Abstract
Nontypeable Haemophilus influenzae (NTHI) bacteria are commensals in the human nasopharynx, as well as pathogens associated with a spectrum of acute and chronic infections. Two important factors that influence NTHI pathogenicity are their ability to adhere to human tissue and their ability to form biofilms. Extracellular polymeric substances (EPS) and bacterial appendages such as pili critically influence cell adhesion and intercellular cohesion during biofilm formation. Structural components in the outer cell membrane, such as lipopolysaccharides, also play a fundamental role in infection of the host organism. In spite of their importance, these pathogenic factors are not yet well characterized at the nanoscale. Here, atomic force microscopy (AFM) was used in aqueous environments to visualize structural details, including probable Hif-type pili, of live NTHI bacteria at the early stages of biofilm formation. Using single-molecule AFM-based spectroscopy, the molecular elasticities of lipooligosaccharides present on NTHI cell surfaces were analyzed and compared between two strains (PittEE and PittGG) with very different pathogenicity profiles. Furthermore, the stiffness of single cells of both strains was measured and subsequently their turgor pressure was estimated.
Haemophilus influenzae is a gram-negative bacterium and a common commensal of the human nasopharynx; however, it can also be responsible for a number of serious infections (17, 28, 64, 65). H. influenzae strains are divided into two groups, according to the presence or absence of six antigenically distinct extracellular polysaccharide capsules (serotypes a to f) (33, 54). In particular, organisms possessing the type b capsule are highly virulent and may cause bacteremia and invasive infections such as meningitis and pneumonia (28, 33, 65). Strains that do not possess one of six antigenically distinct capsules are classified as nontypeable H. influenzae (NTHI) and are associated with colonization in the great majority of healthy individuals (33). The NTHI strains are also associated with acute and chronic infections of the respiratory tract, such as acute otitis media (OM), chronic OM with effusion, otorrhea, sinusitis, bronchitis and chronic obstructive pulmonary disease (28, 42, 68, 81). They are also increasingly linked to invasive diseases such as meningitis and sepsis (66, 67, 74, 75).
NTHI disease occurs when bacteria adhere to and colonize/invade epithelial cells in the respiratory tract or invade into surrounding tissues. The initial interaction between NTHI bacteria and the host is the adherence to epithelial cells (72, 81). Fibrillar appendages, called fimbriae and pili, promote and enhance adherence to epithelial cells and nasal tissue by using adhesins to bind to specific receptors on the host cell surface (28). Pili and fimbriae are present in many gram-negative bacteria, and besides adhesion they also perform other functions such as aiding genetic transfer between bacteria (sex pili) and generation of movement on surfaces via twitching motility (28). Many of the genes encoding these structures appear to be associated with mobile genetic elements and pathogenicity islands (51). Subsequent steps associated with chronic infection might include formation of microcolonies and ultimately a biofilm (11, 26, 35).
H. influenzae lipooligosaccharides (LOS) are important for colonization, bacterial persistence, and survival in the respiratory system. The interaction between bacteria and host cells is influenced by LOS structure, which varies among strains and also among bacterial cells within a strain. In H. influenzae, LOS is comprised of an oligosaccharide, composed mainly of neutral hexose and heptose sugars, linked via a single 2-keto-3-deoxyoctulosonic acid to the membrane-anchoring lipid A moiety (39). To the best of our knowledge, detailed information about the LOS length distribution is not available for NTHI bacteria. H. influenzae bacteria do not produce a true poison or toxin (28). Disease results from the host cell's response to bacterial factors, particularly endotoxin (LOS) (28).
Recent evidence suggests that H. influenzae is capable of forming mucosal biofilms in animals and human patients with middle ear infections (26, 35). Biofilms are surface-attached microbial communities with phenotypic and biochemical properties distinct from those of their free-swimming, planktonic counterparts. Significantly, certain biofilms can develop antibiotic resistance up to 1,000-fold greater than planktonic cells (21, 47).
Fimbriae of different strains of H. influenzae bacteria have been visualized in great detail using electron microscopy (EM), particularly transmission EM (TEM) (10, 20, 33, 41, 52, 69). In addition, their LOS composition has been extensively investigated by electrophoretic and mass spectrometric methods (11, 15, 38, 39, 44). However, none of these studies has been carried out under physiologically relevant conditions. Unlike EM, atomic force microscopy (AFM) is able to operate in liquids (13, 18, 22, 53, 76) and has been increasingly applied in microbiology to observe structural details of microbial cells (55, 57-59), study their elastic properties (6, 73, 78, 82, 83), probe cell-surface interactions (2, 3, 30, 34, 46, 61, 77), and more recently fingerprint individual molecules on the surface of microbial cells (23-25, 29).
In the present study, we employed AFM during the early stages of NTHI biofilm formation to study morphological details as well as nanomechanical properties of two phenotypically and genetically distinct clinical strains (PittEE and PittGG) with very different pathogenicity profiles (17, 37).
MATERIALS AND METHODS
Bacterial preparation.
Two NTHI clinical strains (PittEE and PittGG) were obtained from the Center for Genomic Sciences (Allegheny General Hospital; Pittsburgh, PA) as frozen stock cultures in microcentrifuge tubes (17, 27, 37, 65). Several agar plates of each strain were inoculated, incubated, harvested into replicate 2-ml vials, and frozen at −70°C for future use as additional stock cultures. Generally, H. influenzae cells were grown in BBL brain heart infusion (BHI) medium (37 g/liter; Becton-Dickinson and Company, Sparks, MD) supplemented with 10 ml per liter of hemin solution (1 mg/ml in 4% triethanolamine) and 200 μl per liter of β-NAD solution (10 mg/ml in double-distilled H2O) under 5% CO2 at 37 C in a NAPCO 6100 CO2 incubator (Precision Scientific, Chicago, IL). When agar plates were required, 12 g/liter granulated agar (Fisher Scientific; Pittsburgh, PA) was added to the above medium.
Rotating-disk biofilm reactor.
The spinning-disc biofilm reactor has been described previously and is only briefly described here (56). The reactor system consisted of a 1-liter beaker fitted with an effluent spout and a spinning rotor. The rotor consisted of a star-head magnetic stir bar fitted with a Teflon and neoprene disc which held six removable plugs. Mica discs (Ted Pella, Redding, CA) were attached with epoxy to each of the removable rotor plugs. The reactor system was filled with the appropriate medium and inoculated to a starting optical density (A600) of 0.02. The system was incubated at 37C for 2 h with no spinning. The medium was then drained, and 120 ml of fresh prewarmed medium was added. The system was then placed on a magnetic stir plate within the incubator and set to 250 rpm. The reactor system was operated in a semi-batch mode. The medium was drained at designated sampling intervals and fresh, prewarmed medium was added.
The sample plug, with the attached mica disc, was removed from the neoprene portion of the rotor using sterile forceps. Unattached cells were removed by placing the plug with the attached mica disc in a petri dish with 30 ml of phosphate-buffered saline buffer (8) and incubated at 37°C for 10 min with gentle shaking. The buffer was replaced twice during 30 min of incubation. The sample was then dried in a forced-air incubator at 37°C for 30 min. To remove residual salt crystals, the sample was rinsed once with a dilute phosphate buffer (0.3 mM KH2PO4, 2 mM MgCl2 [pH 7.2]).
Sample preparation for AFM measurements in liquid.
Si(100) wafers (Virginia Semiconductor, Inc., Fredericksburg, Virginia) were cut into small pieces (area of ∼0.5 by 0.5 cm2) and then cleaned by sonication for 15 min in three separate solvents sequentially (acetone, propanol, and methanol; Sigma-Aldrich, St. Louis, MO). This was followed by further cleaning in an ozone/UV chamber (BioForce, Ames, IA) for 30 min. The cleaned silicon surfaces were functionalized with amine groups by exposure to 1% (vol/vol) aminopropyltriethoxysilane (APTES) (Sigma-Aldrich) in 100% methanol for 5 min. The APTES-coated wafers were rinsed three times in methanol for ∼1 min each time and dried with nitrogen gas. For cell attachment and immobilization, one drop of mid-exponential-phase cell suspension (A600, ∼0.05) was placed on the amine-functionalized silicon surfaces for ∼2 h. The droplet was then decanted and replaced with 100 mM HEPES buffer (pH 7.4; Sigma-Aldrich, St. Louis, MO), before the sample was placed on the AFM stage.
AFM.
All measurements were carried out with a Nanoscope III extended-multimode atomic force microscope from Veeco (Santa Barbara, CA) with a 150- by 150-μm2 “J” scanner, using NanoScope III software (version 5,12R3). A multimode AFM liquid cell (Veeco) without an O-ring was used for measurements in buffer solutions. Si3N4 (and Si) AFM tips integrated with cantilevers (Veeco) having nominal spring constants of 0.01 to 0.03 N/m (and 40 N/m) were used for measurements in liquid (and air). Cantilever spring constants were measured as described elsewhere (40, 45). Mechanical properties and unbinding events were measured by acquiring point-by-point force-versus-distance curves over 32-by-32 arrays (force-volume). Further details have been described elsewhere (1, 4, 5, 9, 60). The sensitivity of the photodetector was calibrated by acquiring force-versus-distance curves on clean regions of mica. The tip velocities for the present measurements varied between 0.5 μm/s and 1 μm/s.
Custom MatLab (MathWorks, Natick, MA) routines were written for data analysis. To obtain elasticity maps, force-versus-distance curves were transformed into indentation curves using procedures described elsewhere (3-5, 9, 45). Briefly, the bacterial cell's deformation (penetration depth of the tip), δ, was obtained by subtracting the cantilever deflection from the displacement of the piezo. These values were plotted along the x axis of the indentation curve. Forces were calculated by multiplying the elastic constant of the cantilever by the cantilever deflection. Subsequently, the stiffness for each indentation curve was found by evaluating the derivative of the unloading force with respect to the penetration depth. For data points outside the analyzed bacteria, the stiffness exceeded threshold values, and consequently, a value of zero was assigned to the stiffness in these regions.
Force-extension curves for analysis of unbinding events were obtained using similar transformations to the piezo displacement and cantilever deflection as in indentation curves. Statistical analysis of unbinding events was performed by first identifying local minima in force-versus-distance curves and then evaluating unbinding forces as described previously (3, 9). When unbinding events in a force-extension curve were identified, each event of the curve was fitted to the extended freely jointed chain (m-FJC) model (32, 43, 45, 48-50, 62) to analyze the elasticity of the possible LOS molecule(s) being extended by the AFM tip. In the m-FJC model, the extension (z) of the polymer is related to the force (F) applied by the cantilever by z(F) = Lc{coth[FLk/(kBT)] − kBT/(FLk)}[1 + F/(Lkks)], where Lk is the Kuhn length, Lc is the contour length, coth is the hyperbolic cotangent, ks is the segment elasticity, T is the temperature, and kB is the Boltzmann constant. Lk and Lc were used as fitting parameters. The FJC model is obtained from the m-FJC model when ks is infinite. A value of ks = 2.2 N/m was found suitable for the analyzed force-extension curves after allowing it to vary between 1 and 100 N/m in several fitting attempts. Values of ks have been reported to vary greatly for different polysccharides, depending on the specific polysaccharide and solvent used (32). To discard events unlikely to originate from the extension of LOS molecules, only Kuhn and contour lengths with Lk > 0.09 nm and Lc > 10 nm were considered in the results summarized in the histograms. Events with a shorter Lk were more likely to be due to several molecules being stretched by the AFM tip, and events with a smaller Lc were too small to be considered LOS molecules.
RESULTS AND DISCUSSION
Imaging of biofilm growth.
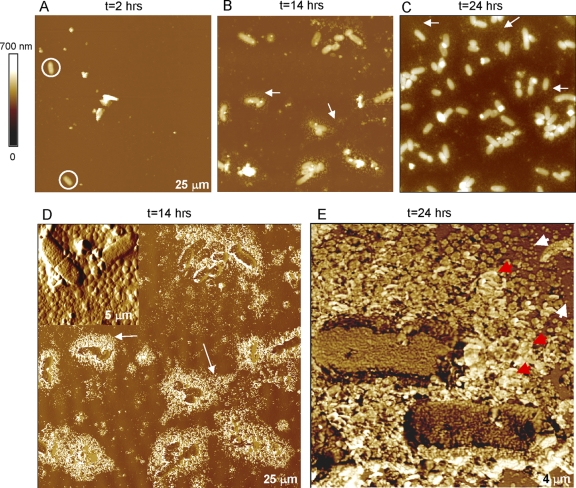
The formation of a stable, robust NTHI biofilm was observed using the rotating-disk reactor (Fig. 1). The coccobacillus-shaped H. influenzae bacteria were clearly visible in the AFM images after a few hours of incubation (Fig. 1A). The number of attached cells consistently and steadily increased with time (Fig. 1B to E). In contrast, biofilms grown without the shear applied in the rotating-disc biofilm reactor usually reorganized or detached during rinsing and thus produced inconsistent AFM images. These results are consistent with earlier reports that demonstrated biofilms were more strongly attached and were cohesively stronger when grown under shear stress (63, 70). As the number of attached bacteria increased, extracellular polymeric substances (EPS) were observed as halo-like features (Fig. 1B to D). These were principally seen in the vicinity of bacteria, thus supporting the bacterial origin of these features. Furthermore, the surface area occupied by EPS increased as the biofilm expanded on the surface (Fig. 1B and C).
FIG. 1.
(A to C) Twenty-five- by 25-μm2 height-tapping-mode AFM images of NTHI (PittGG strain) biofilms grown in a rotating-disk biofilm reactor acquired at different stages of growth. For clarity, white circles and arrows are used to emphasize bacteria and EPS, respectively. (D to E) Phase-tapping-mode AFM images provide more detail for the EPS features shown above. The amplitude-tapping mode AFM image shown in the inset of panel D and the phase image in panel E display granular features with diameters of ∼30 nm. Red arrows highlight the granular structure of EPS from isolated grains. All images were obtained in air.
AFM phase images showed distinctive phase contrast of EPS with respect to bacteria and substratum (Fig. 1D), indicating different viscoelastic properties of the EPS structure. Higher-magnification AFM images revealed a granular structure for the observed EPS (inset in Fig. 1D and E), in agreement with previous work on Pseudomonas putida and Pseudomonas aeruginosa biofilms (7, 12, 36). While the dimensions of the grains ranged from approximately 5 to 40 nm, as determined by their height, the measured widths were larger (approximately hundreds of nanometers) due to tip convolution. The granular structure of EPS is evident from the isolated grains (Fig. 1E), but as the grains grow, the EPS morphology becomes gradually amorphous and continuous. Figure 1E suggests a mechanism in which bacteria release EPS in small discrete amounts and form the more familiar amorphous continuous masses as grains coalesce. NTHI bacteria residing in biofilms are known to release copious amounts of EPS as demonstrated by scanning EM (SEM) imaging (26, 80). The EPS usually appeared as a large amorphous mass in the SEM studies, as mature biofilms were primarily investigated. However, similar granular features were observed in immature biofilms (80).
While the PittGG cells (17, 27, 37, 65) were ∼25% longer than the PittEE cells, PittEE cells were wider and higher by 14% and 19%, respectively (Table 1). This difference in size produces a 15% larger contact area between PittGG cells and a substratum such as host cells. This observation is of interest as the PittGG strain is far more invasive and virulent than the PittEE strain. PittGG was isolated from a patient with a spontaneously perforated tympanic membrane associated with otorrhea, whereas the PittEE strain was isolated from a patient with chronic OM with effusion at the time of myringotomy and tympanostomy and tube placement. Similarly, in the chinchilla model of OM, PittGG was universally associated with systemic spread and induced 100% mortality, but none of the PittEE-infected cohort showed infection beyond the tympanic bullae, and all survived.
TABLE 1.
Average sizes of PittGG and PittEE cells
| NTHI strain | Length (nm) | Width (nm) | Height (nm) | Area occupied (106 nm2)a | Length/width ratio | No. of bacteria |
|---|---|---|---|---|---|---|
| PittGG | 1,936 ± 858 | 757 ± 146 | 211 ± 58 | 1.47 ± 0.93 | 2.54 ± 0.86 | 20 |
| PittEE | 1,425 ± 265 | 881 ± 242 | 255 ± 67 | 1.26 ± 0.58 | 1.76 ± 0.39 | 12 |
Cross-sectional area occupied by a bacterium.
Characterization of biofilms under physiologically relevant conditions. (i) Morphology.
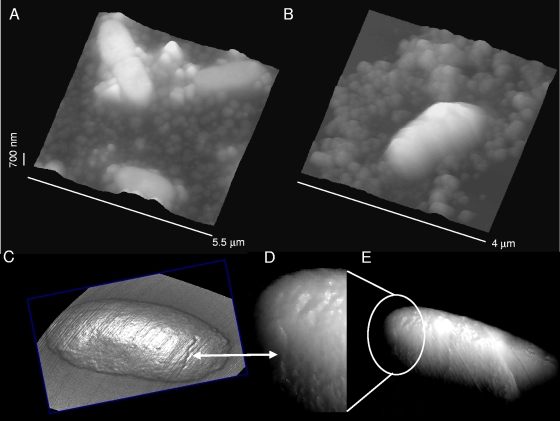
Biofilms of NTHI bacteria that had never been dehydrated were prepared on APTES-modified silicon surfaces and imaged in HEPES solutions (Fig. 2). For these biofilms, we observed similar EPS and bacterial morphological features to those of the air-dried samples. The coccobacillus shape of the bacteria and the granular structure of EPS seen in air (Fig. 2A) were also observed in aqueous solutions (Fig. 2B). Furthermore, cellular scar-like features likely representing the fission plate of the previous division cycle were found on individual bacteria imaged in air (Fig. 2C) or in liquid (Fig. 2D and E).
FIG. 2.
(A) Granular EPS features of NTHI biofilm cultures observed in air. (B) Similar results were obtained in HEPES buffer with samples that were not allowed to dry before images were acquired. Cellular scar-like features (marked with white arrows) seen in the body of single bacteria in air (C) were also observed in aqueous solutions (D and E), although not as clearly. The image in panel D shows a magnified view of the region marked in panel E to illustrate the scar-like feature with more clarity. Images in panels C and D are displayed in the mixed-height/illumination mode to highlight details.
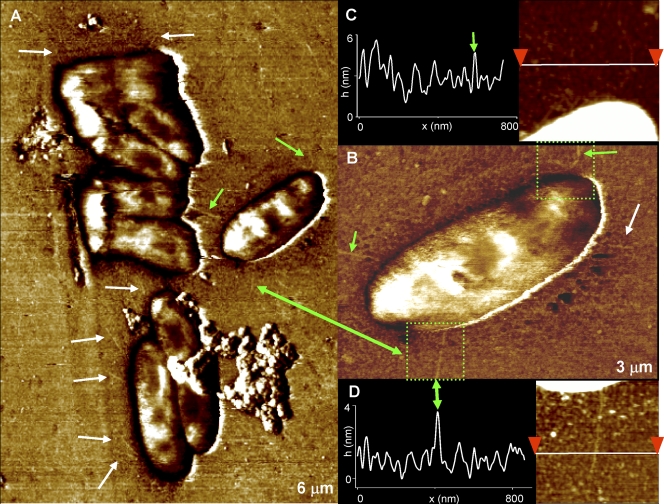
AFM images obtained in aqueous solutions showed (Fig. 3) smoother bacterial surfaces than those imaged in air (Fig. 1E). Significantly, bacterial appendages became apparent for biofilms formed from the PittGG strain. These appendages were faintly visible as filamentous structures (Fig. 3A) and were more clearly observed in phase-tapping-mode images (Fig. 3B) (31, 71), but also became visible in height contact mode (Fig. 3C and D). The appendages appeared to be partially embedded in EPS grains, which produced difficulties in visualizing them clearly (Fig. 3C and D). Their measured lengths were ∼500 to 1,000 nm, and their diameters were ∼5 nm.
FIG. 3.
(A) Filamentous structures (marked with arrows), thought to be Hif-type pili, were identified for the PittGG strain in phase-tapping-mode images. (B) A higher-magnification image of single bacteria displays some of those appendages with more clarity. Green arrows are used as an aid to locate the pili also found in panel A. (C and D) Height images (acquired in contact mode) and cross-sections acquired inside the regions marked by the green squares in panel B. The pilus protrudes only ∼2 nm above the underlying surface, and it appears to be partially buried inside EPS particles on the surface. Its width is ∼5 nm, after tip broadening is taken into account (79). All images were obtained in HEPES buffer.
These PittGG-specific structures are most likely pili and were not observed on the PittEE bacteria. Interestingly, the genomes of both PittGG and PittEE have been completely sequenced and they differ by the possession or absence of 339 orthologous gene clusters (37); however, the Hif locus, inserted between purE and pepN genes in the PittGG genome, is the only annotated locus that could encode such structures. The Hif-type pili are distinct from the larger type IV pili encoded by some NTHI strains associated with mating, DNA transfer, and twitching motility (10, 41) and have been widely associated with virulence (51); neither PittGG nor PittEE contains the genes that encode the type IV pili. Thus, this structural difference between the two strains combined with the supporting genomic data likely plays a role in the demonstrated increased levels of virulence displayed by PittGG compared to PittEE.
(ii) Elasticity of NTHI bacteria.
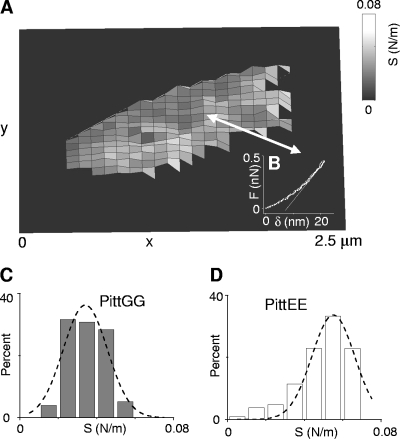
Elasticity maps (Fig. 4A) revealed stiffness (S) values of ∼0.01 to 0.05 N/m (Fig. 4 and Table 2) for the cell walls of NTHI bacteria in 100 mM HEPES buffer. Indentation curves on NTHI cells were generally reversible (Fig. 4B), thus showing an elastic response of the NTHI bacteria to the penetrating AFM tip up to the highest loads (∼0.5 nN) used in our experiments. In the few cases when plastic deformation occurred, this was indicated by a small hysteresis (<20 nm) in the indentation curves. The elastic deformation, δ, of the NTHI bacteria (i.e., indentation depth of the tip) was typically ∼20 to 30 nm for the maximum pressures exerted (∼0.1 to 1 to 1 MPa). For comparison, stiffness values were 3 to 4 orders of magnitude higher for bacteria analyzed in air, thus indicating their considerable hardening, likely due to dehydration.
FIG. 4.
(A) Representative stiffness map of an individual PittGG bacterium obtained from an array of 32 × 32 indentation curves. At each point of the array, the stiffness (S) was determined from the slope of the indentation curve (B) evaluated at a load of ∼0.5 nN. A similar measurement was performed on an individual PittEE cell using the same AFM cantilever cleaned after the first use. A total of n = 250 data points (105 for PittEE) of the stiffness map corresponded to the PittGG cell. Since stiffness values of the silicon substratum are several orders of magnitude larger than those for the bacterium, they were set to zero for figure clarity. (C and D) Histograms of stiffness values for the individual PittGG (C) and PittEE (D) cells described in panel A. Dashed lines are Gaussian fits to the distributions. Stiffness values refer to the center of the Gaussian curves ± their half-width at half-maximum. Values of 0.034 ± 0.014 N/m and 0.054 ± 0.013 N/m, respectively, were determined for these PittGG and PittEE cells. Stiffness values obtained for nine and eight different cells of the PittGG and PittEE strains, respectively, are presented in Table 2 and do not differ significantly from the values found for these cells. All data were acquired in 100 mM HEPES buffer.
TABLE 2.
Stiffness, unbinding forces, Kuhn lengths, and contour lengths for several NTHI cellsa
| NTHI strain | S (N/m) | Fu (nN) | Lk (nm) | Lc (nm) | No. of cells analyzed |
|---|---|---|---|---|---|
| PittGG | 0.035 ± 0.015 | 0.051 ± 0.013 | 0.2 ± 0.1 | 42.5 ± 17.2 | 9 |
| PittEE | 0.053 ± 0.017 | 0.054 ± 0.027 | 0.2 ± 0.1 | 36.8 ± 9.2 | 8 |
S, Fu, Lk, and Lc indicate stiffness, unbinding force, Kuhn length, and contour length, respectively, for the number of cells analyzed. Means ± standard errors of the mean are shown. Results include data collected for groups of bacterial cells in a single measurement. Three different AFM tips with a nominal spring constant (k = 0.01 N/m) were used.
Following previous work by Boulbitch et al. (6, 16), we modeled NTHI bacteria as a thin elastic cylindrical shell deformed locally by the AFM tip. To analyze the total force (F = Ft + Fp) acting against the AFM cantilever, two contributions, Ft and Fp, were taken into account (6). One of them, Ft, originates from the turgor pressure, p, acting against the cell wall, and the second, Fp, stems from the lateral rigidity of the cell wall provided primarily by the covalent bonds that bind the thin peptidoglycan layer (∼1 to 7 nm thick for gram-negative bacteria) (6, 19, 81). Since Fp ≪ Ft (6, 19), the primary contribution to the total force originates from the turgor pressure and this can be determined by the relation P = 2S/(3πRϕ), where R = (W/2, ∼400 nm) (Table 1) is the radius of the analyzed NTHI bacteria, W is the width, and ϕ = ϕ(ρ/d) is a geometric factor that depends on the ratio between ρ, the contact radius of the cantilever tip with the bacterial envelope, and d, the lateral cutoff distance from the tip at which the normal deformation of the membrane vanishes (6). Assuming the same value ϕ ∼ 0.2 for the geometric factor as in reference 6 and using S given in Table 2, turgor pressures of 0.093 ± 0.040 MPa and 0.141 ± 0.045 MPa are found for the PittGG and PittEE strains, respectively (Fig. 4). This correlates reasonably well with published values, considering that turgor pressures commonly encountered in gram-negative bacteria range from 0.08 to 0.5 MPa (81) and that these can be up to 1 order of magnitude smaller in medium than in distilled water (82).
(iii) Elasticity of LOS.
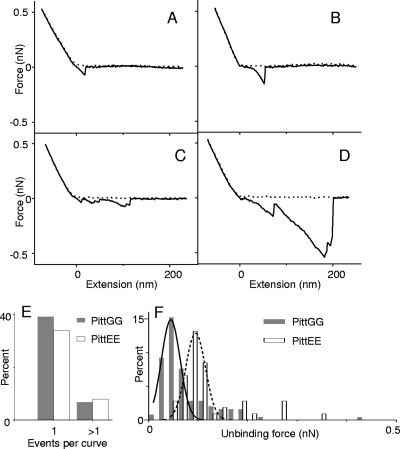
We employed force-extension curves to characterize the extension of polymers on the surface of NTHI cells as a result of the force measured during cantilever retraction from the cell's surface. These curves are marked by characteristic unbinding or rupture events between polymers and the AFM tip. Figure 5A to D display different examples of the force-extension curves obtained on the surface of bacterial cells. Approximately 46% and 42% of force-extension curves on cells of PittGG and PittEE strains, respectively, exhibited unbinding events. In both cases, the majority of events originated from a single event per curve (Fig. 5E). The percentage of events per curve was slightly lower for PittEE cells, but unbinding force values were in general comparable for both strains (Fig. 5F and Table 2). These unbinding forces are lower than the nN range forces reported for Myxococcus xanthus bacteria and diatoms (3, 55). Similarly, the polymer extensions discussed below are also lower than the μm range extensions reported in those studies.
FIG. 5.
(A to D) Examples of force-extension curves acquired on surfaces of NTHI bacteria in HEPES buffer. The dashed lines are approach curves as the AFM tip moves toward a bacterial cell, and the solid lines correspond to retraction curves as the tip moves back to its original position. The zero position in the x axis corresponds approximately to the point of contact between the tip and the cell's surface. The unbinding events, marked by peaks in the retraction curves, are attributed to LOS and extracellular polymers on the cell surface being extended by the tip. (E) Percentage of curves that displayed single or multiple unbinding events acquired on two individual PittGG and PittEE cells. Forty-six percent (42%) of the total number of curves acquired on the PittGG (PittEE) cell had events. Single events were observed more frequently than multiple unbinding events in the curves. Data for both strains were collected using the same AFM tip, which was cleaned with organic solvents (acetone and methanol) and in an ozone chamber after the first use. (F) Histograms for the distribution of unbinding forces between the AFM tip and cell surface polymers for the same NTHI cells as in panel E. For these individual cells, larger unbinding forces were obtained for PittEE (0.091 ± 0.043 nN) than for PittGG (0.049 ± 0.042 nN). However, after results for several cells of each strain were analyzed, values of ∼0.05 nN were found for both strains (Table 2).
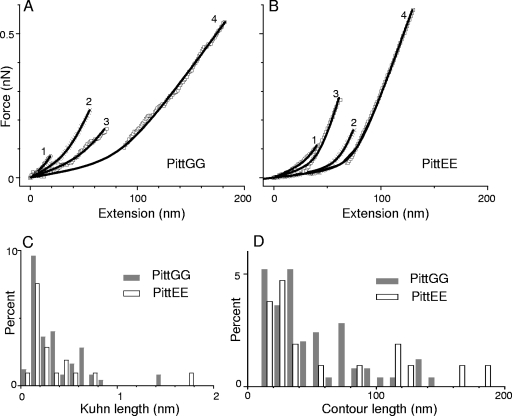
To examine whether these events could possibly originate from stretching of single LOS or EPS molecules, we fitted each unbinding event to the m-FJC model and restricted our analysis to those events obtained on the surface of bacterial cells (Fig. 6A and B). The Kuhn length distributions (Fig. 6C and Table 2) are similar for both strains, and their global maxima lie between 0.1 nm and 0.2 nm. The contour length distributions reveal a larger percentage of events with lengths in the 20- to 60-nm range (Fig. 6D and Table 2), which suggests a larger population of polymers with short chains. Because of their short saccharide chains, the lipopolysaccharides (LPS) of H. influenzae are often referred to as LOS (28) and accordingly, the smaller and more frequently observed single events are tentatively attributed to LOS, while the events with larger extensions and unbinding forces (Fig. 5C and D) are attributed to extracellular polymers. In spite of the restrictions set by the values of Kuhn and contour lengths, both distributions may still be influenced by events, in which the LOS/EPS polymers are only partially elongated, thus making it difficult to attribute all events with small contour lengths entirely to LOS chains. Significantly, TEM images of samples prepared by the freeze-substitution technique for another gram-negative opportunistic pathogen, Pseudomonas aeruginosa (PAO1), have shown that the O-side chains of B-band LPS can extend up to 40 nm from the outer membrane (14). The reasonable agreement between these values and those observed in the contour length distributions; suggest that, at least in some cases, single LOS molecules were getting pulled by the AFM tip.
FIG. 6.
(A and B) Analysis of four possible single-molecule events found in force-extension curves acquired on surfaces of PittGG (A) and PittEE (B) cells in HEPES buffer. The plotted curves are sections of force-extension curves similar to those shown in Fig. 5A to D. Curves 3 and 4 for each strain correspond to different unbinding events observed in the same original force-extension curve, while curves 1 and 2 originate from different force-extension curves with single events. Data points (symbols) were fitted to curves (solid lines) generated using the m-FJC model. The Kuhn length (Lk) and contour length (Lc) for the LOS molecules extended by the AFM tip were determined from this analysis. These values are tabulated in Table 3. (C and D) Statistical distributions for all Kuhn and contour lengths of possible single LOS molecule unbinding events found in the curves collected on individual NTHI cells. A total of 64 (18 for PittEE) possible events were found on the PittGG cell. Data were acquired with the same AFM tip for both strains, as described in the legend to Fig. 5. Results obtained with nine (eight for PittEE) different PittGG cells (Table 2) did not differ significantly from the values obtained from these distributions.
Conclusions.
We have applied AFM to study structural details under physiologically relevant conditions at resolutions comparable to those of EM of two different NTHI clinical isolates with vastly different pathogenicity profiles. Using tapping-mode phase imaging, we visualized appendages believed to be Hif-type pili in the PittGG strain and correlated their presence with the corresponding genes and higher pathogenicity associated with this strain compared with the PittEE strain (17).
By monitoring biofilm formation at its early stages, we observed the formation of granular structures of extracellular polymers, followed by their subsequent aggregation to form larger amorphous masses. AFM-based single-molecule spectroscopy allowed the characterization of NTHI LOS through force-extension curves. The contour length distributions correlated with the expected lengths of LPS observed with TEM for PAO1 bacteria (14). Finally, by measuring the stiffness of single bacterial cells, we determined the turgor pressure of NTHI bacteria and found slightly larger values for the PittEE strain.
TABLE 3.
| Curve no. | PittGG
|
PittEE
|
||
|---|---|---|---|---|
| Lk (nm) | Lc (nm) | Lk (nm) | Lc (nm) | |
| 1 | 0.15 | 21 | 0.17 | 37 |
| 2 | 0.14 | 34 | 0.35 | 65 |
| 3 | 0.14 | 51 | 0.25 | 42 |
| 4 | 0.2 | 82 | 0.36 | 76 |
Lk and Lc indicate the Kuhn length and contour length for the force extension curves and their corresponding m-FJC fits displayed in Fig. 6.
Acknowledgments
We thank Robert Wadowsky for the original strain isolations. We acknowledge John Dutcher (University of Guelph, Canada), Iwona Beech (University of Portsmouth, United Kingdom), and Srinivasan Ramachandran (University of Chicago) for helpful discussions.
This work was supported by Allegheny General Hospital and Allegheny Singer Research Institute and grants from the Health Resources and Services Administration and the NIH-NIDCD: DC02148 and DC04173 (G.D.E.). We also acknowledge partial support by NASA-EPSCOR under grant NCC5-579.
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Almqvist, N., R. Bhatia, G. Primbs, N. Desai, S. Banerjee, and R. Lal. 2004. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys. J. 861753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre, G., K. Leenhouts, P. Hols, and Y. F. Dufrene. 2008. Detection and localization of single LysM-peptidoglycan interactions. J. Bacteriol. 1907079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arce, F. T., R. Avci, I. B. Beech, K. E. Cooksey, and B. Wigglesworth-Cooksey. 2004. A live bioprobe for studying diatom-surface interactions. Biophys. J. 874284-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arce, F. T., R. Avci, I. B. Beech, K. E. Cooksey, and B. Wigglesworth-Cooksey. 2006. Modification of surface properties of a poly(dimethylsiloxane)-based elastomer, RTV11, upon exposure to seawater. Langmuir 227217-7225. [DOI] [PubMed] [Google Scholar]
- 5.Arce, F. T., J. L. Whitlock, A. A. Birukova, K. G. Birukov, M. F. Arnsdorf, R. Lal, J. G. N. Garcia, and S. M. Dudek. 2008. Regulation of the micromechanical properties of pulmonary endothelium by S1P and thrombin: role of cortactin. Biophys. J. 95886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnoldi, M., M. Fritz, E. Bauerlein, M. Radmacher, E. Sackmann, and A. Boulbitch. 2000. Bacterial turgor pressure can be measured by atomic force microscopy. Physiol. Rev. E 621034-1044. [DOI] [PubMed] [Google Scholar]
- 7.Auerbach, I. D., C. Sorensen, H. G. Hansma, and P. A. Holden. 2000. Physical morphology and surface properties of unsaturated Pseudomonas putida biofilms. J. Bacteriol. 1823809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ausubel, F. M. 1992. Short protocols in molecular biology. Greene Publishing Associates, New York, NY.
- 9.Avci, R., M. Schweitzer, R. D. Boyd, J. Wittmeyer, A. Steele, J. Toporski, W. Beech, F. T. Arce, B. Spangler, K. M. Cole, and D. S. McKay. 2004. Comparison of antibody-antigen interactions on collagen measured by conventional immunological techniques and atomic force microscopy. Langmuir 2011053-11063. [DOI] [PubMed] [Google Scholar]
- 10.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 731635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer, S. H. J., M. Mansson, D. W. Hood, J. C. Richards, E. R. Moxon, and E. K. H. Schweda. 2001. A rapid and sensitive procedure for determination of 5-N-acetyl neuraminic acid in lipopolysaccharides of Haemophilus influenzae: a survey of 24 non-typeable H. influenzae strains. Carbohydr. Res. 335251-260. [DOI] [PubMed] [Google Scholar]
- 12.Beech, I. B., J. R. Smith, A. A. Steele, I. Penegar, and S. A. Campbell. 2002. The use of atomic force microscopy for studying interactions of bacterial biofilms with surfaces. Colloids Surf. B 23231-247. [Google Scholar]
- 13.Beech, I. B., J. A. Sunner, and K. Hiraoka. 2005. Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 8157-168. [PubMed] [Google Scholar]
- 14.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1814725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchet, V., D. W. Hood, J. Li, J.-R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. H. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 1008898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulbitch, A. A. 1998. Deflection of a cell membrane under application of a local force. Physiol. Rev. E 572123-2128. [Google Scholar]
- 17.Buchinsky, F. J., M. L. Forbes, J. D. Hayes, K. Shen, S. Ezzo, J. Compliment, J. Hogg, N. L. Hiller, F. Z. Hu, J. C. Post, and G. D. Ehrlich. 2007. Virulence phenotypes of low-passage clinical isolates of nontypeable Haemophilus influenzae assessed using the Chinchilla laniger model of otitis media. BMC Microbiol. 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butt, H. J., B. Cappella, and M. Kappl. 2005. Force measurements with the atomic force microscope: technique, interpretation and applications. Surf. Sci. Rep. 591-152. [Google Scholar]
- 19.Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3601-610. [DOI] [PubMed] [Google Scholar]
- 20.Clemans, D. L., C. F. Marrs, R. J. Bauer, M. Patel, and J. R. Gilsdorf. 2001. Analysis of pilus adhesins from Haemophilus influenzae biotype IV strains. Infect. Immun. 697010-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 22.Dufrene, Y. F. 2004. Using nanotechniques to explore microbial surfaces. Nat. Rev. Microbiol. 2451-460. [DOI] [PubMed] [Google Scholar]
- 23.Dugdale, T. M., R. Dagastine, A. Chiovitti, P. Mulvaney, and R. Wetherbee. 2005. Single adhesive nanofibers from a live diatom have the signature fingerprint of modular proteins. Biophys. J. 894252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dugdale, T. M., R. Dagastine, A. Chiovitti, and R. Wetherbee. 2006. Diatom adhesive mucilage contains distinct supramolecular assemblies of a single modular protein. Biophys. J. 902987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupres, V., F. D. Menozzi, C. Locht, B. H. Clare, N. L. Abbott, S. Cuenot, C. Bompard, D. Raze, and Y. F. Dufrene. 2005. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2515-520. [DOI] [PubMed] [Google Scholar]
- 26.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2871710-1715. [DOI] [PubMed] [Google Scholar]
- 27.Erdos, G., S. Sayeed, P. Antalis, F. Z. Hu, J. Hayes, J. Goodwin, R. Dopico, J. C. Post, and G. D. Ehrlich. 2003. Development and characterization of a pooled Haemophilus influenzae genomic library for the evaluation of gene expression changes associated with mucosal biofilm formation in otitis media. Int. J. Pediatr. Otorhinolaryngol. 67749-755. [DOI] [PubMed] [Google Scholar]
- 28.Erwin, A. L., and A. L. Smith. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 15355-362. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez, J. M. 2005. Fingerprinting single molecules in vivo. Biophys. J. 893676-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francius, G., S. Lebeer, D. Alsteens, L. Wildling, H. J. Gruber, P. Hols, S. De Keersmaecker, J. Vanderleyden, and Y. F. Dufrene. 2008. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano 21921-1929. [DOI] [PubMed] [Google Scholar]
- 31.Garcia, R., R. Magerle, and R. Perez. 2007. Nanoscale compositional mapping with gentle forces. Nat. Mater. 6405-411. [DOI] [PubMed] [Google Scholar]
- 32.Gianotti, M. I., and G. J. Vancso. 2007. Interrogation of single synthetic polymer chains and polysaccharides by AFM-based force spectroscopy. Chemphyschem 92290-2307. [DOI] [PubMed] [Google Scholar]
- 33.Gilsdorf, J. R., K. W. McCrea, and C. F. Marrs. 1997. Role of pili in Haemophilus influenzae adherence and colonization. Infect. Immun. 652997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunning, A. P., S. Chambers, C. Pin, A. L. Man, V. J. Morris, and C. Nicoletti. 2008. Mapping specific adhesive interactions on living human intestinal epithelial cells with atomic force microscopy. FASEB J. 222331-2339. [DOI] [PubMed] [Google Scholar]
- 35.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansma, H. G., L. I. Pietrasanta, I. D. Auerbach, C. Sorenson, R. Golan, and P. A. Holden. 2000. Probing biopolymers with the atomic force microscope: a review. J. Biomater. Sci. Polym. E 11675-683. [DOI] [PubMed] [Google Scholar]
- 37.Hogg, J. S., F. Z. Hu, B. Janto, R. Boissy, J. Hayes, R. Keefe, J. C. Post, and G. D. Ehrlich. 2007. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 8R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hood, D. W., A. D. Cox, M. Gilbert, K. Makepeace, S. Walsh, M. E. Deadman, A. Cody, A. Martin, M. Mansson, E. K. H. Schweda, J.-R. Brisson, J. C. Richards, E. R. Moxon, and W. Wakarchuk. 2001. Identification of a lipopolysaccharide a-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39341-350. [DOI] [PubMed] [Google Scholar]
- 39.Hood, D. W., K. Makepeace, M. E. Deadman, R. F. Rest, P. Thibault, A. Martin, J. C. Richards, and E. R. Moxon. 1999. Sialic acid in the lipopolysaccharide of Haemophilus infuenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33679-692. [DOI] [PubMed] [Google Scholar]
- 40.Hutter, J. L., and I. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 641868-1873. [Google Scholar]
- 41.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 1893868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurcisek, J. A., J. E. Bookwalter, B. D. Baker, S. Fernandez, L. A. Novotny, R. S. Munson, and L. O. Bakaletz. 2007. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 651288-1299. [DOI] [PubMed] [Google Scholar]
- 43.Li, H. B., M. Rief, F. Oesterhelt, H. E. Gaub, X. Zhang, and J. Shen. 1999. Single-molecule force spectroscopy on polysaccharides by AFM—nanomechanical fingerprint of a-(1,4)-linked polysaccharides. Chem. Phys. Lett. 305197-201. [Google Scholar]
- 44.Li, J., A. D. Cox, D. W. Hood, E. K. H. Schweda, E. R. Moxon, and J. C. Richards. 2005. Electrophoretic and mass spectrometric strategies for profiling bacterial lipopolysaccharides. Mol. Biosyst. 146-52. [DOI] [PubMed] [Google Scholar]
- 45.Liu, F., F. T. Arce, S. Ramachandran, and R. Lal. 2006. Nanomechanics of hemichannel conformations. Connexin flexibility underlying channel opening and closing. J. Biol. Chem. 28123207-23217. [DOI] [PubMed] [Google Scholar]
- 46.Lower, S., M. F. Hochella, and T. J. Beveridge. 2001. Bacterial recognition of mineral surfaces: nanoscale interactions between Schewanella and α-FeOOH. Science 2921360-1363. [DOI] [PubMed] [Google Scholar]
- 47.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426306-310. [DOI] [PubMed] [Google Scholar]
- 48.Marszalek, P. E., H. B. Li, and J. M. Fernandez. 2001. Fingerprinting polysaccharides with single-molecule atomic force microscopy. Nat. Biotechnol. 19258-262. [DOI] [PubMed] [Google Scholar]
- 49.Marszalek, P. E., H. Lu, H. B. Li, M. Carrion-Vasquez, A. F. Oberhauser, K. Schulten, and J. M. Fernandez. 1999. Mechanical unfolding intermediates in titin modules. Nature 402100-103. [DOI] [PubMed] [Google Scholar]
- 50.Marszalek, P. E., A. F. Oberhauser, Y.-P. Pang, and J. Fernandez. 1998. Polysaccharide elasticity governed by chair-boat transitions of the glucopyranose ring. Nature 396661-664. [DOI] [PubMed] [Google Scholar]
- 51.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 1804693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu, X.-Q., E. H. Egelman, and E. Bullitt. 2002. Structure and function of Hib pili from Haemophilus influenzae type b. J. Bacteriol. 1844868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunez, M. E., M. O. Martin, P. H. Chan, L. K. Duong, A. R. Sindhurakar, and E. M. Spain. 2005. Atomic force microscopy of bacterial communities. Methods Enzymol. 397256-268. [DOI] [PubMed] [Google Scholar]
- 54.O'Neill, J., J. W. St. Geme III, D. Cutter, E. E. Adderson, J. Anyanwu, R. F. Jacobs, and G. E. Schutze. 2003. Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas. J. Clin. Microbiol. 413064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelling, A. E., Y. N. Li, W. Y. Shi, and J. K. Gimzewski. 2005. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc. Natl. Acad. Sci. USA 1026484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitts, B., M. A. Hamilton, G. A. McFeters, P. S. Stewart, A. Willse, and N. Zelver. 1998. Color measurement as a means of quantifying surface biofouling. J. Microbiol. Methods 34143-149. [DOI] [PubMed] [Google Scholar]
- 57.Plomp, M., T. J. Leighton, K. E. Wheeler, and A. J. Malkin. 2005. Architecture and high-resolution structure of Bacillus thuringiensis and Bacillus cereus spore coat surfaces. Langmuir 217892-7898. [DOI] [PubMed] [Google Scholar]
- 58.Plomp, M., T. J. Leighton, K. E. Wheeler, and A. J. Malkin. 2005. The high-resolution architecture and structural dynamics of Bacillus spores. Biophys. J. 88603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plomp, M., T. J. Leighton, K. E. Wheeler, M. E. Pitesky, and A. J. Malkin. 2005. Bacillus atrophaeus outer spore coat assembly and ultrastructure. Langmuir 2110710-10716. [DOI] [PubMed] [Google Scholar]
- 60.Quist, A. P., S. K. Rhee, H. Lin, and R. Lal. 2000. Physiological role of gap-junctional hemichannels: extracellular calcium-dependent isosmotic volume regulation. J. Cell Biol. 1481063-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Razatos, A., Y.-L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA 9511059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rief, M., F. Oesterhelt, B. Heymann, and H. E. Gaub. 1997. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 2751295-1297. [DOI] [PubMed] [Google Scholar]
- 63.Rupp, C. J., C. A. Fux, and P. Stoodley. 2005. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl. Environ. Microbiol. 712175-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satola, S. W., P. L. Schirmer, and M. M. Farley. 2003. Genetic analysis of the capsule locus of Haemophilus influenzae serotype f. Infect. Immun. 717202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen, K., P. Antalis, J. Gladitz, S. Sayeed, A. Ahmed, S. Yu, J. Hayes, S. Johnson, B. Dice, R. Dopico, R. Keefe, B. Janto, W. Chong, J. Goodwin, R. M. Wadowsky, G. Erdos, J. C. Post, G. D. Ehrlich, and F. Z. Hu. 2005. Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect. Immun. 733479-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sill, M. L., D. K. S. Law, J. W. Zhou, S. Skinner, J. Wylie, and R. S. W. Tsang. 2007. Population genetics and antibiotic susceptibility of invasive Haemophilus influenzae in Manitoba, Canada, from 2000 to 2006. FEMS Immunol. Med. Microbiol. 51270-276. [DOI] [PubMed] [Google Scholar]
- 67.Sill, M. L., and R. S. W. Tsang. 2008. Antibiotic susceptibility of invasive Haemophilus influenzae strains in Canada. Antimicrob. Agents Chemother. 521551-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.St. Geme, J. W. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell. Microbiol. 4191-200. [DOI] [PubMed] [Google Scholar]
- 69.St. Geme, J. W., J. S. Pinkner, G. P. Krasan, J. Heuser, E. Bullitt, A. L. Smith, and S. J. Hultgren. 1996. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc. Natl. Acad. Sci. USA 9311913-11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoodley, P., R. Cargo, C. J. Rupp, S. Wilson, and I. Klapper. 2002. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 29361-367. [DOI] [PubMed] [Google Scholar]
- 71.Suo, Z. Y., X. H. Yang, R. Avci, L. Kellerman, D. W. Pascual, M. Fries, and A. Steele. 2007. HEPES-stabilized encapsulation of Salmonella typhimurium. Langmuir 231365-1374. [DOI] [PubMed] [Google Scholar]
- 72.Touhami, A., M. H. Jericho, J. M. Boyd, and T. J. Beveridge. 2006. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Touhami, A., B. Nysten, and Y. F. Dufrene. 2003. Nanoscale mapping of the elasticity of microbial cells by atomic force microscopy. Langmuir 194539-4543. [Google Scholar]
- 74.Tsang, R. 2008. Changing epidemiology of invasive Haemophilus influenzae disease. Lancet Infect. Dis. 8737. [DOI] [PubMed] [Google Scholar]
- 75.Tsang, R. S. W. 2008. Serotyping and population genetics of invasive Haemophilus influenzae. J. Clin. Microbiol. 461159. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ubbink, J., and P. Schar-Zammaretti. 2005. Probing bacterial interactions: integrated approaches combining atomic force microscopy, electron microscopy and biophysical techniques. Micron 36293-320. [DOI] [PubMed] [Google Scholar]
- 77.Van der Mei, H. C., H. J. Busscher, R. Bos, J. de Vries, C. J. P. Boonaert, and Y. F. Dufrene. 2000. Direct probing by atomic force microscopy of the cell surface softness of a fibrillated and nonfibrillated oral streptococcal strain. Biophys. J. 782668-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velegol, S. B., and B. E. Logan. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir 185256-5262. [Google Scholar]
- 79.Vesenka, J., S. Manne, R. Giberson, T. Marsch, and E. Henderson. 1993. Colloidal gold particles as an incompressible atomic-force microscope imaging standard for assessing the compressibility of biomolecules. Biophysiol. J. 65992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Webster, P., S. Wu, G. Gomez, M. Apicella, A. G. Plaut, and J. W. S. Geme. 2006. Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J. Histochem. Cytochem. 54829-842. [DOI] [PubMed] [Google Scholar]
- 81.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 82.Yao, X., J. Walter, S. Burke, S. Stewart, M. H. Jericho, D. Pink, R. Hunter, and T. J. Beveridge. 2002. Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloid Surf. B 23213-230. [Google Scholar]
- 83.Zhao, L. M., D. Schaefer, H. X. Xu, S. J. Modi, W. R. LaCourse, and M. R. Marten. 2005. Elastic properties of the cell wall of Aspergillus nidulans studied with atomic force microscopy. Biotechnol. Prog. 21292-299. [DOI] [PubMed] [Google Scholar]