Abstract
Rrp2 is the sole σ54-dependent transcriptional activator present in the Borrelia burgdorferi genome. We showed that recombinant Rrp2 binds to DNA in a sequence-nonspecific manner. During infection, Rrp2 activates σ54-dependent rpoS expression without an apparent upstream enhancer element commonly associated with other σ54-dependent transcriptional activators.
Dramatic alteration of surface lipoprotein profiles is a key strategy which the Lyme disease pathogen Borrelia burgdorferi has evolved to persist within its natural enzootic cycle (19, 20). The Rrp2-RpoN-RpoS pathway (or the σ54-σS cascade) plays a central role in modulating the differential gene expression involved in spirochete transmission from the arthropod vector (e.g., Ixodes species ticks) to a mammalian host (e.g., the white-footed mouse) (5, 6, 8, 10, 12, 25). In this pathway, the two-component response regulator Rrp2 acts in concert with the sigma factor RpoN (σ54 or σN) to directly control the production of another sigma factor, RpoS (σS or σ38), which in turn governs expression of more than 10% of the genes annotated in B. burgdorferi (3, 6, 8, 14). This pathway appears to be activated in the spirochete at the onset of nymphal feeding and remains operative as the bacteria replicate in the mammal (6). Therefore, elucidating the mechanism by which the Rrp2-RpoN-RpoS pathway is activated is important to the understanding of host adaptation of B. burgdorferi.
σ54 is a unique alternative sigma factor which differs from the members of the σ70 family both in amino acid sequence and in transcription mechanism (13). σ54 recognizes a conserved −24/−12 promoter sequence, and the activation of the σ54-polymerase holoenzyme requires specialized bacterial enhancer-binding proteins (EBP) (22). All members of the EBP family contain a highly conserved activation domain that interacts with σ54 for transcriptional activation. This domain also possesses ATPase activity that is essential for their activating function (23). In addition, a typical EBP contains a DNA-binding domain that binds to enhancer-like DNA elements and allows transcriptional activation far from the σ54 promoter (0.1 to 1.0 kb) via a DNA-looping mechanism (15, 17).
Whereas most bacterial genomes encode multiple EBP, the B. burgdorferi genome has only one predicted EBP, Rrp2 (9, 25). Rrp2 is an NtrC-like EBP which is comprised of an amino-terminal response regulator domain, a central σ54 activation domain, and a carboxy-terminal helix-turn-helix (HTH)-type DNA-binding domain (25). Like other EBP, Rrp2 was thought to activate rpoS transcription by binding to an enhancer site in the region of the rpoS gene (25). Surprisingly, a recent article by Burtnick et al. (5) reported that a cat transcriptional reporter containing only the minimal −24/−12 σ54-type promoter of rpoS (with an additional 17 bp of upstream sequence) can be activated by endogenous Rrp2 in B. burgdorferi, suggesting that Rrp2 may be an unusual EBP which does not require an enhancer site for activating σ54-dependent transcription. These observations raise several key questions. In particular, is the rpoS gene with the minimal promoter sufficient to fulfill the essential role of RpoS in mammalian infection by B. burgdorferi? If so, what is the function of Rrp2's putative DNA-binding domain?
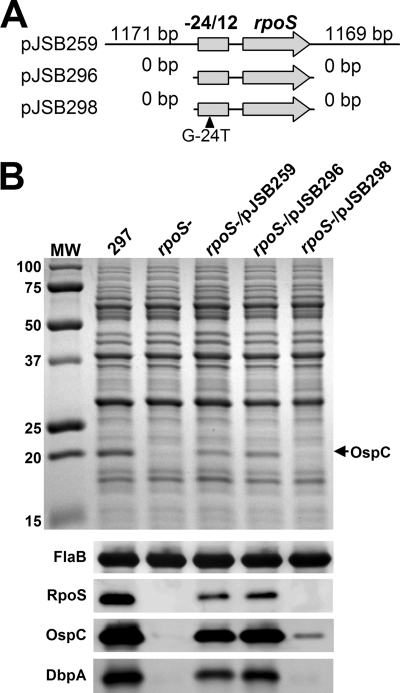
rpoS expressed from its minimal promoter is sufficient for the production of RpoS, OspC, and DbpA.
To study the mechanism of Rrp2-dependent activation of rpoS, a previously described rpoS mutant of B. burgdorferi strain 297 (14) was complemented with the pJD44 shuttle vector (modified from pBSV2 [18]) carrying various rpoS regions (Fig. 1A): rpoS with more than 2 kb of flanking DNA (pJSB259), rpoS with the −24/−12 σ54-type promoter but no flanking regions (pJSB296), or rpoS with a defective −24/−12 promoter (mutation of G-24T) (pJSB298) (18, 21). The primers used to amplify these regions are described in Table S1 of the supplemental material. The abilities of these rpoS constructs to restore the production of RpoS and the RpoS-dependent virulence factors OspC and DbpA in the rpoS mutant were then examined by performing immunoblot analyses on bacteria grown to postexponential phase in pH-adjusted medium (pH 6.8). The antibodies used in the immunoblot analyses have been previously described (2, 21).
FIG. 1.
Complementation of the rpoS mutant with a shuttle vector carrying various versions of the rpoS gene. (A) Diagram of the three shuttle vectors. Only the rpoS portion in each vector is shown. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Coomassie blue stain) (top) and immunoblot assay (bottom) of whole-cell lysates of B. burgdorferi strains (labeled on top). Numbers at the left denote protein molecular mass markers (in kDa). The arrow at the right indicates the band corresponding to OspC. Antibodies used to detect the respective proteins are indicated to the left of the immunoblot results shown at the bottom of the panel.
As expected, complementation of the rpoS mutant with the construct carrying rpoS and 1,171 bp of contiguous upstream DNA (pJSB259), which contains the native −24/−12 region, restored production of RpoS, OspC, and DbpA (Fig. 1B). More importantly, the rpoS mutant complemented with rpoS expressed from only the minimal σ54-dependent promoter (pJSB296) exhibited RpoS, OspC, and DbpA expression levels comparable to those observed in the pJSB259-complemented strain (Fig. 1B). In contrast, complementation with the construct carrying rpoS with the G-24T-mutated σ54-type promoter (pJSB298) failed to restore expression of RpoS, OspC, and DbpA (Fig. 1B). Note that strains complemented with pJSB259 or pJSB296 exhibited levels of RpoS expression that were lower than those observed in the wild-type strain 297; this is likely due to the differences in the DNA topology of the plasmid-encoded and chromosomally encoded copies of rpoS. Alternatively, a possible difference in mRNA stability between the transcripts derived from the two forms of rpoS genes may contribute to the observed variation. It also is worth noting that OspC was detected in the rpoS mutant carrying pJSB298, albeit at a significantly reduced level. This presumably is due to the fact that the G-24T mutation in the promoter did not completely abolish rpoS expression, and therefore a very low level of RpoN-mediated rpoS transcription still occurred in this strain (21). Nevertheless, these results support the conclusion, based on the data from the transcriptional reporter system employed by Burtnick et al. (5), that Rrp2 is capable of activating σ54-dependent rpoS transcription in the absence of a specific upstream DNA cis-element under in vitro growth conditions.
rpoS with the σ54 minimal promoter is sufficient to restore infectivity of the rpoS mutant in mice.
To determine whether rpoS with only the σ54 minimal promoter is sufficient for mammalian infection, groups of five C3H/HeN mice were infected via intradermal needle inoculation with 10, 102, 103, 104, 105, or 106 spirochetes of wild-type strain 297, the rpoS mutant, or the rpoS mutant carrying pJSB296 (rpoS with the minimal promoter region). At 15 days postinoculation, mouse infection was assessed by culturing spirochetes from ear punch biopsy tissues. Whereas the rpoS mutant was completely avirulent in mice, the rpoS mutant complemented with pJSB296 had a 50% infectious dose (ID50) value similar to the wild-type strain (Table 1). The rpoS mutant harboring pJSB296 also was able to cause chronic infection equivalent to the wild-type strain (examined at 28 weeks after infection) (data not shown). These results suggest that Rrp2, in conjunction with RpoN, can activate rpoS expression in the absence of an upstream enhancer under physiological conditions, and such activation is sufficient for spirochetes to establish and maintain infection in mice.
TABLE 1.
The rpoS gene with a minimal −24/−12 σ54-type promoter is sufficient to restore infectivity of the rpoS mutant in micea
| Strain or phenotype | No. infected/no. inoculated with indicated no. of bacteria
|
ID50 | |||||
|---|---|---|---|---|---|---|---|
| 101 | 102 | 103 | 104 | 105 | 106 | ||
| Wild type | 0/6 | 1/8 | 8/8 | 8/8 | ND | ND | 239 |
| RpoS− | ND | ND | ND | 0/8 | 0/8 | 0/8 | >106 |
| RpoS−/pJSB296 | 1/6 | 4/8 | 8/8 | 8/8 | ND | ND | 87 |
The ID50 values were calculated using the method described by Reed and Muench (16). ND, not determined.
Properties of the recombinant Rrp2 proteins.
To determine whether the predicted DNA-binding domain of Rrp2 is capable of binding DNA, we produced recombinant maltose-binding proteins (MBP) fused with either full-length Rrp2 (MBP-Rrp2) or amino acids 391 to 453 of the Rrp2 C-terminal domain (MBP-Rrp2Ct) using the pMAL system (New England Biolabs, Ipswich, MA). Recombinant proteins were expressed in Escherichia coli and sequentially purified from cell lysates using amylose-resin and heparin-Sepharose. Both MBP-Rrp2 and MBP-Rrp2Ct were capable of binding to heparin (data not shown), suggesting that the full-length and C-terminal recombinant Rrp2 protein might be able to bind DNA. Recombinant proteins were then further purified using a Mono Q HR5/5 ion exchange column (GE Healthcare, Chalfont St. Giles, United Kingdom). Purified MBP-Rrp2 and MBP-Rrp2Ct were soluble in solution, and chromatographic gel filtration analyses indicated that both recombinant proteins formed dimers in solution (data not shown). This latter observation suggests that one of the functions of the C-terminal domain (CTD) of Rrp2 likely is dimerization.
To further assess the physiological activities of the purified recombinant MBP-Rrp2 protein, we sought to determine whether purified recombinant MBP-Rrp2 protein had ATPase activity, as all σ54-dependent regulators hydrolyze ATP when activated. Rrp2 is predicted to be activated by phosphorylation of its N-terminal receiver domain. It has been reported that BeF3, but not BeCl2 or NaF, mimics the Asp-phosphate of a receiver domain that activates NtrC and other response regulators (24). Therefore, MBP-Rrp2 (1.4 mM) was incubated in HEPES buffer (pH 7.3) with 0.8 mM ATP and 200 μM BeF3, BeCl2 or NaF at 37°C for 90 min. ATPase activities were measured by monitoring the release of Pi using the PiPer phosphate assay kit (Molecular Probes, Eugene, OR). We found that there was a fivefold increase in Pi release upon addition of BeF3, but not BeCl2 or NaF, to the reaction mixture (data not shown). This observation suggests that Rrp2 has phosphorylation-dependent ATPase activity.
Rrp2 binds DNA in a sequence-nonspecific fashion.
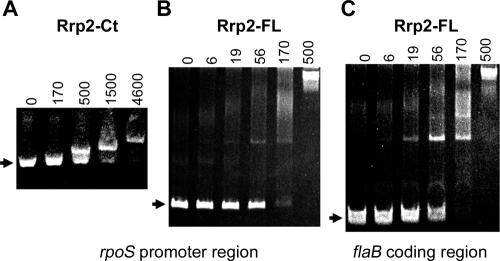
The DNA-binding capacities of recombinant MBP-Rrp2 and MBP-Rrp2Ct were assessed using electrophoresis mobility shift assays. Reaction mixtures containing 10 nM of a 370-bp DNA fragment of the rpoS promoter region (200 bp upstream and 170 bp downstream of ATG), 50 mM HEPES (pH 7.5), 8 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 2 μg/ml poly(dI-dC), and increasing amounts of protein were incubated at room temperature for 20 min. Protein-DNA complexes were resolved from unbound probe by electrophoresis on 5% polyacrylamide-Tris-glycine gels. As shown in Fig. 2A and B, both the C-terminal domain and the full length of Rrp2 were capable of binding to the rpoS promoter region. The minimal concentration of full-length Rrp2 protein required to achieve maximum DNA binding in the presence of competitor poly(dI-dC) was 0.5 μM, which is comparable to that of recombinant E. coli NtrC binding to a single glnAp2 site (0.25 μM) (7, 26). There appeared to be a difference in relative binding activities of MBP-Rrp2 and MBP-Rrp2Ct (approximately 10-fold), suggesting that other domains in Rrp2 may enhance the interaction of the C-terminal domain with DNA. Furthermore, phosphorylation of Rrp2 did not alter the DNA-binding pattern of Rrp2 (data not shown). Interestingly, the DNA-binding activity of Rrp2 was not sequence specific, because Rrp2 also bound to a 227-bp DNA fragment of the flaB coding region (200 bp downstream of ATG) (Fig. 2C) or a 214-bp DNA fragment of the kat gene from Neisseria gonorrhoeae (data not shown).
FIG. 2.
Electrophoretic mobility shift analyses of the DNA-binding activity of Rrp2. Various concentrations of recombinant C-terminal Rrp2 (MBP-Rrp2Ct) or full-length Rrp2 (MBP-Rrp2) (labeled on the top, in nM) were incubated with DNA fragments from either the rpoS promoter region (A and B) or the flaB coding region (labeled at the bottom) (C). Protein-DNA complexes were resolved from unbound DNA fragments on polyacrylamide gels and stained with ethidium bromide (0.5 μg/ml). The arrows indicate the positions of unbound DNA fragments.
Although the promiscuous DNA-binding activity of Rrp2 in vitro might be interpreted as a nonspecific interaction, several lines of evidence support the conclusion that Rrp2 possesses DNA-binding activity. First, Rrp2 has a predicted HTH DNA-binding domain (25). Second, recombinant Rrp2 binds heparin. Third, both recombinant full-length Rrp2 and the Rrp2 C-terminal domain bind to DNA in the presence of the nonspecific competitor poly(dI-dC). Lastly, the relative DNA-binding affinity of full-length recombinant Rrp2 is comparable to that of recombinant E. coli NtrC binding to a single site (7, 26). Taken together, these results suggest that, although a specific enhancer-binding site is not required upstream of the −24/−12 region for Rrp2 to activate rpoS transcription, the predicted C-terminal domain of Rrp2 indeed possesses DNA-binding activity and that the interaction with DNA does not appear to be dependent upon a specific DNA sequence.
In an effort to determine the contribution of the DNA-binding activity of Rrp2 to transcriptional activation, we sought to take a genetic approach by constructing an rrp2 B. burgdorferi mutant strain that expressed either Rrp2 lacking the entire C-terminal domain of Rrp2 or Rrp2 with a truncated HTH DNA-binding motif. Following multiple rounds of transformation and recombination of the mutant construct into B. burgdorferi, no clones have been identified which carry a mutated rrp2 allele that is missing either the entire CTD or the last 10 amino acids (corresponding to the last α-helix of the HTH motif). Because an analogous approach was used to generate our previously described Rrp2 G239C mutant of B. burgdorferi (3, 27), the inability to generate a strain carrying the CTD truncation or HTH mutation suggests that the CTD may provide a function that is essential for cell survival. This notion is consistent with the previous observation that deletion of rrp2 appears to be lethal (5, 25).
In conclusion, the results from this study along with previous findings suggest that the rpoS gene, containing only the minimal σ54-type promoter, is essential and sufficient to produce RpoS for mammalian infection of B. burgdorferi. This notion further supports the hypothesis that Rrp2 is an unusual σ54-dependent activator that does not require an enhancer site for transcriptional activation. To this end, recent genomic sequence analyses have identified a group of σ54-dependent activators that lack an apparent DNA-binding domain: FlgR in Helicobacter pylori, FlgR in Campylobacter jejuni, and CtcC in Chlamydia trachomatis (1, 11). Subsequent experimentation has confirmed that the FlgR proteins of H. pylori and C. jejuni are functional activators of σ54-dependent genes (1, 4, 11). One common theme among FlgR, CtcC, and Rrp2 is that they are the sole σ54-dependent activator present in each corresponding genome and, therefore, are responsible for activating all genes with a σ54-type promoter in that genome. As a result, no enhancer site is required for these activators to discriminate σ54-dependent genes for activation within the cell. The mechanism by which these activators that do not bind an enhancer site are recruited to σ54-dependent promoter regions is not clear. One possibility is that they are capable of interacting with unbound σ54 prior to activation (e.g., in solution) and are subsequently recruited to the promoter site by σ54. However, Rrp2 has the predicted C-terminal DNA-binding domain that is not found in FlgR and CtcC. The sequence-nonspecific DNA-binding nature may be advantageous to Rrp2-dependent activation, because once recruited to the promoter by σ54, Rrp2 would be capable of binding to any DNA sequence adjacent to the −24/−12 promoter site, which provides further stabilization to the Rrp2-σ54-holoenzyme-DNA complex for transcriptional activation. One caveat of this study is that, despite intense efforts, the mutation of the C-terminal domain of Rrp2 to assess its role in the activation of rpoS transcription could not be achieved, potentially as a result of the essential nature of this domain to spirochetal survival. Further work, possibly through in vitro transcription analyses, is needed to study the role of the DNA-binding function of Rrp2 in transcriptional activation and the mechanism of recruitment of Rrp2 to the rpoS promoter region, which will provide insight into this novel form of σ54-dependent transcriptional activation.
Supplementary Material
Acknowledgments
The work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR-54023; X.F.Y.) and the National Institute of Allergy and Infectious Diseases (AI-72144 [X.F.Y.] and AI-59602 [M.V.N.]) and an American Heart Association Scientist Development Grant (X.F.Y.).
Footnotes
Published ahead of print on 6 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Beck, L. L., T. G. Smith, and T. R. Hoover. 2007. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 15530-537. [DOI] [PubMed] [Google Scholar]
- 2.Blevins, J., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boardman, B. K., M. He, Z. Ouyang, H. Xu, X. Pang, and X. F. Yang. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 763844-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmachary, P., M. G. Dashti, J. W. Olson, and T. R. Hoover. 2004. Helicobacter pylori FlgR is an enhancer-independent activator of σ54-RNA polymerase holoenzyme. J. Bacteriol. 1864535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burtnick, M. N., J. S. Downey, P. J. Brett, J. A. Boylan, J. G. Frye, T. R. Hoover, and F. C. Gherardini. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65277-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P., and L. J. Reitzer. 1995. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J. Bacteriol. 1772490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 1025162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 10.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 9812724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joslin, S. N., and D. R. Hendrixson. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-Like protein. J. Bacteriol. 1902422-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lybecker, M. C., and D. S. Samuels. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 641075-1089. [DOI] [PubMed] [Google Scholar]
- 13.Merrick, M. J. 1993. In a class of its own: the RNA polymerase sigma factor σN (σ54). Mol. Microbiol. 10903-909. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang, Z., J. S. Blevins, and M. V. Norgard. 2008. Transcriptional interplay among the regulators Rrp2, RpoN, and RpoS in Borrelia burgdorferi. Microbiology 1542641-2658. [DOI] [PubMed] [Google Scholar]
- 15.Popham, D. L., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243629-635. [DOI] [PubMed] [Google Scholar]
- 16.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 17.Reitzer, L. J., and B. Magasanik. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45785-792. [DOI] [PubMed] [Google Scholar]
- 18.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 1026972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3129-143. [DOI] [PubMed] [Google Scholar]
- 20.Singh, S. K., and H. J. Girschick. 2004. Molecular survival strategies of the Lyme disease spirochete Borrelia burgdorferi. Lancet Infect. Dis. 4575-583. [DOI] [PubMed] [Google Scholar]
- 21.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 1892139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 1851757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wedel, A., and S. Kustu. 1995. The bacterial enhancer-binding protein NtrC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 92042-2052. [DOI] [PubMed] [Google Scholar]
- 24.Yan, D., H. S. Cho, C. A. Hastings, M. M. Igo, S. Y. Lee, J. G. Pelton, V. Stewart, D. E. Wemmer, and S. Kustu. 1999. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA 9614789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, X. F., S. M. Alani, and M. V. Norgard. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 10011001-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, X. F., Y. Ji, B. L. Schneider, and L. Reitzer. 2004. Phosphorylation-independent dimer-dimer interactions by the enhancer-binding activator NtrC of Escherichia coli: a third function for the C-terminal domain. J. Biol. Chem. 27936708-36714. [DOI] [PubMed] [Google Scholar]
- 27.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.