate antibiotics at 37°C for 48 h and restreaked 16 h prior to use (28, 30). Bacteria were suspended from the agar plates in MH broth, and total RNA was extracted from the bacteria with Trizol reagent (Invitrogen). The RNA was then treated with DNase prior to analysis. The final concentration of RNA used in a Sybr green PCR master mixture was 50 ng/μl. Real-time reverse transcription (RT)-PCR was performed using a 7500 real-time PCR system (Applied Biosystems). Detection of mRNA for gyrA, encoding DNA gyrase, served as an endogenous control, and the transcript levels of flgS and flgR in mutants (lacking flhA, flhB, or fliP) were compared to those in the wild-type strain (DRH212). The following primer pairs were used for real-time RT-PCR analysis: flgS RT#1 (5′-GCTACAGATATTAGCGATGAAAAACG-3′) and flgS RT#2 (5′-TAGGATTTCTTATCTCATGTGCCAAAT-3′), flgR RT#3 (5′-TCAAGCCAAACTTTTAAGAGCTTTG-3′) and flgR RT#4 (5′-CTATTTTGATGCTTTTCGTACTTCCA-3′), and gyrA F (5′-CGACTTACACGGCCGATTTC-3′) and gyrA R (5′-ATGCTCTTTGCAGTAACCAAAAAA-3′).
Transposon mutagenesis.
Chromosomal DNA from C. jejuni 81-176 ΔastA ΔflhA flgDE2::nemo (DRH1021 [30]), 81-176 ΔastA ΔflhB flgD::astA (SNJ331), and 81-176 ΔastA ΔfliP flaB::astA (DRH1178 [30]) was purified and subjected to in vitro random transposon mutagenesis with the darkhelment transposon by using previously published protocols (27-30). Twelve in vitro transposon mutagenesis reactions were performed with DNA from each strain. Each reaction mixture contained 2 μg of chromosomal DNA, 1 μg of pSpaceball1, and 250 ng of Himar1 C9 transposase purified from DH5α/pMalC9 (1). After transposition, the mutagenized DNA was repaired and transformed into each strain as previously described (28). Transposon mutants were recovered after growth on MH agar containing chloramphenicol and 5-bromo-4-chloro-3-indolyl sulfate and then examined for blue or white colony phenotypes.
RESULTS
FlgS is a cytoplasmic protein.
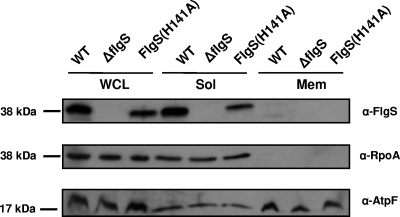
Bioinformatic analysis suggests that unlike most sensor kinases that are localized to the bacterial inner membrane, the C. jejuni FlgS sensor kinase is a cytoplasmic protein. This protein lacks both a predicted signal sequence that would target it for secretion and hypothetical spans of hydrophobic residues that would be indicative of a protein associated with the inner membrane. To determine if FlgS is localized to the cytoplasm, we fractionated wild-type C. jejuni 81-176 Smr (DRH212 [28]) to obtain a soluble fraction containing cytoplasmic and periplasmic proteins and an insoluble fraction containing proteins associated with the outer and inner membranes. As shown in Fig. 1, FlgS was found only in WCLs and the soluble fraction of wild-type bacteria. In a comparison with the control proteins, FlgS was present in the same fraction as the soluble cytoplasmic protein RpoA and absent in the fraction containing the insoluble inner membrane protein AtpF. Considering both the bioinformatic and biochemical analyses, we concluded that FlgS is a cytoplasmic protein (Fig. 1).
FIG. 1.
Localization and stability of FlgS proteins in C. jejuni. Wild-type strain C. jejuni 81-176 Smr (DRH212) (WT), 81-176 Smr ΔflgS (DRH460), and 81-176 Smr flgS(H141A) (DRH1323) were grown, and protein samples were obtained from the WCL, the soluble fraction (Sol), and the insoluble membrane fraction (Mem) after sonication. Anti-FlgS Rab11 antiserum (α-FlgS) was used to detect FlgS proteins (25). Anti-RpoA M59 antiserum (α-RpoA) and anti-AtpF M3 antiserum (α-AtpF) were used to detect the soluble cytoplasmic RpoA protein and the insoluble inner membrane protein AtpF, respectively (4).
Autophosphorylation of residue H141 is required for FlgS activity.
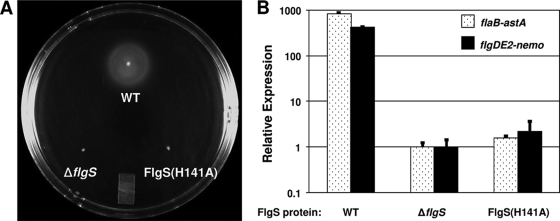
It has been shown that the autophosphorylation site of the NtrB sensor kinase is residue H139 (52). Alignment of FlgS to NtrB indicated that this phosphorylated residue likely corresponds to H141 of FlgS, an amino acid located within the putative phosphotransfer domain (spanning amino acids 131 to 195) that receives the phosphate group upon autophosphorylation of other kinases. To determine if H141 is essential for FlgS activity as a kinase and for flagellar gene expression, the wild-type flgS allele of C. jejuni was replaced with flgS(H141A), which results in production of FlgS with alanine at position 141 instead of histidine. The resulting mutant was first examined for a potential defect in FlgS stability. We found that while FlgS(H141A) appears to lack any detectable degradation products, the levels of the FlgS(H141A) protein present in WCLs and the soluble fraction were about one-half the levels of the wild-type FlgS (Fig. 1). By comparing the phenotypes of the wild-type and mutant strains, we found that the flgS(H141A) mutation affected motility, flagellar biosynthesis, and σ54-dependent flagellar gene expression (Fig. 2 and data not shown). The nonmotile phenotype of the flgS(H141A) mutant on semisolid agar plates at 24 h after inoculation was similar to that observed for a ΔflgS strain in which flgS had been deleted from the chromosome (Fig. 2A and data not shown) (30). This lack of motility in the flgS(H141A) mutant correlated with a complete absence of flagella as analyzed by transmission electron microscopy (data not shown). We then analyzed expression of flgDE2- and flaB-astA transcriptional fusions in strains producing the FlgS(H141A) protein and found that the level of σ54-dependent flagellar gene expression in an flgS(H141A) mutant was equivalent to that in a ΔflgS mutant (Fig. 2B), indicating that H141 is critical for proper function of FlgS in C. jejuni.
FIG. 2.
Phenotypic analyses of C. jejuni wild-type and flgS(H141A) mutant strains. (A) Motility phenotypes of C. jejuni strains producing wild-type or mutant FlgS proteins in MH semisolid agar 24 h after inoculation. The strains used included wild-type strain 81-176 Smr (DRH212) (WT), 81-176 Smr ΔflgS (DRH460), and 81-176 Smr flgS(H141A) (DRH1323). (B) Arylsulfatase assays for analysis of expression of flaB::astA and flgDE2::nemo in C. jejuni 81-176 derivatives producing wild-type and FlgS mutant proteins. The results are the results of a typical assay in which each strain was tested in triplicate. The values reported for each strain are the average arylsulfatase activity ± standard deviation relative to the level of expression of each transcriptional fusion in 81-176 Smr ΔastA ΔflgS, which was defined as 1 arylsulfatase unit. For expression of flaB::astA, the strains used included wild-type strain DRH665 (81-176 Smr ΔastA flaB::astA) (WT), DRH939 (81-176 Smr ΔastA ΔflgS flaB::astA), and SNJ958 [81-176 Smr ΔastA flgS(H141A) flaB::astA]. For expression of flgDE2::nemo, the strains used included wild-type strain DRH533 (81-176 Smr ΔastA flgDE2::nemo) (WT), DRH936 (81-176 Smr ΔastA ΔflgS flgDE2::nemo), and SNJ956 [81-176 Smr ΔastA flgS(H141A) flgDE2::nemo]. The FlgS proteins produced by the strains are indicated below the graph.
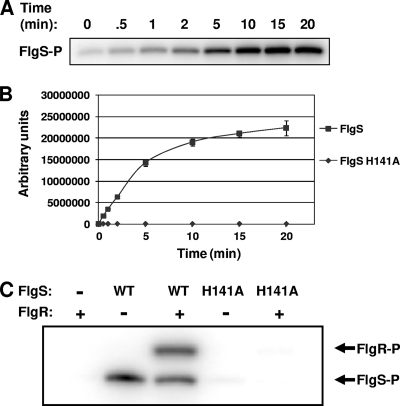
Since H141 is the predicted site of phosphorylation, we performed autophosphorylation assays with purified His6-tagged versions of FlgS and FlgS(H141A). Whereas FlgS autophosphorylated and accumulated radiolabeled phosphate over time, FlgS(H141A) remained unphosphorylated (Fig. 3A and 3B). In previous work, we showed that the FlgR response regulator is modified by phosphorylation in the presence of purified FlgS in vitro (35). We performed similar experiments to determine if phosphotransfer to FlgR was abolished in the presence of FlgS(H141A). In these experiments, we observed phosphorelay to FlgR in the presence of wild-type FlgS but not in the presence of the FlgS(H141A) protein (Fig. 3C), consistent with the hypothesis that autophosphorylation of FlgS on H141 contributes to phosphotransfer to FlgR. Thus, we believe that H141A is the most likely site of autophosphorylation and is essential for proper function of the protein.
FIG. 3.
Autophosphorylation of FlgS proteins and phosphorelay to FlgR. (A and B) Analysis of autophosphorylation of His6-FlgS and His6-FlgS(H141A) over time after incubation of proteins with [γ-32P]ATP. (A) Representative gel analyzed by autoradiography from an FlgS autophosphorylation assay. (B) Relative quantification of autophosphorylation of FlgS proteins as determined by densitometry after autoradiography of gels. Three separate FlgS and FlgS(H141A) autophosphorylation assays were performed, and the results of these assays were averaged. The amount of incorporation of 32P is expressed in arbitrary units based on the densitometric analysis. (C) Analysis of phosphorelay to His6-FlgR from His6-tagged FlgS or FlgS(H141A) protein. FlgS proteins were preincubated with [γ-32P]ATP before addition of His6-FlgR. A representative gel analyzed by autoradiography from a phosphotransfer assay is shown. The presence (+) or absence (−) of FlgR and the FlgS protein used in each reaction are indicated above the lanes. WT, wild type.
Production of FlgS and FlgR is not dependent on the presence of the FEA.
The FEA is a multiprotein complex that translocates flagellar subunits across the inner membrane for incorporation into a functional organelle (for a review, see reference 44). As mentioned above, many of the FEA components (e.g., FlhA, FlhB, FliP, and FliR) in addition to FlgS and FlgR are required for σ54-dependent flagellar gene expression in C. jejuni (30). We next performed experiments to determine if the FEA and FlgSR systems are linked together in a regulatory cascade that terminates in activation of expression of σ54-dependent flagellar genes. More specifically, we investigated whether the FEA influences the production or activity of the FlgSR two-component system.
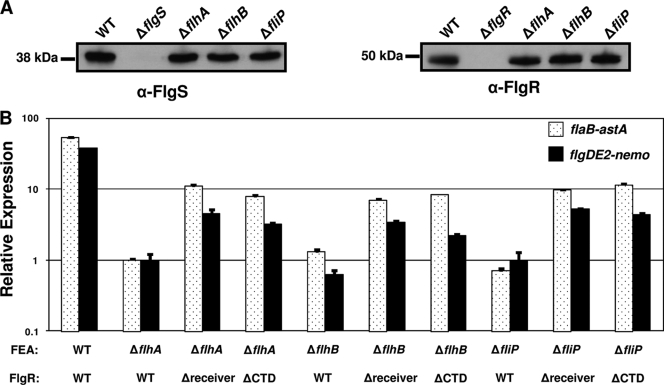
To examine if production of FlgS or FlgR is dependent on the FEA, we performed an immunoblot analysis of cell lysates from the wild-type strain and mutant strains lacking flhA, flhB, and fliP, which encode some of the proteins comprising the FEA. We observed similar levels of FlgS and FlgR in the wild-type strain and the FEA mutants (Fig. 4A), indicating that production of FlgS and FlgR is independent of the FEA. As additional verification that the FEA does not affect production of FlgS and FlgR, we compared the levels of the flgS and flgR mRNA transcripts in mutants lacking flhA, flhB, and fliP to the levels in the wild-type strain by real-time RT-PCR analysis. We did not detect significant changes in the levels of the flgS or flgR mRNAs in the mutant strains compared to wild-type bacteria (data not shown). Therefore, FEA mutants of C. jejuni appear to produce normal levels of the FlgS and FlgR proteins but have defects in signaling pathways for stimulation of σ54-dependent flagellar gene expression.
FIG. 4.
Production of FlgS and FlgR and activity of FlgR proteins in FEA mutants of C. jejuni. (A) Production of FlgS and FlgR proteins in mutants of C. jejuni lacking one component of the FEA. WCLs of wild-type and C. jejuni mutant strains were prepared for immunoblot analysis. Anti-FlgS Rab11 (α-FlgS) and anti-FlgR (α-FlgR) Rab13 antisera were used to detect the FlgS (left gel) and FlgR (right gel) proteins (25). The strains used for analysis included wild-type strain DRH212 (81-176 Smr) (WT), DRH460 (81-176 Smr ΔflgS), DRH737 (81-176 Smr ΔflgR), DRH946 (81-176 Smr ΔflhA), SNJ471 (81-176 Smr ΔflhB), and DRH1065 (81-176 Smr ΔfliP). (B) Arylsulfatase assays for analysis of expression of flaB::astA and flgDE2::nemo in the C. jejuni 81-176 Smr wild-type strain and mutant strains lacking a component of the FEA and producing wild-type and FlgR mutant proteins. The results are the results of a typical assay in which each strain was tested in triplicate. The values reported for each strain are the average arylsulfatase activity ± standard deviation relative to the level of expression of each transcriptional fusion in 81-176 Smr ΔastA ΔflhA, which was defined as 1 arylsulfatase unit. For expression of flaB::astA, the strains used included (from left to right) wild-type strain DRH665, DRH1049, SNJ112, SNJ273, DRH1827, SNJ109, SNJ1021, DRH1178, SNJ261, and SNJ1015. For expression of flgDE2::nemo, the strains used included (from left to right) wild-type strain DRH533, DRH1021, SNJ115, SNJ274, DRH1827, SNJ113, SNJ1017, DRH1204, SNJ358, and SNJ1012. The FEA mutation and the type of FlgR protein produced in each strain are indicated below the graph. WT, wild type.
We next analyzed C. jejuni to determine if the FlgSR system functions downstream of the FEA in a regulatory cascade to activate expression of σ54-dependent flagellar genes. Previous work in our laboratory generated flgR alleles encoding proteins lacking the N-terminal receiver or C-terminal domain of the response regulator (35). These proteins were shown to have partial constitutive activity in the absence of the FlgS sensor kinase, indicating that FlgR functions downstream of FlgS (35). We used these flgR alleles (flgRΔreceiver and flgRΔCTD) to replace wild-type flgR on the chromosome of mutants lacking flhA, flhB, or fliP to determine if these partially constitutively active FlgR proteins suppress the phenotype of the FEA mutants for expression of flagellar genes. As shown previously (30) and in Fig. 4B, flhA, flhB, or fliP mutants containing wild-type flgR and producing the wild-type protein expressed 40- to 50-fold less of the σ54-dependent flaB- and flgDE2-astA transcriptional fusions. When flgR in these FEA mutants was replaced with the flgR alleles encoding FlgRΔreceiver and FlgRΔCTD, partial restoration of σ54-dependent flagellar gene expression was observed (Fig. 4B). Although the levels of expression were not restored to wild-type levels, they were approximately 5- to 10-fold higher than those in the FEA mutants that produced wild-type FlgR. These analyses suggest that FlgSR functions downstream of the FEA and that activation of FlgSR is dependent in some manner on the FEA of C. jejuni.
Formation of the FEA likely initiates activation of the FlgSR system.
Considering our data, we speculated that the FEA may contribute an essential signal to activate the FlgSR system to terminate in expression of σ54-dependent flagellar genes. We hypothesized that either formation of the FEA or the secretory activity of the FEA may comprise the signal to activate the FlgS sensor kinase. If the former hypothesis is correct, it is possible that positioning one component of the FEA or the whole FEA complex in the inner membrane may directly provide the signal sensed directly by the cytoplasmic FlgS protein, leading to autophosphorylation of the kinase. Alternatively, formation of the FEA may be required for production of a downstream signal sensed by FlgS. The latter hypothesis includes the possibility that the secretory activity of a formed FEA may influence activation of FlgS. For instance, a negative regulator that represses activity of FlgS may be present in the cell before the FEA is competent for secretion, and the secretory activity of the FEA may be required to inactivate or remove this protein from the cytoplasm, relieving FlgS from repression and allowing autophosphorylation and phosphorelay to FlgR to occur.
To distinguish between these possibilities, we generated mutants with FEA complexes that are predicted to assemble in the inner membrane but are hindered for secretion of flagellar substrates. For this approach, we targeted fliI and flhB for mutation. FliI functions as an ATPase that dissociates export substrates (e.g., flagellins) from their chaperones in S. enterica serovar Typhimurium (49, 55). While FliI is not absolutely required for secretion of flagellar substrates, its absence substantially reduces the efficiency of this process. Due to the significant homology between the FliI proteins of C. jejuni and S. enterica serovar Typhimurium strain LT2 (43% identity and 62% similarity over 424 amino acids), we hypothesize that FliI serves a similar function in C. jejuni in increasing the efficiency of secretion of flagellar proteins. Therefore, we deleted fliI from the C. jejuni genome to create a mutant with possibly impaired efficiency of FEA-mediated secretion of flagellar proteins.
Previous analysis with S. enterica serovar Typhimurium revealed that defined mutations can also be made in flhB so that the FEA assembles in the inner membrane, but secretion of substrates through the FEA is reduced or blocked (21). These mutations include mutations that result in small, in-frame deletions and point mutations in the FlhB protein. By aligning the sequences of the S. enterica serovar Typhimurium and C. jejuni strain 81-176 proteins (which are 36% identical and 60% similar across 351 amino acids), we identified regions of the FlhB protein of C. jejuni that may be deleted or mutated, resulting in FEA mutants that form but do not secrete efficiently. To this end, we constructed flhB mutant alleles that encoded FlhBΔ214-218, FlhBΔ224-228, FlhBΔ244-253, and FlhB(N267A) mutant proteins. The deletions and mutations in the C. jejuni FlhB protein correspond to types of domain deletions and point mutations resulting in the FlhBΔ2, FlhBΔ4, FlhBΔ8-9, and FlhB(N269A) proteins of S. enterica serovar Typhimurium constructed by Fraser et al. (21), respectively.
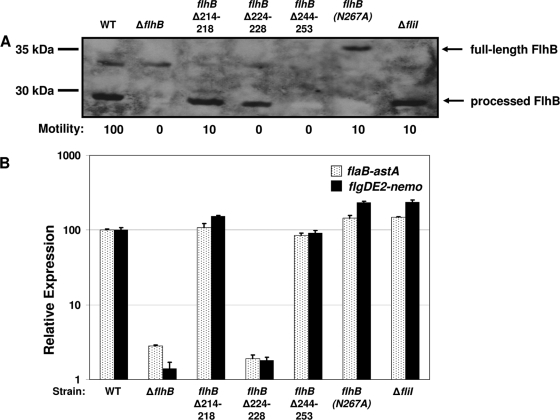
After construction of fliI and flhB mutants of C. jejuni, we first analyzed the strains to determine stability of the FlhB protein produced in each mutant by immunoblot analysis. FlhB is produced as a 42-kDa protein in S. enterica serovar Typhimurium that is cleaved to a 31-kDa protein by autoproteolysis of the peptide bond between positions N269 and P270 (19, 21, 48). Although flhB of C. jejuni appears to encode a 37-kDa protein, we predict that similar processing may occur between N267 and P268, resulting in a 30-kDa FlhB protein. Immunoblot analysis of the total membrane fraction of wild-type C. jejuni revealed that FlhB appeared as the processed 30-kDa protein (Fig. 5A). In three of the four fliI and flhB mutants, we observed similar levels of processed FlhB proteins, indicating that the mutant FlhB proteins were stable. The flhB(N267A) mutant was expected to produce an FlhB protein that is not able to undergo autoproteolytic processing. Indeed, we observed only the full-length 37-kDa protein in this mutant (Fig. 5A). In the flhBΔ244-253 mutant, we could not detect any mutant FlhB protein. The reason for the lack of detection of this mutant form of FlhB remains unknown, but it may be due to the method used to generate the anti-FlhB antiserum. The antigen that was used to make the anti-FlhB antiserum contained amino acids 209 to 367 of FlhB, which form the complete cytoplasmic domain of the protein before processing. Due to predicted processing of FlhB at position 267 in C. jejuni, ultimately only a maximum of 58 amino acids (amino acids 209 to 267) in processed FlhB proteins are the same as the amino acids in the antigen that was used to generate the anti-FlhB antiserum. Since FlhBΔ244-253 lacks 10 of the 58 amino acids of the antigen, the epitope that the anti-FlhB antiserum recognizes may have been destroyed or deleted in this protein, resulting in its lack of detection. Because the mutant producing FlhBΔ244-253 stimulated expression of σ54-dependent flagellar genes (see below), we believe that this protein is made and is stable but is undetectable with current reagents.
FIG. 5.
Phenotypic analyses of C. jejuni strains with formed but secretion-impaired FEA complexes. (A) Immunoblot analysis of FlhB proteins and motility phenotypes of C. jejuni wild-type and flhB or fliI mutant strains. Total membrane proteins were isolated from wild-type and mutant strains of C. jejuni. Equal amounts of proteins from the strains were analyzed. Anti-FlhB Rab476 antiserum was used to detect the FlhB proteins. The arrows indicate the positions of the 37-kDa full-length, unprocessed FlhB protein and the 30-kDa processed FlhB protein. The motility phenotypes of wild-type and mutant strains are indicated below the blot. The diameter of the motile ring around the point of inoculation in MH semisolid agar was measured after 36 h of incubation at 37°C under microaerobic conditions. The level of motility of each mutants is expressed relative to the level of motility of the wild-type strain, which was defined as 100%. The strains used for both analyses included (from left to right) wild-type strain DRH461 (WT), DRH1734, SNJ464, SNJ428, SNJ475, and SNJ438. (B) Arylsulfatase assays for analysis of expression of flaB::astA and flgDE2::nemo in C. jejuni 81-176 Smr wild-type or mutant strains containing a secretion-impaired FEA. The results are the results of a typical assay in which each strain was tested in triplicate. The values reported for each strain are the average arylsulfatase activity ± standard deviation relative to the level of expression of each transcriptional fusion in wild-type strain 81-176 Smr ΔastA, which was defined as 100 arylsulfatase units. For expression of flaB::astA, the strains used included (from left to right) wild-type strain DRH665 (WT), DRH1830, SNJ467, SNJ434, SNJ508, SNJ442, and SNJ422. For expression of flgDE2::nemo, the strains used included wild-type strain DRH533 (WT), DRH1827, SNJ466, SNJ433, SNJ504, SNJ439, and SNJ457. The type of mutation in the FEA of each strain is indicated below the graph.
We next determined if the secretion of the flhB and fliI mutants was impaired. To do this, we performed two different analyses. We first determined if motility was reduced since motility is directly dependent on FEA-mediated secretion of flagellar proteins out of the cytoplasm to construct a flagellar organelle. For all the flhB and fliI mutants, we observed that the level of motility was ≤10% of that of the wild-type strain, indicating that flagellar motility and presumably secretion through the FEA had been severely impaired (Fig. 5A).
We next performed a more direct analysis of the secretion competence of the FEA in the derived mutants by monitoring FEA-dependent secretion of the FlaA flagellin protein to the outer membrane of C. jejuni strains. Unlike the situation in S. enterica serovar Typhimurium, the complete regulatory pathways that govern flaA expression in C. jejuni are not completely understood. In S. enterica serovar Typhimurium, σ28-dependent expression of fliC encoding the major flagellin is repressed in FEA mutants due to cytoplasmic retention of the anti-σ28 factor FlgM (32, 38). In C. jejuni, flaA is expressed by a σ28-dependent promoter (10, 28, 30, 66). However, evidence for expression of flaA and secretion of the encoded protein via the FEA to form a truncated flagellum with partial motility has been obtained for an fliA (encoding σ28) mutant, indicating that a σ28-independent promoter likely exists (28, 30, 37). Also unlike the situation in S. enterica serovar Typhimurium, there is evidence that flaA expression is only moderately decreased in certain FEA mutants of C. jejuni 81-176, indicating that some expression of flaA is independent of the FEA status of the bacterium (30). Furthermore, any existing translation controls for flaA mRNAs in C. jejuni have not been characterized. Since evidence that flaA expression and FlaA production are not entirely dependent on the status of the FEA in C. jejuni, as they are in other bacteria, we analyzed FEA-dependent secretion of FlaA in our defined flhB and fliI mutants.
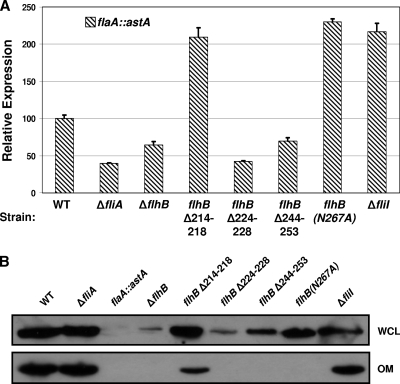
We first ensured that flaA was expressed in the mutants by monitoring expression of flaA::astA in the flhB and fliI mutants. We found that flaA::astA expression was not defective in three of the mutants [flhBΔ214-218, flhB(N267A), and ΔfliI]. Rather, the expression of flaA::astA in these mutants was approximately twofold higher than that in the wild-type strain (Fig. 6A). Expression of flaA::astA was slightly reduced in the flhBΔ244-253 mutant, to approximately 75% of that in the wild-type strain. The remaining mutant, flhBΔ224-228, expressed flaA::astA at a level that was 50% less than the level in the wild-type strain (Fig. 6A). The level of expression of flaA::astA in this mutant was similar to that in ΔflhB or ΔfliA (lacking σ28) mutants. With the exception of the expression in the flhBΔ224-228 muant, flaA::astA expression in the mutants was mostly intact or the level was higher than the level in the wild-type strain.
FIG. 6.
Analysis of flaA expression and FlaA secretion mediated by the FEA. (A) Arylsulfatase assays for analysis of expression of flaA::astA in the C. jejuni 81-176 Smr wild-type strain or strains with a secretion-impaired FEA. The results are the results of a typical assay in which each strain was tested in triplicate. The values for each strain are the average arylsulfatase activity ± standard deviation relative to the level of expression of each transcriptional fusion in wild-type strain 81-176 Smr ΔastA, which was defined as 100 arylsulfatase units. For expression of flaA::astA, the strains used included (from left to right) wild-type strain DRH655 (WT), DRH1070, SNJ365, SNJ427, SNJ1033, SNJ1034, SNJ1038, and SNJ1042. The type of mutation in each strain is indicated below the graph. (B) Immunoblot analysis of FlaA production in WCLs and secretion to the outer membrane of wild-type and FEA mutant strains. WCL and outer membrane (OM) fractions were isolated from wild-type and mutant strains of C. jejuni. Anti-FlaA LL-1 antiserum was used to detect the FlaA proteins (42). The strains used included (from left to right) wild-type strain DRH212 (WT), DRH724, DRH655, SNJ471, SNJ464, SNJ428, SNJ475, SNJ438, and DRH2257.
We next monitored FEA-mediated secretion of FlaA by comparing the levels of FlaA associated with outer membranes of wild-type and mutant strains of C. jejuni. As shown in Fig. 6B, the flhBΔ214-218, flhB(N267A), and ΔfliI mutants produced comparable levels of FlaA in WCLs, but reduced levels of the protein were associated with the outer membrane compared to the outer membrane of wild-type bacteria. The most severe mutation was flhB(N267A), which caused complete lack of FlaA in the outer membrane. The other two mutants, the flhBΔ214-218 and ΔfliI mutants, had approximately two- to fivefold reductions in the level of of FlaA associated with the outer membrane, suggesting that secretion had been impaired in these mutants. For the flhBΔ244-253 mutant there was about threefold less FlaA in WCLs, but this mutant completely lacked FlaA in the outer membrane. Only in one mutant, the flhBΔ224-228 mutant, did FlaA production appear to be greatly hindered, similar to a ΔflhB mutant.
Considering that four of the five mutants that we created appeared to have FEAs with greatly diminished secretion abilities, we then analyzed expression of σ54-dependent flagellar genes in these mutants. In the same four mutants [flhBΔ214-218, flhBΔ244-253, flhB(N267A), and ΔfliI], expression of the flaB- and flgDE2-astA transcriptional fusions was equal to or slightly higher than the expression in the wild-type strain (Fig. 5B). These results indicate that completely blocking or hindering secretion through the FEA did not affect expression of σ54-dependent flagellar genes. This analysis provided evidence that formation of the FEA, rather than secretory activity of the apparatus, is required and may be the key element to activate the FlgSR system for expression of σ54-dependent flagellar genes.
Only in the mutant that produced the FlhBΔ224-228 protein did we observe reduced expression of flaB::astA and flgDE2::nemo comparable to that of the ΔflhB mutant (Fig. 5B). Considering that this mutant also behaved similar to the ΔflhB mutant in terms of expression of flaA::astA and secretion of the FlaA protein, we believe that, like the ΔflhB mutant, this mutant may not form a complete FEA. Thus, this mutant may not actually be germane to our goal of creating secretion-incompetent but correctly formed FEAs. However, if an FEA forms in this mutant, then our alternative hypothesis that a negative regulator may be active and inhibit the FlgSR system in a nonsecreting bacterium may have some credence. To investigate this hypothesis, we performed transposon mutagenesis with the darkhelment transposon (27) in C. jejuni 81-176 ΔastA ΔflhA flgDE2::nemo, 81-176 ΔastA ΔflhB flgD::astA, and 81-176 ΔastA ΔfliP flaB::astA. These mutants do not express the σ54-dependent transcriptional astA fusions due to the lack of a complete FEA. Disruption of a gene encoding a putative repressor would allow expression of the transcriptional reporters in the FEA mutants. Such a transposon mutant could be identified by recovering mutants on media containing a chromomeric substrate for arylsulfatase and observing a switch from a white colony phenotype to a blue colony phenotype. Despite screening over 65,000 transposon mutants, we were unable to identify any mutant with a transposon that disrupted a gene for such a negative regulator, suggesting that such a gene may not exist or is an essential gene. Considering these data as a whole, we propose that FlgSR activation likely depends on proper assembly of the FEA. While we cannot entirely exclude the possibility that the secretory activity is required for FlgSR activation, our results indicating that four of five flhB or fliI mutants were impaired for secretion but had mutations that did not affect expression of σ54-dependent flagellar genes, coupled with the results of our transposon mutagenesis screen, weaken this hypothesis.
DISCUSSION
Previous studies in our laboratory have found that the proteins of the FEA, the putative FlhF GTPase, and the FlgSR two-component system are required for full expression of σ54-dependent flagellar genes in C. jejuni (30, 35). In the current study, we obtained evidence that links the FEA to stimulation of the FlgSR two-component regulatory system. We found that activation rather than production of the FlgSR system is dependent on the FEA. Furthermore, we believe that formation of the apparatus rather than the secretory function of the apparatus is key to producing the signal detected by FlgS leading to its activation and subsequent expression of σ54-dependent flagellar genes. Analysis of the genomic sequences of various C. jejuni strains indicates that the consensus σ54-binding site is in the promoters of most genes that encode the flagellar proteins that are external to the cytoplasm and likely secreted by the FEA (20, 31, 54). Because gene expression and protein production are energetically expensive processes, it is likely that the introduction of a level of transcriptional control by the FEA allows C. jejuni to ensure that σ54-dependent flagellar genes are expressed and the secreted proteins are produced only after the apparatus has formed.
The flagellar regulatory cascade of C. jejuni appears to bear some resemblance to the cascades utilized by species of Helicobacter, Vibrio, and Pseudomonas (2, 16, 34, 39, 40, 47, 51, 56, 60). First, all the cascades are known to require σ54 and a two-component regulatory system with functional similarity to FlgSR for expression of a subset of flagellar genes. In addition, Vibrio and Pseudomonas species require the activity of a master regulator protein to initiate transcription of genes encoding FEA proteins and these flagellar two-component regulatory systems (2, 15, 16, 36, 40, 56). However, in C. jejuni and Helicobacter pylori, no master regulator of flagellar biosynthesis has been found, and one current hypothesis is that the expression of genes encoding components of the FEA and FlgSR is largely constitutive (26, 51). In all these bacteria, activation of the flagellar two-component regulatory system leads to the σ54-dependent expression of genes encoding flagellar proteins that are secreted by the FEA (16, 25, 27, 30, 35, 40, 51, 56). Considering the similarity of the compositions of these flagellar regulatory cascades, our findings may suggest that the formation of the FEA could influence σ54-dependent flagellar gene expression in a number of bacterial species. Further analysis of each of these organisms is required to determine if this relationship is shared across multiple genera of motile bacteria.
The analysis presented in this work allowed us to more precisely clarify the relationship between the FEA and the FlgSR system in σ54-dependent flagellar gene expression. We constructed C. jejuni mutants whose mutations impaired FEA-mediated secretion to determine if formation of the export apparatus or its secretory activity was required for FlgS activation. Based on our finding that three of four flhB mutations and a fliI mutation reduced or blocked secretion of the FlaA flagellin but did not negatively affect σ54-dependent gene expression, we concluded that the formation of the FEA in the inner membrane could be the signal detected by FlgS that directly leads to activation of the kinase. Alternatively, formation of the FEA may be indirectly involved by being required for the production of a downstream activating signal. Although the data alone do not define the nature of the communication between the FEA and FlgSR, we have provided a foundation for future studies to understand activation of the system. Characterization of additional FEA proteins and structures such as the inner membrane MS ring and the cytoplasmic C ring that are associated with the FEA (43, 44), along with better reagents to detect complete FEA formation, may allow us to further define the activating signal emanating from this secretory apparatus.
If our hypothesis that FlgS detects formation of the FEA for autoactivation is correct, the cytoplasmic localization of FlgS may provide insight into the origin of the signal relative to the FEA structure. Since FlgS is a cytoplasmic protein, FlgS may detect a signal originating on the cytoplasmic face of the inner membrane-localized FEA complex. For instance, FlgS may detect a completed FEA structure by monitoring whether certain proteins with large cytoplasmic domains are in the FEA. Possible candidates for this type of signal include the cytoplasmic domains of FlhA and FlhB. To find evidence supporting this hypothesis, we attempted to use numerous approaches to directly detect interactions that may occur between FlgS and FEA proteins, including affinity chromatography, affinity blotting, and in vivo chemical cross-linking. However, the results of these assays were inconsistent and inconclusive. New and better reagents and protocols have to be developed to extend these types of analyses. In vivo detection of an FlgS interaction with a member of the FEA may be difficult, due to the fact that flagellated C. jejuni assembles only one or two of these secretory apparatuses per bacterium. Thus, the number of interactions of FlgS with the FEA or an FEA component may be small and the interactions may be temporally transient.
As mentioned above, our results strongly support the hypothesis that formation of the FEA either directly comprises the signal or is required to produce the signal to activate FlgSR and expression of σ54-dependent flagellar genes. An alternative hypothesis that we considered suggested that the secretory activity of the FEA could be the activating signal, with a cytoplasmic repressor hindering the FlgSR regulatory cascade prior to formation of and secretion by the FEA. However, four of the five flhB or fliI mutants whose mutations were shown to hinder or block secretion of flagellar proteins were not affected for σ54-dependent expression of flagellar genes. Only the flhBΔ226-230 mutant showed decreased expression of these genes, but analysis of this mutant suggested that it behaved most like a ΔflhB mutant, which does not form a complete FEA. Thus, we cannot confidently conclude that the flhBΔ224-228 mutant makes a fully formed but secretion-incompetent apparatus. Second, our transposon mutagenesis screen did not reveal any transposon insertions in FEA mutants that relieved repression of expression of σ54-dependent flagellar genes. These combined results greatly weaken the hypothesis that the secretory activity of the FEA alone forms the FlgS-activating signal. Thus, the results of this study strongly favor the hypothesis that that formation of the FEA is a requirement for and quite possibly a component of the essential signal for activating the FlgSR system that results in expression of σ54-dependent flagellar genes.
Our work also suggests a new function in the signaling mediated by the FEA in flagellar regulatory cascades. In the well-characterized pathways observed in E. coli and Salmonella, formation of the FEA ultimately controls the activity of the alternative sigma factor σ28 involved in expression of genes encoding the major flagellins and some motor proteins (41). The FEA is responsible for secretion of flagellar proteins and the anti-σ factor, FlgM, which represses the activity of σ28 until the cell has completed formation of the FEA, basal body, and hook structures required to secrete flagellins to build a filament (32, 38). In this study, we found that the FEA is intimately involved in creating a signal that activates the FlgSR two-component system, leading to activation of σ54. Therefore, the FEA plays a different role in influencing signaling for σ54-dependent expression of flagellar genes in C. jejuni. This finding may also be applicable to other motile bacteria that utilize σ54 in flagellar gene regulation and biosynthesis, including species of Vibrio, Pseudomonas, and Helicobacter. This work expands the known mechanisms of regulating flagellar gene expression and suggests that there are more complex functions associated with the FEA beyond protein secretion.
Future analyses of FlgS will involve determining the domain and residues of the protein required for sensing an autoactivating signal. In analyzing the sequence of FlgS, we found that the central and C-terminal portions of the protein contain the histidine-containing phosphotransfer domain and the ATP-catalytic domain (61, 65). These domains are required for accepting a phosphate group on a conserved histidine and for ATP hydrolysis, respectively, for autophosphorylation. Indeed, we found that H141 in the phosphotransfer domain is required for modification by phosphorylation and for functioning of the active FlgS to stimulate expression of σ54-dependent flagellar gene expression. In a comparison of the amino acid sequence of FlgS to those of other sensor kinases, the predominant homology with the latter kinases is localized almost exclusively to the phosphoacceptor and ATP hydrolysis domains. Only limited homology between the initial 130 amino acids of FlgS and other sensor kinases is apparent. The sensor kinases that share the most homology to this region of the C. jejuni FlgS protein are other FlgS homologues in Campylobacter species (almost 100% identity), the FlgS orthologue in Helicobacter species (31 to 37% identity and 57 to 66% similarity), and the FlrB sensor kinase of Vibrio cholerae (26% identity and 54% similarity). The N-terminal regions of these proteins have no obvious motifs that suggest a function or how they may sense a specific factor. Since these N-terminal domains are unique to the group of FlgS orthologues, it is likely that this region of these proteins may function in specifically recognizing the signal necessary to culminate in expression of σ54-dependent flagellar genes. Future studies will focus on further characterizing this domain of the protein.
Previous work in our laboratory focused on understanding the activation and function of the FlgR response regulator (25, 30, 35). In this study, we describe work that provides a foundation for understanding the activation of the cognate sensor kinase, FlgS, and how the FEA influences activation of this two-component regulatory system. To date, we have linked activation of the FlgSR system to the FEA and have characterized a previously undescribed mechanism for controlling activation of flagellar gene expression. In addition, FlgSR appears to be an unusual two-component system in which expression of both components is controlled by phase-variable mechanisms (25, 27), a trait unique among well-characterized bacterial two-component systems. Thus, there appears to be at least two mechanisms for controlling σ54-mediated expression via the FlgSR proteins. Future analyses will focus on further defining the nature of the activating signal emanating from the FEA and how it influences expression of σ54-dependent flagellar genes.
Acknowledgments
This work was supported by NIH grant R01 AI065539, by National Research Initiative Grant 2006-35201-17382 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program, and by start-up funds from the University of Texas Southwestern Medical Center. S.N.J. was supported by NIH training grant T32 AI007520 from the Molecular Microbiology Graduate Program.
We thank Kevin Gardner for helpful discussions regarding autophosphorylation experiments and analyses.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Akerley, B. J., and D. J. Lampe. 2002. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 358100-108. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1795574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 289-22. [DOI] [PubMed] [Google Scholar]
- 4.Bingham-Ramos, L. K., and D. R. Hendrixson. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 761105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157472-479. [DOI] [PubMed] [Google Scholar]
- 6.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49489-522. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J., and J. Engberg. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections, p. 99-121. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 8.Blaser, M. J., and L. B. Reller. 1981. Campylobacter enteritis. N. Engl. J. Med. 3051444-1452. [DOI] [PubMed] [Google Scholar]
- 9.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104269-279. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 27920327-20338. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, United States, 2005. MMWR Morb. Mortal. Wkly. Rep. 55392-395. [PubMed] [Google Scholar]
- 12.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coker, A. O., R. D. Isokpehi, B. N. Thomas, K. O. Amisu, and C. L. Obi. 2002. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35743-755. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta, N., E. P. Ferrell, K. J. Kanack, S. E. West, and R. Ramphal. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 1845240-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 17.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferris, H. U., Y. Furukawa, T. Minamino, M. B. Kroetz, M. Kihara, K. Namba, and R. M. Macnab. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 28041236-41242. [DOI] [PubMed] [Google Scholar]
- 20.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser, G. M., T. Hirano, H. U. Ferris, L. L. Devgan, M. Kihara, and R. M. Macnab. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 481043-1057. [DOI] [PubMed] [Google Scholar]
- 22.Goon, S., C. P. Ewing, M. Lorenzo, D. Pattarini, G. Majam, and P. Guerry. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect. Immun. 74769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerry, P., R. A. Alm, M. E. Power, S. M. Logan, and T. J. Trust. 1991. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 1734757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 13980-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 611646-1659. [DOI] [PubMed] [Google Scholar]
- 26.Hendrixson, D. R. 2008. Regulation of flagellar gene expression, p. 545-558. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 27.Hendrixson, D. R. 2008. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70519-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40214-224. [DOI] [PubMed] [Google Scholar]
- 29.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52471-484. [DOI] [PubMed] [Google Scholar]
- 30.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50687-702. [DOI] [PubMed] [Google Scholar]
- 31.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 744694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 2621277-1280. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, P., and A. J. Ninfa. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 1811906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43307-322. [DOI] [PubMed] [Google Scholar]
- 35.Joslin, S. N., and D. R. Hendrixson. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol. 1902422-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jyot, J., N. Dasgupta, and R. Ramphal. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 1845251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalmokoff, M., P. Lanthier, T. L. Tremblay, M. Foss, P. C. Lau, G. Sanders, J. Austin, J. Kelly, and C. M. Szymanski. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 1884312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlinsey, J. E., S. Tanaka, V. Bettenworth, S. Yamaguchi, W. Boos, S.-I. Aizawa, and K. T. Hughes. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 371220-1231. [DOI] [PubMed] [Google Scholar]
- 39.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28501-520. [DOI] [PubMed] [Google Scholar]
- 41.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, L. H., E. Burg III, S. Baqar, A. L. Bourgeois, D. H. Burr, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evaluation of a truncated recombinant flagellin subunit vaccine against Campylobacter jejuni. Infect. Immun. 675799-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 5777-100. [DOI] [PubMed] [Google Scholar]
- 44.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694207-217. [DOI] [PubMed] [Google Scholar]
- 45.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29970-972. [DOI] [PubMed] [Google Scholar]
- 46.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 1824906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minamino, T., and K. Namba. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451485-488. [DOI] [PubMed] [Google Scholar]
- 50.Nachamkin, I., X.-H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 591269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52947-961. [DOI] [PubMed] [Google Scholar]
- 52.Ninfa, A. J., and R. L. Bennett. 1991. Identification of the site of autophosphorylation of the bacterial protein kinase/phosphatase NRII. J. Biol. Chem. 2666888-6893. [PubMed] [Google Scholar]
- 53.Olson, C. K., S. Ethelberg, W. van Pelt, and R. V. Tauxe. 2008. Epidemiology of Campylobacter jejuni infections in industralized nations, p. 163-189. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 54.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 55.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451489-492. [DOI] [PubMed] [Google Scholar]
- 56.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 391595-1609. [DOI] [PubMed] [Google Scholar]
- 57.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 634868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejewski, A. D. Grossman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13689-701. [DOI] [PubMed] [Google Scholar]
- 59.Sommerlad, S. M., and D. R. Hendrixson. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterzenbach, T., L. Bartonickova, W. Behrens, B. Brenneke, J. Schulze, F. Kops, E. Y. Chin, E. Katzowitsch, D. B. Schauer, J. G. Fox, S. Suerbaum, and C. Josenhans. 2008. Role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J. Bacteriol. 1906398-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 62.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, L., R. Grau, M. Perego, and J. A. Hoch. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 112569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wassenaar, T. M., B. A. M. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 1391171-1175. [DOI] [PubMed] [Google Scholar]
- 65.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3REVIEWS301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wosten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 27916214-16222. [DOI] [PubMed] [Google Scholar]
- 67.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 1783335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]