Abstract
Myxococcus xanthus is a gram-negative soil bacterium that undergoes multicellular development upon nutrient limitation. Intercellular signals control cell movements and regulate gene expression during the developmental process. C-signal is a short-range signal essential for aggregation and sporulation. C-signaling regulates the fmgA gene by a novel mechanism involving cooperative binding of the response regulator FruA and the transcription factor/antitoxin MrpC2. Here, we demonstrate that regulation of the C-signal-dependent fmgBC operon is under similar combinatorial control by FruA and MrpC2, but the arrangement of binding sites is different than in the fmgA promoter region. MrpC2 was shown to bind to a crucial cis-regulatory sequence in the fmgBC promoter region. FruA was required for MrpC and/or MrpC2 to associate with the fmgBC promoter region in vivo, and expression of an fmgB-lacZ fusion was abolished in a fruA mutant. Recombinant FruA was shown to bind to an essential regulatory sequence located slightly downstream of the MrpC2-binding site in the fmgBC promoter region. Full-length FruA, but not its C-terminal DNA-binding domain, enhanced the formation of complexes with fmgBC promoter region DNA, when combined with MrpC2. This effect was nearly abolished with fmgBC DNA fragments having a mutation in either the MrpC2- or FruA-binding site, indicating that binding of both proteins to DNA is important for enhancement of complex formation. These results are similar to those observed for fmgA, where FruA and MrpC2 bind cooperatively upstream of the promoter, except that in the fmgA promoter region the FruA-binding site is located slightly upstream of the MrpC2-binding site. Cooperative binding of FruA and MrpC2 appears to be a conserved mechanism of gene regulation that allows a flexible arrangement of binding sites and coordinates multiple signaling pathways.
Myxococcus xanthus is a rod-shaped bacterium that glides on solid surfaces, forming a single-species biofilm that provides an attractive model to study how signaling couples gene expression to environmental and cellular cues (64). M. xanthus cells in the biofilm grow and divide when nutrients are available but, upon starvation, a multicellular developmental process ensues, during which the cells move into aggregates and form mound-shaped structures called fruiting bodies. Approximately 105 cells participate in forming a fruiting body, in which a portion of the cells differentiate into dormant, stress-resistant, spherical spores. Other cells undergo programmed cell death (37) or autolysis caused by siblings in the developing biofilm (41, 66), and some cells remain outside of fruiting bodies as peripheral rods (40). These fates are met by different proportions of cells in the biofilm, depending on genetic and environmental factors (3, 41). The spores in a fruiting body can germinate and resume growth and division when nutrients become available.
Signals act at different times during the developmental process to coordinate cell behavior and determine cell fate. Nutrient limitation causes a stringent response that results in production of (p)ppGpp and the induction of early developmental genes (12). A mixture of amino acids and peptides, known as the A-signal, is generated by secreted proteases and is believed to allow quorum sensing (27). A-signal-dependent genes are expressed, and cells alter their pattern of movement so that aggregates begin to form. Subsequent gene expression, and the maturation of aggregates into spore-filled fruiting bodies, depends on C-signaling, which is mediated by the product of the csgA gene (51). CsgA is associated with the outer membrane of the cell, where it is processed by a secreted protease to a 17-kDa form that appears to act as a short-range signal (21, 30, 45). C-signal transduction requires cell alignment (19) and possibly end-to-end contact between cells (46), so it communicates positional information. Cells become aligned as aggregates transform into nascent fruiting bodies, and the resulting high level of C-signaling has been proposed to trigger the expression of genes required for sporulation (48). Indeed, the expression of C-signal-dependent genes that are important for sporulation is restricted to nascent fruiting bodies (16, 47), and many studies support a model in which an increasing level of C-signaling controls gene expression to coordinate aggregation and sporulation during development (10, 20, 26, 29).
How does C-signaling regulate expression of target genes? FruA plays a key role in the C-signal transduction pathway (5, 42). It is similar to response regulators of two-component signal transduction systems and has been proposed to be phosphorylated in its N-terminal regulatory domain in response to C-signal and perhaps other signals (5, 15), but a cognate histidine protein kinase has not been identified, and several types of evidence suggest that FruA might function without being phosphorylated (35). The C-terminal domain of FruA is similar to the C-terminal DNA-binding domain of the NarL/FixJ subfamily of response regulators (63). The C-terminal domain of FruA has been shown to bind to sites in the promoter regions of developmentally regulated genes that fail to be expressed in fruA mutant cells, suggesting that FruA is a transcriptional activator (56, 57, 62, 68). Recently, FruA was shown to bind cooperatively with MrpC2 to the promoter region of the C-signal-dependent fmgA (for FruA- and MrpC2-regulated gene A) gene (35), revealing a novel mechanism of combinatorial control, as cooperative binding of a response regulator (FruA) and a distinct transcription factor (MrpC2) had not been observed previously.
MrpC2 is a smaller form of MrpC (58), which is similar to the cyclic AMP receptor protein (CRP) family of transcriptional regulators (54). MrpC is expressed during vegetative growth and is phosphorylated by a cytoplasmic serine/threonine protein kinase (STPK) called Pkn14 (38, 39). Pkn14 is in turn phosphorylated by a membrane STPK called Pkn8. Phosphorylation of MrpC by the Pkn8/Pkn14 cascade results in weaker binding of MrpC to DNA and also appears to inhibit proteolytic cleavage of MrpC to MrpC2 (39), which lacks the 25 N-terminal residues of MrpC (58). The STPK cascade is counteracted by an unknown mechanism early in development, allowing MrpC and MrpC2 concentrations to rise. MrpC2 binds to DNA with higher affinity than MrpC, and appears to play a key role as a transcriptional activator during development (39). Recently, MrpC was shown to function as an antitoxin by interacting directly with the toxin MazF, an mRNA interferase that mediates programmed cell death during development (37). MrpC also binds to the mazF promoter region and activates expression. Binding of MrpC2 to the mazF promoter region and MazF has not been tested. The dual functions of MrpC, and possibly MrpC2, as an antitoxin and a transcription factor make it an important determinant of cell fate. The finding that MrpC2 and FruA bind cooperatively to crucial cis-regulatory sequences upstream of the fmgA promoter suggests that these transcription factors coordinate starvation signaling and cell death with positional information via short-range C-signaling to govern gene expression and cell fate during M. xanthus development (35). This novel mechanism of fmgA combinatorial control was predicted to be conserved because similar cis-regulatory sequences have been found upstream of other developmentally regulated M. xanthus promoters (6, 31, 52, 59-61, 67).
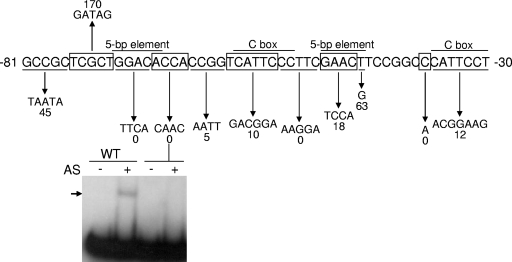
The promoter region of a putative operon (named here fmgBC for FruA- and MrpC2-regulated genes B and C) at the Ω4499 locus in the M. xanthus chromosome has cis-regulatory sequences similar to those bound by MrpC2 in the fmgA promoter region. The fmgBC operon was identified by an insertion of the transposon Tn5 lac into fmgC (25). FmgB and FmgC are similar to reductase and oxidase components, respectively, of bacterial cytochrome P-450 systems, which typically are involved in catabolism or anabolism of unusual compounds (6). M. xanthus DNA upstream of fmgBC was cloned, a putative transcriptional start site was mapped, and the region from positions −100 to +50 was shown to encompass the promoter (6, 67). Expression from the fmgBC promoter was reduced in a csgA mutant but was restored upon codevelopment of the csgA mutant with wild-type cells, which supply C-signal, demonstrating that promoter activity is partially dependent on C-signaling (6, 24). Mutational analysis identified critical cis-regulatory sequences at positions −71 to −45 upstream of the promoter (67). This region contains two C boxes (consensus CAYYCCY; Y means C or T) and two 5-bp elements (consensus GAACA) (Fig. 1), which are sequence motifs found in the promoter regions of several developmentally regulated genes (6, 31, 52, 59-61, 67). In the fmgA promoter region, between positions −63 and −46, a 5-bp element is located 6 bp upstream of a C box, and this region is bound by MrpC2, while FruA binds cooperatively to a site located slightly upstream (35).
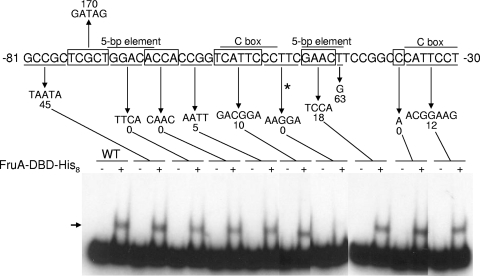
FIG. 1.
Effects of mutations on fmgBC promoter activity in vivo and on DNA binding in vitro. The top part of the figure shows a summary of mutational effects on developmental fmgB-lacZ expression (67). The wild-type fmgBC upstream sequence is alternately boxed or underlined to indicate changed sequences, which are shown below the downward arrows. The number beneath each mutant sequence indicates the maximum β-galactosidase activity during development, expressed as a percentage of the maximum activity observed for the wild-type promoter. The bottom part shows EMSAs performed with 32P-labeled fmgBC DNA (12 nM) spanning from positions −104 to −29 and proteins in the AS fraction (0.7 μg/μl). The arrow indicates the shifted complex produced by incubating the wild-type (WT) DNA fragment with the AS fraction. No complex was observed with a DNA fragment bearing the indicated mutation at positions −67 to −64.
Here, we report that MrpC2 and FruA bind to sequences between positions −71 and −45 upstream of the fmgBC promoter, but the arrangement of binding sites is the reverse of that found in the fmgA promoter region. Nevertheless, the association of MrpC and/or MrpC2 with the fmgBC promoter region in vivo required FruA. Furthermore, there appeared to be cooperative binding of MrpC2 and FruA to fmgBC promoter region DNA in vitro. Our results demonstrate combinatorial control by MrpC2 and FruA at a second promoter and reveal surprising flexibility in the arrangement of the binding sites.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in the present study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm with DE3, a λ prophage carrying the T7 RNA polymerase gene | Novagen |
| SMhisMrpC2 | BL21(DE3) containing pET16b/His10-MrpC2 | 35 |
| SMFruAhis | BL21(DE3) containing pET11km/FruA-His6 | 35 |
| EDYFruA | BL21(DE3) containing pET11a/FDBD-H8 | 68 |
| M. xanthus | ||
| DK1622 | Wild type | 17 |
| DK4499 | Tn5 lac Ω4499 | 25 |
| MSM1727.DZF1 | sglA1 attB::pREG1727 | This study |
| MDY1727.FA | sglA1 fruA::TnV Ω786 attB::pREG1727 | 68 |
| MSM4499.DZF1 | sglA1 attB::pDY52 | This study |
| MSM4499.FA | sglA1 fruA::TnV Ω786 attB::pDY52 | This study |
| Plasmids | ||
| pET11a/FDBD-H8 | pET11a with a gene encoding FruA-DBD-His8 under the control of a T7 RNA polymerase promoter | 57 |
| pET16b/His10-MrpC2 | pET16b with a gene encoding His10-MrpC2 under the control of a T7 RNA polymerase promoter | 39 |
| pET11km/FruA-His6 | pET11km with a gene encoding FruA-His6 under the control of a T7 RNA polymerase promoter | S. Inouye |
| pREG1727 | Apr Kmr P1-inc attP 'lacZ | 7 |
| pDY52 | pREG1727 with 150-bp XhoI-BamHI fragment from pDY51 | 67 |
| pDY100 | pGEM7Zf with fmgBC DNA from positions −218 to +50 | 67 |
| pDY133 | pDY100 with a GCCGC-to-TAATA mutation at positions −81 to −77 | 67 |
| pDY129 | pDY100 with a GGAC-to-TTCA mutation at positions −71 to −68 | 67 |
| pDY127 | pDY100 with a ACCA-to-CAAC mutation at positions −67 to −64 | 67 |
| pDY125 | pDY100 with a CCGG-to-AATT mutation at positions −63 to −60 | 67 |
| pDY49 | pDY100 with a TCATTC-to-GACGGA mutation at positions −59 to −54 | 67 |
| pDY121 | pDY100 with a CCTTC-to-AAGGA mutation at positions −53 to −49 | 67 |
| pDY47 | pDY100 with a GAAC-to-TCCA mutation at positions −48 to −45 | 67 |
| pDY117 | pDY100 with a C-to-A mutation at position −37 | 67 |
| pDY45 | pDY100 with a CATTCCT-to-ACGGAAG mutation at positions −36 to 30 | 67 |
Growth and development.
Escherichia coli BL21(DE3) containing plasmids were grown at 37°C in Luria-Bertani (LB) medium (49) containing 200 μg of ampicillin per ml. M. xanthus strains were grown at 32°C in CTT (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4, [final pH 7.6]) medium (14) or on CTT agar (1.5%) plates. When required, 40 μg of kanamycin sulfate per ml was added. Fruiting body development was performed on 1.5% TPM agar plates (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4, [final pH 7.6]) as described previously (25).
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing pREG1727 or its derivatives integrated at the Mx8 phage attachment site, attB, were constructed by electroporation (18) of M. xanthus, and transformants were selected on CTT agar plates containing kanamycin sulfate. Transformants were screened on TPM agar plates containing 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml in order to avoid rare transformants with unusual developmental lacZ expression (60). Three transformants were chosen for further analysis, and the β-galactosidase activity was measured as described previously (25).
Preparation of DNA fragments.
DNA fragments spanning the fmgBC promoter region from positions −104 to −29 were generated by PCR using wild-type or mutant plasmid (Table 1) as the template and the oligonucleotide primers 5′-GCGCGAGGAGATTGCGTTCATAC-3′ (for −104) and 5′-GAGGAATGGGCCGGAAGTTC-3′ (for −29). For the electrophoretic mobility shift assays (EMSAs), 32P-labeled DNA was synthesized by PCR after labeling the primers with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) and the DNA fragment was purified after 15% polyacrylamide gel electrophoresis (49).
EMSAs.
EMSAs were performed as described previously (68), except that binding reaction mixtures were incubated at 25°C for 15 min.
DNA-affinity chromatography.
An fmgBC DNA fragment (positions −104 to −29) was synthesized by PCR with a 5′-biotin label at position −104, bound to streptavidin beads, and DNA-affinity chromatography was performed with the AS fraction as described previously (62).
Preparation of His10-MrpC2, FruA-His6, and FruA-DBD-His8.
Recombinant proteins were expressed in E. coli and purified as described previously (35, 39, 68).
ChIP.
M. xanthus strains MSM1727.DZF1, MSM4499.DZF1, and MSM4499.FA were used for chromatin immunoprecipitation (ChIP) as described previously (68). The primers used for PCR of the fmgBC promoter region integrated ectopically were 5′-CTGCCAGGAATTGGGGATC-3′ (the upstream primer in the vector) and 5′-CGGATCCAGCGGGTGAGGTCGACGACG-3′ (the downstream primer with its 5′ end at position +50 of fmgBC). The primers used for PCR of the vector alone integrated ectopically were the same upstream primer as described above and 5′-CGGGCCATCCGCCAGTGG-3′ (downstream primer in the vector). The primers used for PCR of the rpoC coding region were described previously (68).
RESULTS
An insertion in fmgC reduces spore formation.
M. xanthus strain DK4499 contains Tn5 lac Ω4499 inserted in fmgC, which was predicted previously to encode an oxidase of a cytochrome P-450 system (6). fmgC corresponds to MXAN4127 in the annotation of the genomic sequence (9). Only 59 bp upstream of fmgC is fmgB (MXAN4126), which was predicted previously to code for a reductase likely to function in the same P-450 system as FmgC, although the substrate and products of the system are unknown (6). The short distance between fmgB and fmgC, and the finding that their products are likely components of a P-450 system, suggested that the two genes might be cotranscribed. In agreement, 5′-deletion analysis and mapping of an mRNA 5′ end located a promoter upstream of fmgB capable of driving expression of lacZ during development similar to that observed for DK4499 containing Tn5 lac Ω4499 (6). The gene upstream of fmgB is in the opposite orientation (9). The gene downstream of fmgC is in the same orientation but is separated from the end of fmgC by an intergenic region of at least 243 bp and is predicted to encode a transposase, so it is unlikely to be cotranscribed with the putative fmgBC operon.
M. xanthus DK4499 bearing Tn5 lac Ω4499 aggregated normally under conditions that induce development, but the number of heat- and sonication-resistant spores that were able to germinate and form a colony was sixfold lower than observed for wild-type DK1622. The reduced sporulation of DK4499 is likely due to loss of FmgC, although we cannot rule out an effect of the Tn5 lac insertion on expression of fmgB (e.g., due to altered mRNA stability) or a gene downstream of fmgC (i.e., if transcription from the fmgBC promoter normally reads through a downstream gene). In any case, our results suggest that transcription from the fmgBC promoter is important for sporulation.
MrpC2 binds to a key cis-regulatory sequence in the fmgBC promoter region.
Mutational analysis of the fmgBC promoter region was performed previously (67) and showed that sequences upstream of the promoter are important for its activity (Fig. 1). These regulatory sequences include two 5-bp elements and two C boxes, which are found in the promoter regions of several developmentally regulated genes (6, 31, 52, 59-61, 67). To identify putative transcription factors, we performed EMSAs with a DNA fragment from the fmgBC promoter region and partially purified DNA-binding proteins (AS fraction) from M. xanthus cells that had undergone 12 h of development, since fmgBC is expressed at this time (25). A single shifted complex was observed with a DNA fragment spanning from positions −104 to −29, but no complex was observed when the DNA fragment contained a mutation in the sequence from positions −67 to −64 (Fig. 1). Since this mutation was shown previously to eliminate fmgBC promoter activity in vivo (67), these results showed that a protein in the AS fraction binds to a crucial cis-regulatory sequence upstream of the fmgBC promoter.
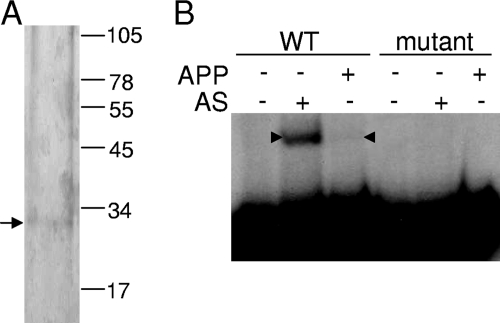
To purify the putative activator protein from the AS fraction, DNA affinity chromatography was performed with the fmgBC DNA fragment (−104 to −29). The major species after purification was ∼30 kDa in size (Fig. 2A). The affinity-purified protein (APP) generated a shifted complex of similar mobility, as observed with the AS fraction when the fmgBC DNA fragment with the wild-type sequence was used in EMSAs, and no complex was observed with the APP and the mutant (−67 to −64) fmgBC promoter region (Fig. 2B). It appeared that APP contained the putative activator protein from the AS fraction.
FIG. 2.
DNA-affinity purification of protein that binds to the fmgBC promoter region. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of protein purified from the AS fraction using fmgBC DNA (positions −104 to −29). The arrow indicates the major species in the APP after staining with silver. The numbers indicate the migration positions of molecular mass (in kilodaltons) standards. (B) EMSAs with 32P-labeled fmgBC DNA (12 nM) spanning from positions −104 to −29 and proteins in the AS fraction or the APP. Arrowheads indicate the shifted complexes produced with the wild-type (WT) DNA fragment. No complex was observed with a DNA fragment bearing the ACCA to CAAC mutation at positions −67 to −64 (mutant).
To identify the putative activator protein, the APP was subjected to mass spectrometry analysis after protease digestion. The peptide sequences primarily matched MrpC, a protein that is ∼30 kDa in size, which is consistent with the size of the major species in the APP (Fig. 2A). MrpC is similar to CRP family transcription factors and is essential for M. xanthus development (54). MrpC2, a shortened form of MrpC that lacks the 25 N-terminal residues, is produced during development, and was identified in an AS fraction previously by DNA-affinity chromatography with the fruA promoter region (58). We infer that MrpC2 in the AS fraction and in the APP is responsible for the shifted complex we observed with fmgBC promoter region DNA.
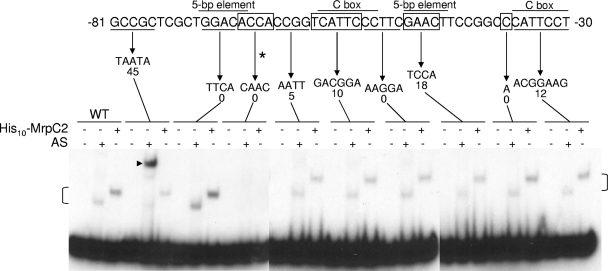
To confirm that MrpC2 binds to the fmgBC promoter region fragment, N-terminally His-tagged MrpC2 (His10-MrpC2) was expressed in E. coli and purified. His10-MrpC2 displayed a pattern of binding to wild-type and mutant fmgBC DNA fragments similar to that of the AS fraction (Fig. 3). The mutation from positions −67 to −64 that resulted in the loss of shifted complex formation with the AS fraction (Fig. 1) and the APP (Fig. 2B) also caused the loss of shifted complex formation with His10-MrpC2 (Fig. 3). This mutation includes 1 bp of a 5-bp element (Fig. 1); however, an adjacent mutation at positions −71 to −68, which changes the remaining 4 bp of the 5-bp element, did not markedly impair formation of shifted complexes with the AS fraction or with His10-MrpC2 (Fig. 3). Likewise, none of the other mutations between positions −63 and −30 markedly impaired complex formation. The slower migration of the complex produced by His10-MrpC2, compared to the complex produced by the AS fraction, is presumably due to the 10 His residues plus 8 additional residues present in the His10-MrpC2 fusion protein. The mutation from positions −81 to −77 resulted in diminished formation of the complex that we believe contains MrpC2, by the AS fraction, and the appearance of a novel shifted complex. The novel complex appears to be due to an unknown protein in the AS fraction that is capable of binding to this mutant fmgBC DNA fragment, since purified His10-MrpC2 did not show this effect. Rather, His10-MrpC2 formed a complex that migrated at the expected position, suggesting that formation of the novel complex by the AS fraction might account for its diminished ability to form the complex that we believe contains MrpC2. We conclude that MrpC2 binds to an important cis-regulatory sequence at positions −67 to −64 in the fmgBC promoter region.
FIG. 3.
Comparison of purified His10-MrpC2 and the AS fraction for binding to the fmgBC promoter region. EMSAs with 32P-labeled fmgBC DNA (2 nM) spanning from positions −104 to −29, wild-type (WT), or mutant as indicated and His10-MrpC2 (1 μM) or the AS fraction (0.7 μg/μl). An asterisk indicates the mutation from positions −67 to −64 that impairs shifted complex formation. Brackets indicate the shifted complexes produced by the AS fraction and His10-MrpC2 upon addition to wild-type fmgBC DNA and to most of the mutant DNA fragments. The mutation from positions −81 to −77 causes a novel shifted complex to form with the AS fraction (arrowhead). The image is a composite from three experiments, and in each experiment the wild-type fmgBC DNA served as a control, and the signal intensity of the shifted complexes was comparable to that shown. The results shown are representative of results observed in at least two experiments.
Since MrpC2 is similar to CRP family transcription factors, and cyclic nucleotides affect DNA binding by some family members (22), we examined His10-MrpC2 binding to the fmgBC promoter region (−104 to −29) in the presence of different nucleotides. At concentrations designed to reflect physiological conditions, no effect of cyclic AMP (4 to 8 μM), cyclic GMP (20 to 40 nM), cyclic di-GMP (1 to 10 μM), ppGpp (50 to 400 μM), nucleoside triphosphates (400 μM), or deoxynucleoside triphosphates (200 μM) was observed (data not shown).
MrpC and/or MrpC2 associates with the fmgBC promoter region in vivo and this depends on FruA.
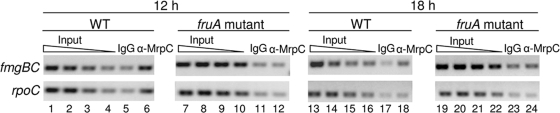
ChIP assays were performed with polyclonal antibodies to MrpC, which also recognize MrpC2 (39), to determine whether MrpC and/or MrpC2 associate with the fmgBC promoter region during development. M. xanthus cells with the fmgBC promoter region (−100 to +50) integrated ectopically at a phage attachment site via site-specific recombination were collected after 12 or 18 h of development and subjected to ChIP with antibodies to MrpC or, as a control, immunoglobulin G (IgG). DNA recovered after ChIP was analyzed by PCR with primers designed to amplify the ectopic copy of the fmgBC promoter region. The PCR analysis showed that the fmgBC promoter region was enriched by ChIP with the anti-MrpC antibodies relative to the IgG control at 12 h (Fig. 4, lanes 5 and 6, top panel) and 18 h (Fig. 4, lanes 17 and 18, top panel) into development. PCR analysis with primers designed to amplify the rpoC coding region showed no enrichment of this region by ChIP with anti-MrpC antibodies relative to control antibodies at 18 h (Fig. 4, lanes 17 and 18, bottom panel), as reported previously (35), but at 12 h we unexpectedly yet reproducibly observed enrichment of the rpoC coding region by ChIP with anti-MrpC antibodies relative to control antibodies (Fig. 4, lanes 5 and 6, bottom panel). These results indicate that MrpC and/or MrpC2 is present in the vicinity of the rpoC coding region at 12 h into development, but not at 18 h, and that MrpC and/or MrpC2 is associated with the fmgBC promoter region at both times.
FIG. 4.
Association of MrpC and/or MrpC2 with the fmgBC promoter region during development of wild-type and fruA mutant cells. ChIP analysis of M. xanthus with the fmgBC promoter region (−100 to +50) integrated ectopically in otherwise wild-type (WT) or fruA mutant backgrounds. At 12 and 18 h into development, cells were treated with formaldehyde and lysed, and cross-linked chromatin was immunoprecipitated with anti-MrpC antibodies or IgG as a control. DNA was amplified with appropriate primers for the fmgBC promoter region at the ectopic chromosomal site or with appropriate primers for the rpoC coding region as a control. A twofold dilution series of input DNA purified from 0.25, 0.125, 0.0625, or 0.03125% of the total cellular extract prior to immunoprecipitation was used as a template in parallel PCRs to show that the PCR conditions were in the linear range of amplification for each primer set.
Recently, regulation of the fmgA gene was shown to be under combinatorial control by MrpC2 and FruA (35). Since the expression of fmgA occurs with similar timing during development as fmgBC (25) and the expression of both genes depends partially on C-signaling (4, 6, 24), to which FruA has been proposed to respond (5), we hypothesized that fmgBC is also under direct control by FruA. In the case of fmgA, association of MrpC and/or MrpC2 with the promoter region in vivo, as measured by ChIP-PCR analysis, was dependent on FruA (35). We carried out a similar analysis for fmgBC by performing ChIP-PCR analysis of a fruA mutant with the fmgBC promoter region (−100 to +50) integrated ectopically as described above. In contrast to the wild-type strain, no enrichment of the fmgBC promoter region was observed with anti-MrpC antibodies relative to control antibodies at 12 h or 18 h into development (Fig. 4, lanes 11, 12, 23, and 24 [top panel]). Likewise, no enrichment of the rpoC coding region was observed with anti-MrpC antibodies relative to control antibodies (Fig. 4, lanes 11, 12, 23, and 24 [bottom panel]). We conclude that FruA is necessary for the association of MrpC and/or MrpC2 with the fmgBC promoter region during development and for the association of MrpC and/or MrpC2 with the rpoC coding region at 12 h into development.
FruA associates with the fmgBC promoter region in vivo and governs expression.
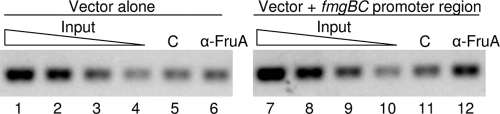
If FruA plays a direct role in recruitment of MrpC and/or MrpC2 to the fmgBC promoter region, as observed previously for fmgA (35), it should be possible to detect FruA at the fmgBC promoter region by ChIP with antibodies against FruA. To test this expectation, ChIP was performed on the wild-type strain with the fmgBC promoter region (−100 to +50) integrated ectopically. At 12 h into development, enrichment of the fmgBC promoter region was observed with anti-FruA antibodies compared to control preimmune serum (Fig. 5, lanes 11 and 12). No enrichment was observed for a strain with vector lacking the fmgBC promoter region integrated ectopically (Fig. 5, lanes 5 and 6). We conclude that FruA associates with the fmgBC promoter region in vivo, which is consistent with the notion that it directly recruits MrpC and/or MrpC2.
FIG. 5.
Association of FruA with the fmgBC promoter region in vivo. ChIP analysis of M. xanthus with the vector alone or with the fmgBC promoter region (−100 to +50) integrated ectopically. At 12 h into development, cells were treated with formaldehyde and lysed, and cross-linked chromatin was immunoprecipitated with anti-FruA antibodies or preimmune serum as a control (lane C). A twofold dilution series of input DNA purified from 0.25, 0.125, 0.0625, or 0.03125% of the total cellular extract prior to immunoprecipitation was used as a template in parallel PCRs to show that the PCR conditions were in the linear range of amplification.
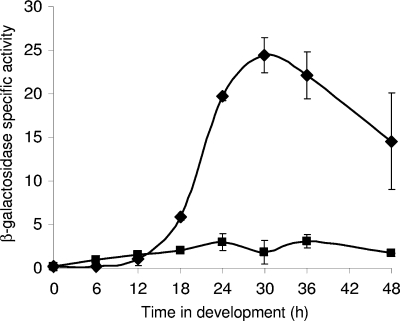
If FruA plays a key role in regulation of fmgBC, expression of fmgBC is predicted to be impaired in a fruA mutant, as observed previously for fmgA (68). To test this prediction, fruA mutant and wild-type M. xanthus cells were transformed with a plasmid containing the −100 to +50 region of the fmgBC promoter transcriptionally fused to the E. coli lacZ gene. The plasmid integrates into the M. xanthus genome ectopically via site-specific recombination at a phage attachment site. As negative controls, strains bearing the vector with promoterless lacZ were also constructed. β-Galactosidase specific activity was measured in cell extracts at different times during development. The activity of each negative control strain was subtracted from that of the corresponding promoter-containing strain. The fruA mutation abolished developmental lacZ expression from the fmgBC promoter region (Fig. 6). This demonstrates that FruA governs fmgBC expression and, together with our other data, strongly suggests that FruA binds to the fmgBC promoter region and recruits MrpC and/or MrpC2, activating transcription.
FIG. 6.
Developmental expression from fmgB-lacZ. The fmgBC promoter region from positions −100 to +50 was fused to lacZ, and the β-galactosidase specific activity was measured during the development of M. xanthus wild-type (⧫) and fruA mutant (▪) cells. In each background, the activity from the vector with no promoter was measured as a negative control. Points show the average of three transformants, after subtracting the average of three transformants with the promoterless vector. The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Error bars depict one standard deviation of the data.
The FruA DNA-binding domain binds to a key cis-regulatory sequence in the fmgBC promoter region.
To determine whether FruA binds to the fmgBC promoter region, the C-terminally His-tagged FruA DNA-binding domain (FruA-DBD-His8) was overexpressed in E. coli, purified, and used in EMSAs. FruA-DBD-His8 generated a single shifted complex with a DNA fragment spanning from positions −104 to −29 of the fmgBC promoter region (Fig. 7). EMSAs with mutant probes localized the binding to positions −53 to −49, since a mutation in this region abolished the FruA-DBD-His8 binding. This region was shown previously to be critical for fmgBC promoter activity (67). It includes part of a C box and lies immediately upstream of a 5-bp element.
FIG. 7.
Effects of mutations on binding of FruA-DBD-His8 to fmgBC promoter region DNA. EMSAs with 32P-labeled fmgBC DNA (2 nM) spanning from positions −104 to −29, wild type (WT), or mutant as indicated and FruA-DBD-His8 (14 μM). A horizontal arrow indicates the shifted complex produced with wild-type DNA. An asterisk indicates the mutation from positions −53 to −49 that impairs shifted complex formation. The image is a composite from three experiments, and intervening lanes were removed from one of the images. In each experiment, the wild-type fmgBC DNA served as a control, and the signal intensity of the shifted complex was comparable to that shown.
Enhanced complex formation in the presence of FruA-His6 andHis10-MrpC2.
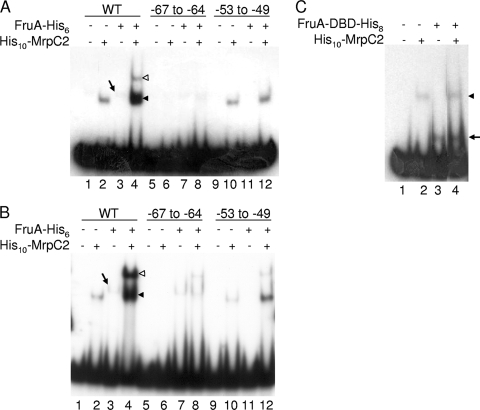
The combination of FruA and MrpC2 led to enhanced formation of shifted complexes with fmgA promoter region DNA, due to cooperative binding of the two proteins to adjacent (possibly overlapping) sites (35). Both sites were required for the enhancement of shifted complex formation, as was full-length FruA (i.e., FruA-DBD-His8 was insufficient), suggesting that the N-terminal domain of FruA might directly interact with MrpC2 (35). To test whether similar effects could be observed with fmgBC promoter region DNA, FruA-His6 was overexpressed in E. coli and purified. As observed previously with fmgA promoter region DNA (35), the fmgBC promoter region (−104 to −29) was bound weakly by FruA-His6 in EMSAs (Fig. 8A, lane 3), but the combination of FruA-His6 and His10-MrpC2 resulted in the formation of an abundant lower complex (LC) and a faint upper complex (UC) when analyzed on 5% polyacrylamide gels (Fig. 8A, lane 4). Migration of the LC was similar to that of complexes formed by either protein alone, suggesting that the LC is composed of DNA bound by His10-MrpC2 or FruA-His6. The slower migration of the UC was suggestive of DNA bound by both proteins simultaneously. When analyzed on 8% polyacrylamide gels, more UC was observed when both proteins were added to the fmgBC DNA fragment (Fig. 8B, lane 4), and more complex was observed when only FruA-His6 was added (Fig. 8B, lane 3). The 8% polyacrylamide gel seems to stabilize FruA-His6 binding to DNA under the conditions of the EMSAs, as observed previously with fmgA promoter region DNA (35).
FIG. 8.
EMSAs with MrpC2 and full-length FruA or just the DNA-binding domain of FruA. (A) Shifted complex formation with His10-MrpC2 and full-length FruA-His6 and the effect of mutations. EMSAs with 32P-labeled fmgBC DNA (2 nM) spanning from positions −104 to −29, wild-type (WT), or mutant as indicated and no protein, His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM) as indicated, electrophoresed on a 5% polyacrylamide gel. A slanted arrow indicates the faint shifted complex produced by FruA-His6 alone. The unfilled and filled arrowheads indicate the UCs and LCs, respectively, produced by the combination of proteins. (B) Same as in panel A except electrophoresed on an 8% polyacrylamide gel. (C) Shifted complex formation with His10-MrpC2 and FruA-DBD-His8. EMSAs with 32P-labeled fmgBC DNA (2 nM) spanning from positions −104 to −29 and no protein, His10-MrpC2 (1 μM), FruA-DBD-His8 (14 μM), or both His10-MrpC2 (1 μM) and FruA-DBD-His8 (14 μM) as indicated. The arrowhead indicates the complex produced by His10-MrpC2, and the arrow indicates the complex produced by FruA-DBD-His8. Intervening lanes were removed from the image.
To determine whether the binding of both proteins to DNA is required for the observed enhancement of complex formation, EMSAs were performed with mutant DNA fragments. A mutation at positions −67 to −64 that abolished His10-MrpC2 binding (Fig. 3) also abolished enhancement of complex formation by the combination of proteins on 5% polyacrylamide gels; the UC was undetectable, and the faint LC was comparable in intensity to that formed by FruA-His6 alone (Fig. 8A, lanes 7 and 8). On 8% polyacrylamide gels, a similar result was observed, except that a small amount of UC was detected (Fig. 8B, lane 8), perhaps indicating that FruA-His6 facilitates weak binding of His10-MrpC2 to the mutant site. Similarly, a mutation at positions −53 to −49 that abolished detectable binding of FruA-DBD-His8 (Fig. 7) or FruA-His6 (Fig. 8A, lane 11) resulted in no detectable UC on 5% polyacrylamide gels, and LC of an intensity comparable to that formed by His10-MrpC2 alone (Fig. 8A, lanes 10 and 12). On 8% polyacrylamide gels, there was slight enhancement of LC and a small amount of UC (Fig. 8B, lanes 10 and 12). The small amount of UC might indicate that His10-MrpC2 facilitates weak binding of FruA-His6 to the mutant site. The slight enhancement of LC might result from the initial binding of both proteins, followed by dissociation of FruA-His6. In any case, much less of the shifted complexes is observed with the DNA fragment containing the mutation at positions −53 to −49 (Fig. 8B, lane 12) than with the wild-type fragment (Fig. 8B, lane 4). MrpC2 and FruA appear to bind cooperatively to the fmgBC promoter region, as seen previously for the fmgA promoter region, although the arrangement of binding sites relative to the promoter is different. FruA binds upstream of MrpC2 in the fmgA promoter region (35), whereas FruA binds downstream of MrpC2 in the fmgBC promoter region (Fig. 3 and 7).
Despite the different arrangement of binding sites, we found that the fmgA and fmgBC promoter regions share the characteristic that FruA-DBD-His8 is insufficient to enhance complex formation in combination with His10-MrpC2 (Fig. 8C, lane 4). The complexes formed by the combination of proteins were similar to the complexes formed by His10-MrpC2 or FruA-DBD-His8 alone (Fig. 8C, lanes 2 and 3). We propose that the N-terminal regulatory domain of FruA interacts with MrpC2 at the fmgBC promoter region, mediating cooperative binding of the two transcription factors and subjecting fmgBC expression to combinatorial control similar to that observed for fmgA.
DISCUSSION
Our results demonstrate that MrpC2 and FruA bind to key cis-regulatory sequences upstream of the fmgBC promoter, placing it under similar combinatorial control as observed previously for fmgA (35). Surprisingly, the arrangement of binding sites for MrpC2 and FruA is different in the two promoter regions. FruA binds downstream of MrpC2 in the fmgBC promoter region (Fig. 3 and 7), whereas FruA binds upstream of MrpC2 in the fmgA promoter region (35). In both cases, FruA is required for promoter activity and for recruitment of MrpC and/or MrpC2 to the promoter region in vivo. In vitro, FruA and MrpC2 appear to bind cooperatively to both promoter regions, and this depends on the N-terminal regulatory domain of the FruA response regulator. Preliminary results, described below, indicate that cooperative binding by FruA and MrpC2 is a common mechanism of gene regulation during M. xanthus development. This mechanism is proposed to allow integration of positional information via short-range C-signaling with starvation signaling and cell death, controlling spatiotemporal gene expression and determining cell fate.
Combinatorial control of C-signal-dependent genes involving cooperative binding of FruA and MrpC2 appears to be a common mechanism of gene regulation during M. xanthus development. In addition to fmgA and fmgBC, the promoter region of the dev operon appears to utilize this mechanism. MrpC2 binds to a region that includes a 5-bp element and two C box-like sequences, and the addition of FruA greatly enhances complex formation in EMSAs (S. Mittal, P. Viswanathan, and L. Kroos, unpublished data). Expression of the dev operon is confined to fruiting bodies (16, 47) and has been proposed to be a crucial step in commitment of cells to differentiate into spores (23, 35). The gene identified by Tn5 lac Ω4403 encodes a putative serine protease whose role in development is unknown, but whose expression depends absolutely on C-signaling (7, 24). The promoter region contains two 5-bp elements in inverted orientation that are bound by MrpC2, and FruA appears to bind cooperatively (J. Lee, S. Mittal, and L. Kroos, unpublished data). Therefore, at least four promoter regions appear to be bound cooperatively by MrpC2 and FruA, since the combination of proteins greatly enhances formation of shifted complexes in EMSAs, and this was shown to correlate with cooperative binding at the fmgA promoter region by DNase I footprinting (35). Moreover, enhancement of shifted complex formation was shown to require the binding sites for both MrpC2 and FruA at both the fmgA (35) and the fmgBC (Fig. 8) promoter regions.
Although the combination of MrpC2 and FruA produces a strikingly similar enhancement of shifted complex formation in EMSAs with fmgA or fmgBC promoter region DNA, the arrangement of the MrpC2 and FruA binding sites is different in the two promoter regions. In the fmgA promoter region, mutations from positions −86 to −77 impaired the binding of FruA-DBD-His8 (68) and mutations from positions −76 to −46 affected the binding of His10-MrpC2 (35). In addition, DNA upstream of position −76 was found to be required for His10-MrpC2 binding, suggesting that the MrpC2- and FruA-binding sites might partially overlap, with the two proteins presumably interacting with opposite faces of the DNA in the region of overlap (35). In contrast, FruA-DBD-His8 and His10-MrpC2 binding to the fmgBC promoter region was impaired only by mutations from positions −53 to −49 and from positions −67 to −64, respectively (Fig. 3 and 7). Adjacent mutations did not impair the binding of either protein, although these mutations had previously been shown to reduce promoter activity (67), suggesting that sequences important for binding in vivo might be missed under the in vitro conditions of the EMSAs. Alternatively, other transcription factors might bind to the adjacent sequences. In any case, FruA binds downstream of MrpC2 in the fmgBC promoter region, whereas FruA binds upstream of MrpC2 in the fmgA promoter region.
The different arrangement of FruA and MrpC2 binding sites in the fmgA and fmgBC promoter regions suggests a somewhat different mechanism of transcriptional activation from the two promoters. As noted previously, in the fmgA promoter region, the two proteins occupy a location typical for class I activators (35), which contact the C-terminal domain of the α subunits of RNA polymerase (1). In the fmgBC promoter region, FruA and MrpC2 occupy a similar location, but their positions relative to the promoter are reversed, so presumably a different contact(s) with the C-terminal domain of the α subunits of RNA polymerase would be involved in the activation of transcription. Two activators can contact the C-terminal domain of the α subunits of RNA polymerase at the same promoter, based on studies of both synthetic (28, 55) and natural promoters (2).
Despite the different arrangement of FruA and MrpC2 binding sites with respect to the fmgA and fmgBC promoters, the two proteins might interact with each other similarly at the two promoter regions. Our results show that the N-terminal regulatory domain of FruA is required for enhancement of shifted complex formation in combination with MrpC2 at both promoter regions (35) (Fig. 8C). This domain is similar to receiver domains of response regulators that are phosphorylated by histidine protein kinases (5, 42); however, it lacks two aspartate residues that are highly conserved in receiver domains and normally play an important role in phosphorylation of a third aspartate residue (5, 63). Moreover, several lines of evidence suggest that FruA might function without phosphorylation (35). Here, we showed that recombinant (presumably unphosphorylated) FruA-His6 greatly enhances formation of shifted complexes in combination with His10-MrpC2 at the fmgBC promoter region, and the receiver domain of FruA is required for enhancement (Fig. 8). Therefore, the unphosphorylated receiver domain of FruA might interact directly with MrpC2 to mediate cooperative DNA binding. Receiver domains that cannot or need not be phosphorylated have been described in bacterial DNA-binding proteins (11, 44, 50) and in proteins that regulate circadian rhythms in bacteria (36, 65) and plants (53). These proteins are sometimes called pseudo-response regulators. Whether FruA is a pseudo-response regulator (i.e., whether its receiver domain is phosphorylated in vivo) remains an open question but, to our knowledge, cooperative binding of a response regulator-like protein and an independent transcription factor (MrpC2) is a novel mechanism of gene regulation (35).
Consistent with the idea that FruA and MrpC2 interact similarly with each other at the fmgA and fmgBC promoter regions, the combination of proteins produces a strikingly similar enhancement of shifted complex formation in EMSAs with DNA from either promoter region (35) (Fig. 8). In both cases, the percentage of polyacrylamide in gels used in the EMSAs influenced the shifted complexes that were observed, with 8% gels (compared to 5% gels) facilitating the detection of FruA binding and the detection of UC that presumably represents FruA and MrpC2 bound to DNA. We infer that the two proteins bind cooperatively to DNA in solution, as demonstrated by DNase I footprinting in the case of fmgA (35), but FruA binding is less stable than MrpC2 binding, especially when analyzed on 5% gels, so LC is predominantly MrpC2 bound to DNA. Since their invention, it has been known that the gel matrix can influence the stability of protein-DNA complexes during EMSAs (8).
Another observation consistent with the idea that FruA and MrpC2 might interact similarly with each other at the fmgA and fmgBC promoter regions is that sequences matching the consensus binding site for FruA are in the opposite orientation in the two promoters. The consensus sequence for binding of FruA-DBD-His8 is GGGC/TA/G(N4-6)C/TGGG (62). The sequence GGGTG(N5)TGGG from positions −81 to −68 in the fmgA promoter region matches the consensus perfectly, and some mutations in this sequence impair FruA-DBD-His8 binding in vitro (68). In the fmgBC promoter region, in the opposite orientation, the sequence GGGAA(N4)CGGT from positions −52 to −64 matches the consensus except at two positions, and the mutation at positions −53 to −49 that impaired FruA-DBD-His8 binding in vitro overlaps this sequence (Fig. 7). MrpC is dimeric, and one type of site to which MrpC and MrpC2 bind is palindromic, with a consensus sequence of GTGTC(N8)GACAC (39). Presumably, a dimer of MrpC or MrpC2 bound to such a palindromic site could present the same surface to FruA bound upstream or downstream. In the fmgA promoter region, the sequence GAGCG(N8)CACAT from positions −67 to −50 is the best match to the consensus between positions −76 and −46, where mutations affected His10-MrpC2 binding (35). In the fmgBC promoter region, the sequence ACGCC(N8)GACAC from positions −83 to −66 matches half the consensus perfectly, and the mutation at positions −67 to −64 that impaired His10-MrpC2 binding in vitro overlaps this sequence (Fig. 3). We hypothesize that the N-terminal domain of FruA can interact directly with dimeric MrpC2 to permit cooperative DNA binding, whether FruA binds upstream of MrpC2 (as at the fmgA promoter region) or whether FruA binds to a site in the opposite orientation downstream of MrpC2 (as at the fmgBC promoter region). This flexibility in the arrangement of FruA and MrpC2 at different promoters would presumably result in a different contact(s) with RNA polymerase and different levels of transcriptional activation.
Our finding that the response regulator-like FruA and the transcription factor/antitoxin MrpC bind cooperatively in different arrangements in the promoter regions of C-signal-dependent genes has important implications for M. xanthus development. Since MrpC2 appears to activate fruA transcription (58), combinatorial regulation of target genes by MrpC2 and FruA constitutes a coherent feed-forward loop, which is a motif found commonly in regulatory networks since it has beneficial characteristics (33, 34). One characteristic is that the expression of target genes is delayed until both transcription factors reach a sufficient concentration. Full expression of partially C-signal-dependent target genes such as fmgBC and the dev operon, which are important for sporulation, may be delayed until cell alignment in the nascent fruiting body causes a high level of C-signaling, which could affect FruA and/or MrpC2. Since a mutant defective in C-signaling accumulates FruA normally during development, it has been proposed that one or more histidine protein kinases alter the activity of FruA via phosphorylation in response to C-signaling (5, 15, 58). However, if FruA is not phosphorylated, perhaps C-signaling affects the concentration of MrpC2 and/or its precursor, MrpC. The accumulation of MrpC and MrpC2 is inhibited by the STPK cascade that leads to phosphorylation of MrpC during growth (39). Starvation triggers accumulation of both forms of the protein by counteracting the STPK cascade (39); however, the EspA signal transduction pathway appears to delay their accumulation in response to an unknown signal (13). Therefore, the concentrations of MrpC and MrpC2 appear to be linked to starvation and perhaps other developmental signals via several pathways. Only if starvation persists and the other putative signals, including C-signal, are received, would the MrpC2 concentration rise to a threshold that permits full expression of target genes in combination with FruA, committing the cell to form a spore. In its role as an antitoxin, binding of MrpC to the MazF toxin would prevent programmed cell death in cells destined to form spores (37). In cells destined to undergo programmed cell death, binding of MrpC to the mazF promoter region would activate transcription, leading to increased MazF. According to this model, MrpC is a key determinant of cell fate, and determining whether MrpC2 binds to MazF and/or the mazF promoter region is an important goal. Also, determining whether the effects of FruA on target gene expression depend solely on the strength of its binding sites and their position relative to MrpC2 binding sites, or whether FruA integrates additional signal inputs, is an important question for future studies. Signal-responsive auxiliary regulatory proteins have been shown to interact with the response regulator RcsB (32) and the pseudo-response regulator AmiR (43), so perhaps FruA interacts with other partners in addition to binding cooperatively to DNA with MrpC2.
Acknowledgments
We are grateful to Sumiko Inouye for providing plasmids, protocols, and antibodies. We thank Chris Waters for the gift of cyclic di-GMP.
This research was supported by NSF grant MCB-0744343 and by the Michigan Agricultural Experiment Station.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7102-108. [DOI] [PubMed] [Google Scholar]
- 2.Beatty, C. M., D. F. Browning, S. J. Busby, and A. J. Wolfe. 2003. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 1855148-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berleman, J. E., and J. R. Kirby. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J. Bacteriol. 1895675-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 1801995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30807-817. [DOI] [PubMed] [Google Scholar]
- 6.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 1815467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisseha, M., M. Gloudemans, R. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 1782539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried, M., and D. M. Crothers. 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 96505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 10315200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gronewold, T. M., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40744-756. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie, E. P., C. S. Flaxman, J. White, D. A. Hodgson, M. J. Bibb, and K. F. Chater. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144727-738. [DOI] [PubMed] [Google Scholar]
- 12.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 121022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgs, P. I., S. Jagadeesan, P. Mann, and D. R. Zusman. 2008. EspA, an orphan hybrid histidine protein kinase, regulates the timing of expression of key developmental proteins of Myxococcus xanthus. J. Bacteriol. 1904416-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 742938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelsbak, L., M. Givskov, and D. Kaiser. 2005. Enhancer-binding proteins with an FHA domain and the σ54 regulon in Myxococcus xanthus fruiting body development. Proc. Natl. Acad. Sci. USA 1023010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 979098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 765952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashefi, K., and P. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15483-494. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249926-928. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 1731722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of Myxococcus xanthus. Cell 6119-26. [DOI] [PubMed] [Google Scholar]
- 22.Kolb, A., S. Busby, I. I. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62749-797. [DOI] [PubMed] [Google Scholar]
- 23.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 4113-39. [DOI] [PubMed] [Google Scholar]
- 24.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1840-854. [DOI] [PubMed] [Google Scholar]
- 25.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117252-266. [DOI] [PubMed] [Google Scholar]
- 26.Kruse, T., S. Lobedanz, N. M. Berthelsen, and L. Sogaard-Andersen. 2001. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 40156-168. [DOI] [PubMed] [Google Scholar]
- 27.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signaling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 1747360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langdon, R. C., and A. Hochschild. 1999. A genetic method for dissecting the mechanism of transcriptional activator synergy by identical activators. Proc. Natl. Acad. Sci. USA 9612673-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, S.-F., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6401-410. [DOI] [PubMed] [Google Scholar]
- 30.Lobedanz, S., and L. Sogaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 172151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loconto, J., P. Viswanathan, S. J. Nowak, M. Gloudemans, and L. Kroos. 2005. Identification of the Ω4406 regulatory region, a developmental promoter of Myxococcus xanthus, and a DNA segment responsible for chromosomal position-dependent inhibition of gene expression. J. Bacteriol. 1874149-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59379-405. [DOI] [PubMed] [Google Scholar]
- 33.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feed-forward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334197-204. [DOI] [PubMed] [Google Scholar]
- 34.Milo, R., S. Shen-Orr, S. Itzkovitz, N. Kashtan, D. Chklovskii, and U. Alon. 2002. Network motifs: simple building blocks of complex networks. Science 298824-827. [DOI] [PubMed] [Google Scholar]
- 35.Mittal, S., and L. Kroos. 2009. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc. Natl. Acad. Sci. USA 1061965-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutsuda, M., K. P. Michel, X. Zhang, B. L. Montgomery, and S. S. Golden. 2003. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 27819102-19110. [DOI] [PubMed] [Google Scholar]
- 37.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 13255-66. [DOI] [PubMed] [Google Scholar]
- 38.Nariya, H., and S. Inouye. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol. Microbiol. 58367-379. [DOI] [PubMed] [Google Scholar]
- 39.Nariya, H., and S. Inouye. 2006. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol. Microbiol. 601205-1217. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 1733318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor, K. A., and D. R. Zusman. 1988. Reexamination of the role of autolysis in the development of Myxococcus xanthus. J. Bacteriol. 1704103-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22757-767. [DOI] [PubMed] [Google Scholar]
- 43.O'Hara, B. P., R. A. Norman, P. T. Wan, S. M. Roe, T. E. Barrett, R. E. Drew, and L. H. Pearl. 1999. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 185175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otten, S. L., C. Olano, and C. R. Hutchinson. 2000. The dnrO gene encodes a DNA-binding protein that regulates daunorubicin production in Streptomyces peucetius by controlling expression of the dnrN pseudo response regulator gene. Microbiology 1461457-1468. [DOI] [PubMed] [Google Scholar]
- 45.Rolbetzki, A., M. Ammon, V. Jakovljevic, A. Konovalova, and L. Sogaard-Andersen. 2008. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in Myxococcus xanthus. Dev. Cell 15627-634. [DOI] [PubMed] [Google Scholar]
- 46.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 82793-2804. [DOI] [PubMed] [Google Scholar]
- 47.Sager, B., and D. Kaiser. 1993. Spatial restriction of cellular differentiation. Genes Dev. 71645-1653. [DOI] [PubMed] [Google Scholar]
- 48.Sager, B., and D. Kaiser. 1993. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc. Natl. Acad. Sci. USA 903690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Schar, J., A. Sickmann, and D. Beier. 2005. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 1873100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimkets, L. J., R. E. Gill, and D. Kaiser. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. USA 801406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan, D., and L. Kroos. 2004. Mutational analysis of the fruA promoter region demonstrates that C-box and 5-base-pair elements are important for expression of an essential developmental gene of Myxococcus xanthus. J. Bacteriol. 1865961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strayer, C., T. Oyama, T. F. Schultz, R. Raman, D. E. Somers, P. Mas, S. Panda, J. A. Kreps, and S. A. Kay. 2000. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289768-771. [DOI] [PubMed] [Google Scholar]
- 54.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 1834786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tebbutt, J., V. A. Rhodius, C. L. Webster, and S. J. Busby. 2002. Architectural requirements for optimal activation by tandem CRP molecules at a class I CRP-dependent promoter. FEMS Microbiol. Lett. 21055-60. [DOI] [PubMed] [Google Scholar]
- 56.Ueki, T., and S. Inouye. 2005. Activation of a development-specific gene, dofA, by FruA, an essential transcription factor for development of Myxococcus xanthus. J. Bacteriol. 1878504-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueki, T., and S. Inouye. 2005. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J. Biol. Chem. 28032279-32284. [DOI] [PubMed] [Google Scholar]
- 58.Ueki, T., and S. Inouye. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 1008782-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viswanathan, K., P. Viswanathan, and L. Kroos. 2006. Mutational analysis of the Myxococcus xanthus Ω4406 promoter region reveals an upstream negative regulatory element that mediates C-signal dependence. J. Bacteriol. 188515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 1851405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viswanathan, P., K. Murphy, B. Julien, A. G. Garza, and L. Kroos. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 1893738-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viswanathan, P., T. Ueki, S. Inouye, and L. Kroos. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc. Natl. Acad. Sci. USA 1047969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 64.Whitworth, D. E. (ed.). 2008. Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 65.Williams, S. B., I. Vakonakis, S. S. Golden, and A. C. LiWang. 2002. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc. Natl. Acad. Sci. USA 9915357-15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 1863766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoder-Himes, D., and L. Kroos. 2006. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J. Bacteriol. 1885167-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]