Abstract
The stringent response is a mechanism by which bacteria adapt to environmental stresses and nutritional deficiencies through the synthesis and hydrolysis of (p)ppGpp by RelA/SpoT enzymes. Alphaproteobacteria and plants contain a single Rsh enzyme (named for RelA/SpoT homolog) that is bifunctional. Here we report the identification of a new species of bacteria belonging to the genus Novosphingobium and characterization of an rsh mutation in this plant tumor-associated isolate. Isolate Rr 2-17, from a grapevine crown gall tumor, is a member of the Novosphingobium genus that produces the N-acyl-homoserine lactone (AHL) quorum-sensing (QS) signals. A Tn5 mutant, Hx 699, deficient in AHL production was found to have an insertion in an rsh gene. The Rsh protein showed significant percent sequence identity to Rsh proteins of alphaproteobacteria. The Novosphingobium sp. rsh gene (rshNsp) complemented the multiple amino acid requirements of the Escherichia coli relA spoT double mutant by restoring the growth on selection media. Besides QS signal production, the rsh mutation also affects soluble polysaccharide production and cell aggregation. Genetic complementation of the Hx 699 mutant with the rshNsp gene restored these phenotypes. This is the first discovery of a functional rsh gene in a member of the Novosphingobium genus.
Members of the Sphingomonas genera are sphingolipid-containing organisms belonging to the α-4 subclass of proteobacteria (56, 61). The type species, Sphingomonas paucimobilis, is a gram-negative, yellow-pigmented obligate aerobe (45). Even though members of Sphingomonadaceae have membranes with an unusual structure, they are widely distributed in nature and have metabolic capabilities that can be exploited for bioremediation (56). Members of the Sphingomonadaceae family are commonly isolated from marine, soil, and plant environments (34, 37). Based on phylogenetic, chemotaxonomic, and phenotypic observations, the Sphingomonas genus has been expanded to include three new genera, Sphingobium, Novosphingobium, and Sphingopyxis (45), although this expansion has been questioned (60).
By utilizing a gene regulatory mechanism known as quorum sensing (QS), bacteria can regulate gene expression according to cell density (51, 55, 57). In one QS system, bacteria produce and secrete chemical signals called N-acyl-homoserine lactones (AHLs) to their surroundings (51, 57). Upon reaching a concentration threshold, the AHL signal can activate population-wide responses leading to the coordination of gene activation or repression (56). In response to changing environmental conditions, bacteria have evolved many cellular regulatory mechanisms. One such mechanism is the stringent response for the control of gene expression in bacterial cells subject to starvation for amino acids, carbon, nitrogen, phosphate, fatty acids, or UV light exposure (6, 9, 26, 29, 38, 42). The hallmark of the stringent response is the accumulation of the effector molecule or alarmone, guanosine 3′,5′-(bis)pyrophosphate (ppGpp), derived from guanosine 3′-diphosphate 5′-triphosphate (pppGpp) by hydrolysis. Together, pppGpp and ppGpp are termed (p)ppGpp (38). Changes in (p)ppGpp levels affect gene expression by directly and indirectly regulating transcription (27, 28). In Escherichia coli, two proteins are involved in stress-induced (p)ppGpp accumulation: RelA and SpoT. RelA is a ribosome-associated (p)ppGpp synthetase responding to uncharged tRNAs that accumulate as a result of amino acid limitation. SpoT is a cytostolic protein which functions as a bifunctional (p)ppGpp synthetase and hydrolase. The (p)ppGpp synthetase activity of SpoT is triggered by carbon and fatty acid starvation (38). Bacterial and plant RelA/SpoT homologs with (p)ppGpp synthetase and hydrolase activities are widespread and are referred to as Rsh for RelA/SpoT homologs (2, 9, 33, 38).
Even though the genomes of Sphingopyxis alaskensis RB2256 and Sphingomonas wittichii RW1 have been sequenced, to our knowledge no genes that affect QS signal synthesis, perception, or response have been characterized in the Sphingomondaceae family (5, 56). Using bioinformatic analyses, the QS core proteins, LuxI- and LuxR-type homologs, have been identified in these two species (5). A recent study of soil bacteria showed that Sphingomonas agrestis produces an AHL similar to that of Agrobacterium tumefaciens, the N-(3-hydroxyoctanoyl)-l-homoserine lactone signal (11). The involvement of the stringent response in QS gene expression has also been described for plant-associated pathogenic (A. tumefaciens, Erwinia carotovora, and Pseudomonas aeruginosa) and symbiotic (Rhizobium meliloti and Rhizobium etli) bacteria (4, 16, 35, 50, 52, 54, 63).
The gram-negative bacteria A. tumefaciens and Agrobacterium vitis are the causal agents of crown gall disease on a number of dicotyledonous plants and grapevine, respectively (3, 36, 51, 55). The N-(3-oxo-octanoyl)-L-HL (3-O-C8-HL) signal of A. tumefaciens is involved in the regulation of replication and conjugal transfer of the tumor-inducing (Ti) plasmid to Ti plasmidless A. tumefaciens cells (36, 55, 57). The identification of AHL-producing nonagrobacterial isolates from crown gall tumor tissues may indicate that AHL signals in planta can be produced by one bacterial species and sensed by another to elicit a physiological response. In order to investigate the role of the stringent response in AHL QS systems in nonagrobacteria colonizing grapevine tumors, we have chosen a newly identified species belonging to the Sphingomonadaceae family because of its ability to activate a response in AHL-dependent biosensors. This species was subjected to transposon mutagenesis and screened for mutants defective in AHL signal synthesis. A Tn5 mutant deficient in AHL production was shown to have an insertion into the rsh gene. The rshNsp gene was cloned and used in genetic analyses. The rsh gene in Rr 2-17 was found to control the formation of soluble polysaccharides and cell aggregations in addition to AHL QS signals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. The Novosphingobium strains were grown in tryptone yeast extract (TYE), potato dextrose (PD) (Difco Laboratories, Detroit), and AB minimal media at 28°C (10). E. coli strains were grown on Luria-Bertani broth (LB) medium at 37°C. Agrobacterium biosensor strain NTL4(pZLR4) (7) was grown in AB minimal medium (10) supplemented with 0.2% dextrose and 0.01% yeast extract. Antibiotics used for Novosphingobium sp. strains were kanamycin (25 μg/ml) and tetracycline (3 μg/ml for broth and 5 μg/ml for agar media). For E. coli, antibiotics used were ampicillin (100 μg/ml), kanamycin (50 μg/ml), and tetracycline (10 μg/ml). For plate assays of ppGpp production, we used AT medium, which is M9 dextrose (32) supplemented with all amino acids except histidine, either with or without 15 mM of 3-amino-1,2,4-triazole (Sigma-Aldrich) (20), and SMG plates consisting of M9 dextrose medium supplemented with 1 mM of the amino acids serine, methionine, and glycine (49).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CF1648 | Wild-type K-12 strain (MG1655) | 59 |
| CF1652 | ΔrelA251::kan derivative of CR1648 | 59 |
| CF1693 | ΔspoT207::cat derivative of CF1652 | 59 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15ΔlacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (Strr) endA1 nupG | Invitrogen |
| EC100D pir+ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL nupG pir+ (DHFR) | Epicentre |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 62 |
| Sphingomonas sp. | ||
| Rr 2-17 | Wild-type isolate from nopaline grapevine tumor | This work |
| Hx 699 | Mutant isolate 699, rsh::EZ-Tn5, Kmr derivative of Rr2-17, deficient in AHL synthesis | This work |
| A. tumefaciens NTL4(pZLR4) | Indicator strain for detection of AHLs | 7 |
| Chromobacterium violaceum | ||
| Wild type | ATTC 31532, produces violacein pigment and C6-HL signal | ATCC |
| CV026 | Indicator strain for detection of alkanoyl-AHLs, derivative of 31532 with mini-Tn5, Kmr, cviI | 31 |
| Plasmids | ||
| pHG1 | 12-kb EcoRI fragment from digestion of Hx 699 genomic DNA containing EZ-Tn5 transposon, Kmr | This work |
| pCR2.1 | pUC ori, Kmr Ampr | Invitrogen |
| pHG2 | Same as pCR2.1 but with EcoRI deletion | This work |
| pHG3 | pCR2.1 with rsh and native promoter, Kmr Ampr | This work |
| pRK290 | Broad host range, oriT and oriV, Tcr | 14 |
| pHG4 | pRK290 with rsh and native promoter, Tcr | This work |
| pSB401 | luxR+ PluxI-luxCDABE Tcr p15A ori | 58 |
16S rRNA sequencing and transposon mutagenesis.
Total genomic DNA of Rr 2-17 was isolated using the QIAamp DNA minikit (Qiagen). Primers FD1 and RD1 (see Table S1 in the supplemental materials) (53) were used to amplify the 16S rRNA sequence of Rr 2-17. PCR conditions included an initial denaturation for 3 min at 95°C followed by 40 cycles of 1 min at 94°C, 30 s at 49°C, and 2 min at 72°C. The amplicons were purified and sequenced. To prepare electrocompetent cells of Rr 2-17, an overnight culture in TYE was diluted (1:10, vol/vol) and cultured for 6 h to reach early log phase (optical density [OD] at 600 nm of 0.3), and the culture was washed three times with 10% glycerol. The cells in glycerol were electroporated with the EZ-Tn5 R6Kγori/KAN-2>Tnp transposon (Epicentre, Madison WI) using a Bio-Rad Gene Pulser at Ec3 mode yielding 3.0 kV. After 2 hours of incubation in SOC (superoptimal broth with catabolite repression) medium, the culture was plated on PD agar medium supplemented with 25 μg/ml of kanamycin.
Screening of mutants for AHL production and signal separation assays using TLC.
Rr 2-17 transposon mutants were tested for AHL production using reporter strain CV026 on TYE-PD agar medium (1:1, vol/vol). Twenty-times-concentrated acidified ethyl acetate (aEtOAc) cell extracts were used for thin-layer chromatography (TLC) analysis and quantitative bioluminescence assays (see below). Concentrated (aEtOAc) extracts were spotted onto the TLC plate origin in 2-μl volumes equal to 40 μl supernatant equivalents unless otherwise noted. The loaded C18 RP-TLC plate (EMD Chemicals, Inc. Gibbstown, NJ) was developed in a 60% methanol-water mobile phase (41). The dried TLC plate was then overlaid with AB minimal medium seeded with early-log-phase suspensions of NTL4(pZLR4) supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (50 μg/ml) and incubated for 24 to 48 h (40). AHL signals from unknown strains can be tentatively identified by the retardation factor (Rf) values of appropriate reference compounds prepared from known bacterial strains that produce previously identified AHL(s) and pure AHL standards on the same TLC plate (31, 40, 41).
Quantification of AHL signals using a bioluminescent AHL biosensor.
AHLs from the bacterial extract samples were detected and quantified using aliquots of an overnight culture of E. coli JM109(pSB401) (58). We used the protocol previously established in our laboratory (40).
Determination of the transposon-disrupted gene in the mutant designated Hx 699.
Total genomic DNA of Hx 699 was isolated using the QIAamp DNA minikit, cut with endonuclease EcoRI, and recircularized using quick T4 ligase (NEB). DNAs were electroporated into TransforMax EC100D pir+ electrocompetent E. coli (Epicentre) and selected on LB agar with kanamycin. DNA sequencing of the insertion site was done using the KAN-2 FP-1 forward primer 5′-ACCTACAACAAAGCTCTCATCAACC-3′ (Epicentre, Madison, WI) and TR1 5′-CTGGCTACCCTGTGGAACAC-3′ (this study). The sequences were then compared with other DNA sequences available online using BLAST. For Southern blot analysis, the labeling and detection were carried out using the digoxigenin High Prime DNA labeling and detection starter kit 1 (Roche Applied Science). The template for the probe was constructed via PCR with mosaic end primers (see Table S1 in the supplemental material).
Construction of plasmids for the expression of rsh with a native promoter in E. coli, Rr 2-17, and Hx 699.
The 2,094-nucleotide rsh open reading frame, 599 nucleotides upstream of the initiation codon, and 46 nucleotides downstream of the stop codon were amplified with primers rshF and rshR (see Table S2 in the supplemental material) and cloned into vector pCR2.1 (Invitrogen) to give pHG3. To express rsh in strain Rr 2-17, we subcloned the 2.8-kb EcoRI rsh fragment with the native promoter into the broad-host-range vector pRK290 to give pHG4 (14). The empty vector control for Rr 2-17 was pRK290. Plasmid DNA from a blue transformant from the rsh cloning experiment with pCR2.1 was digested with EcoRI and separated on an agarose gel, and the 3.9-kb vector-only fragment was gel extracted, recircularized, and transformed into E. coli Top10 (Invitrogen) to give E. coli Top10(pHG2), our empty vector control plasmid for E. coli.
Determination of soluble polysaccharide and cell aggregation.
Three-day-old cultures grown on PD agar medium were scraped from the plates and weighed. The cells were then resuspended in 2 ml of pure water, vortexed, and pelleted by centrifugation. The cell-free supernatant was used to estimate total soluble polysaccharides using the anthrone reaction after a 15-min incubation at 95°C and measuring absorbance at 578 nm (19). Samples were quantified using a standard curve constructed from different concentrations of glucose solution. To measure the development of cell aggregation, cultures were grown in TYE broth with shaking at 150 rpm. After 1 week, the absorbance of the upper suspension of the culture, lacking visible cell-cell aggregates, was measured at 600 nm. The upper suspension was added back to the aggregates and vortexed, and absorbance was determined at 600 nm. The cell aggregation percentage is expressed as 1 − (OD of upper suspension/OD of total bacterial suspension) × 100 (13).
Phylogenetic analyses of Rr2-17 16S rRNA gene and rsh gene sequences.
Phylogenetic analyses of the 16S rRNA gene was performed using the parsimony, minimum-evolution, and neighbor-joining methods (39). A bootstrap consensus tree was inferred from 1,000 pseudoreplicates (17). Branches corresponding to partitions reproduced in fewer than 50% of the bootstrap replicates are collapsed on the consensus tree. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (17). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances shown on the consensus trees were computed using the maximum-composite-likelihood method (46). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (the pairwise deletion option). There were a total of 1,356 positions in the final data set for the 16S rRNA gene. Phylogenetic analyses were performed with PAUP* 4.0 and MEGA 4 (44, 46, 47). The phylogenic analysis of the rsh gene was identical to that of the 16S rRNA gene except for the following modifications. The evolutionary distances were computed using the maximum-composite-likelihood method (46) and are in units of number of base substitutions per site. Codon positions included were first, second, third, and noncoding. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,927 positions in the final data set for the rsh gene.
Nucleotide sequence accession numbers.
The full-length 16S rRNA sequence and entire rsh coding sequence of Rr 2-17 were deposited in GenBank and assigned accession numbers EU984513 and EU984514, respectively.
RESULTS
A previous screen of 128 bacterial isolates from eight field grapevine tumors producing octopine, nopaline, or vitopine led to the isolation of several 3-ketolactose, nopaline, and tartrate utilization-negative and β-galactosidase-positive yellowish colonies which commonly occurred in crown galls in addition to the characteristic A. vitis colonies. They activated reporter gene expression both in the short-chain alkanoyl-AHL sensitive biosensor CV026 and in the broad-range AHL detection biosensor NTL4(pZLR4) (data not shown). Of these a Riesling, nopaline tumor isolate named Rr 2-17 was chosen for further studies.
Identification of Rr 2-17 and chromatographic profile of AHL signals.
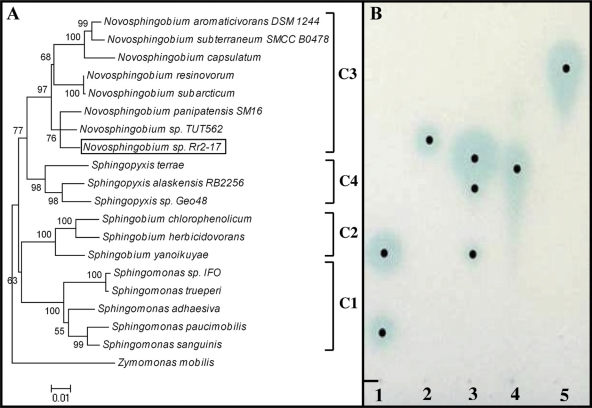
The 16S rRNA sequence was compared with sequences deposited in the NCBI database. The phylogenetic analyses confirm the monophyly of the four ingroup genera, Novosphingobium, Sphingopyxis, Sphingobium, and Sphingomonas, with >90% bootstrap support. The analyses also strongly nest Rr 2-17 within the genus Novosphingobium of the Sphingomonadales order (Fig. 1A) (21). The amplified Rr 2-17 16S rRNA gene is 1,481 nucleotides in length and is 98% identical to the Novosphingobium sp. strain TUT562 16S rRNA gene, 97% identical to the uncultured Novosphingobium sp. clone AV_7R-N-H03 16S rRNA gene (partial sequence), 97% identical to the Sphingomonas sp. strain KAI 16S rRNA gene, and 96% identical to the Novosphingobium aromaticivorans 16S rRNA gene (Fig. 1A).
FIG. 1.
Riesling grapevine nopaline tumor isolate Rr 2-17 is a member of the Novosphingobium genus and produces cell-to-cell communication signals. (A) Phylogenetic position of Rr 2-17 among 18 taxa of the family Sphingomonadaceae based on the 16S rRNA gene. Monophyly of each ingroup genus is confirmed with >90% bootstrap support (Sphingomonas, cluster 1 [C1]; Sphingobium, cluster 2; Novosphingobium, cluster 3; and Sphingopyxis, cluster 4) using parsimony, minimum-evolution, and neighbor-joining methods. These results support the revision of the genus Sphingomonas (60). A member of the Sphingomonadaceae family, Zymomonas mobilis, was used as the outgroup (22). The accession numbers of the sequences used for the construction of the phylogenic tree are presented in Table S3 in the supplemental material. (B) AHL signal profile. Chromatography of extracts and standards with biosensor strain NTL4(pZLR4) overlays was performed with 10 μl of Rr 2-17 supernatant equivalent (lane 3), 1 nmol of C8-HL (lane 1), 10 nmol of C10-HL (lane 1), 250 pmol of C6-HL (lane 2), 3-O-C8-HL produced from A. tumefaciens strain NT1(pTiC58ΔaccR) (1 μl of a 20× preparation) (lane 4), or 10 pmol of 3-O-C6-HL (lane 5). The black dots represent the centers of signals to determine the Rf values. The loading origin is indicated.
Analysis of AHLs by TLC showed that Rr 2-17 produces at least three different AHL signals when grown in liquid media (Fig. 1B). We were able to putatively identify one of the AHLs as C8-HL, which gave an Rf value of 0.27, identical to the Rf value of a pure C8-HL standard (Fig. 1B, lanes 1 and 3). Another AHL signal detected gave an Rf value of 0.47, followed by another at 0.39 (Fig. 1A, lane 3). Neither of these Rf values was identical or near identical to those of the standards of C6-HL (Rf of 0.51, lane 2), 3-O-C8-HL (Rf of 0.45, lane 4), 3-O-C6-HL (Rf of 0.66, lane 5), or C10-HL (Rf of 0.10, lane 1). The structural analysis of the signals produced by Rr 2-17 is currently in progress.
Transposon mutagenesis of strain Rr2-17, screening for an AHL-deficient mutant, and gene disruption in mutant Hx 699.
A mutant deficient in AHL signal production by Rr 2-17 was identified following transposon mutagenesis and named mutant Hx 699. Southern blot analysis showed that Hx 699 contained only one copy of Tn5. The deficiency in AHL signal production by Hx 699 led to the lack of violacein pigment synthesis in the AHL-dependent biosensor strain CVO26 (data not shown).
Cell extracts of Rr 2-17 displayed about 10-fold-higher induction of bioluminescence in JM109(pSB401) (58) than those of Hx 699 (Fig. 2A). Pure C6-HSL (100 nM) activated bioluminescence production to levels similar to those for extracts from Rr 2-17, whereas Hx 699 activation was equivalent to that for the ethyl acetate solvent (Fig. 2A). Hx 699 grown on PD agar medium for 5 days also showed a distinct hypomucoid phenotype in comparison to Rr 2-17 (Fig. 2B). This hypomucoid phenotype of Hx 699 could not be complemented with exogenous AHLs or signals secreted from Rr 2-17 growing adjacent to Hx 699 (data not shown).
FIG. 2.
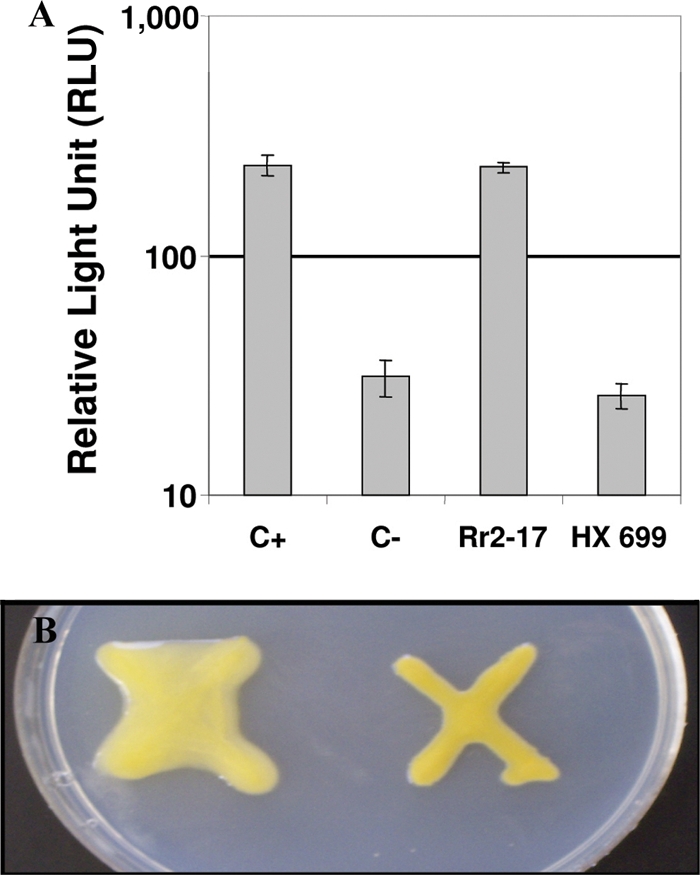
Identification of a mutant deficient in AHL signal synthesis and the hypomucoid phenotype. (A) Identification of AHL deficiency in extracts of Hx 699. Extracts of Rr 2-17 and Hx 699 (10 μl) were assayed. C−, aEtOAc-only negative control; C+, C6-HSL in aEtOAc at 100 nmol (positive control). Treatments were assayed using the LuxR-dependent E. coli biosensor JM109(pSB401). The experiment was repeated three times, and the mean values of replications and standard errors are shown. (B) Hypomucoid phenotype of Hx 699 compared to Rr 2-17. Rr 2-17 and Hx 699 were grown on PD agar medium for 5 days.
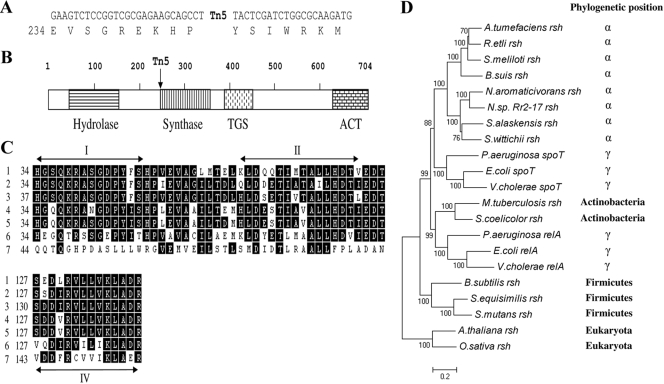
Two E. coli transformants selected on LB supplemented with kanamycin were generated through transformation with recircularized EcoRI-digested Hx 699 genomic DNA. Each transformant contained the same plasmid, and one, named pHG1, was chosen and sequenced. DNA sequencing revealed that an rsh gene was disrupted (Fig. 3A and B). The presence of a transposon mosaic end sequence (see Table S1 in the supplemental material, and data not shown) confirmed that the forward primer annealed to the correct position and started extending toward the disrupted gene.
FIG. 3.
Site of Tn5 insertion; distinct domains of RshNsp; partial alignment of conserved motifs I, II, and IV of the histidine-aspartate domain of the superfamily of metal-dependent phosphohydrolases (1); and phylogenetic analysis. (A) The site of Tn5 insertion is between the proline codon CCT (residue 249) and the tyrosine codon TAC near the beginning of the synthase domain (vertical stripes in panel B). (B) The full-length Rr 2-17 RelA/SpoT homolog (Rsh) contains four conserved superfamily domains, i.e., the histidine-aspartate (HD) or hydrolase (horizontally striped box), synthase (vertically striped box), TGS (vertically dashed box), and ACT (bricked box) domains. The arrow indicates the approximate site of Tn5 insertion in Hx 699. (C) Partial amino acid alignment of the Rsh proteins from Rr 2-17 (line 1; accession number EU984514), Sphingomonas wittichii (line 2; accession number ABQ69873), Sphingopyxis alaskensis (line 3; accession number ABF54382), Agrobacterium tumefaciens (line 4; accession number AAK86838), Rhizobium etli (line 5; accession number ABC90188), and E. coli SpoT (line 6, accession number AAC76674) and RelA (line 7; accession number AAC75826). The alignment was constructed using the program MegAlign (DNAstar) with the Clustal W algorithm. This alignment was against the conserved motifs (I, II, and IV) of the entire superfamily of metal-dependent phosphohydrolases (1). The number before the first amino acid in each sequence indicates the amino acid residue at the beginning of the partial alignments. Motif I shows the conserved region potentially involved in the ppGpp degradation activity (residues RASGKPYF) flanked by conserved histidine (H) residues. Motif II shows the conserved histidine and aspartate (HD) and glutamate and aspartate (ED) signatures, and motif IV shows the conserved aspartate (D) residue. No conserved residues in the Rsh (lines 1 to 5) and SpoT (line 6) proteins are conserved in the RelA protein of E. coli (line 7). (D) Phylogenetic tree based on the nucleotide sequences of relA/spoT and relA/spoT-like homologs (rsh genes), showing relationships of rsh sequences from 20 taxa, including the Novosphingobium sp. strain Rr 2-17 Rsh protein. The rsh gene sequences from the higher plants Arabidopsis thaliana and Oryza sativa were used as outgroups to root the tree. The new sequence is confidently (100% bootstrap) nested within the alphaproteobacterial group. The results confirm a close sister relationship of the rsh sequence of the new isolate Rr 2-17 to that from N. aromaticivorans using parsimony, minimum-evolution, and neighbor-joining methods with 100% bootstrap support. The accession numbers of the sequences used for the construction of the phylogenetic tree are presented in Table S2 in the supplemental material.
The open reading frame of the full-length Novosphingobium sp. rsh gene (rshNsp) is 2,094 nucleotides in length. Phylogenic analysis of rsh was performed (Fig. 3D; see Table S3 in the supplemental material). The Rr 2-17 rsh gene encodes a protein of 704 amino acids that is 37% identical to E. coli SpoT and 31% identical to E. coli RelA. The deduced Rr 2-17 Rsh protein sequence also displays high similarity to Rsh protein sequences from certain alphaproteobacteria, particularly those of the closely related Sphingomonadaceae family (79% to Novosphingobium aromaticivorans, 66% to Sphingopyxis alaskensis, and 61% to Sphingomonas wittichii) and plant-associated alphaproteobacteria (43% to Agrobacterium tumefaciens and 44% to Rhizobium etli).
Like the E. coli SpoT protein, RshNsp contains the N-terminal phosphohydrolase (HD) domain and the (p)ppGpp synthase domain and thus has a proposed bifunctional role (Fig. 3B). RshNsp also retains the TGS and ACT domains associated with RelA, SpoT, and Rsh proteins of most other microorganisms (1, 33). The TGS domain plays a role in the acyl carrier protein, while the C-terminal half of the protein contains the ACT domain, which is predicted to act in regulation of the opposing catalytic functions, (p)ppGpp synthesis and degradation, of SpoT and Rsh proteins (Fig. 3B). Unlike RelA, Rsh contains a domain called the HD domain which is conserved in a superfamily of metal-dependent phosphohydrolases (1, 33). The sequences of RelA and SpoT from E. coli are related, and it is challenging to assign (p)ppGpp hydrolase or (p)ppGpp synthase activities based on sequence analyses. The histidine (H) and aspartate (D) residues in motif II of the HD domain are thought to be involved in (p)ppGpp degradation (1, 23). RshNsp was compared with the Conserved Domain Database and shown to have a region that can align with the HD domains of metal-dependent phosphohydrolases. Partial alignment of RshNsp with its homologs from Sphingomonadaceae, A. tumefaciens, R. etli, and E. coli showed that motifs I, II, and IV of the HD domain are highly conserved in Rsh and SpoT but not in RelA (Fig. 3C). The presence of only one relA/spoT homolog (rsh) in the five completely sequenced genomes of the Sphingomonadales order (21) and the similarity of their products to RshNsp suggest that these homologs have synthetase and hydrolase activities. The five genomes of the Sphingomonadales sequenced to date include those of N. aromaticivorans DSM 12444 (accession number CP000248), S. alaskensis RB2256 (accession number CP000356), S. wittichii RW1 (accession number CP000699), Erythrobacter litoralis HTCC2594 (accession number NC_007722), and Zymomonas mobilis subsp. mobilis ZM4 (accession number AE008692).
Functionality of rshNsp in E. coli.
The rshNsp gene was unable to complement the E. coli strain relA deletion in CF1652(pHG3) to restore growth on SMG medium. However, when rshNsp in pHG3 was transformed into CF1693, an E. coli relA spoT double mutant, restoration of the growth phenotype was observed on SMG medium (Fig. 4A, B, and C). This implies that (p)ppGpp levels due to the balance of the synthetase/hydrolase of rshNsp are not high enough to saturate the hydrolase activity of CF1652, raising basal levels sufficiently for SMG resistance. However, this is not the case for the hydrolase-deficient CF1693, and growth on SMG is obtained. CF1693 grows well in rich medium but not on SMG agar medium due to a deficiency in (p)ppGpp production resulting in the inability to overcome isoleucine starvation. The ability of rshNsp to restore growth of CF1693 on SMG medium requires both the reversal of multiple amino acid auxotrophy due to the accumulation of adequate levels of (p)ppGpp and induction of ile synthase genes to allow growth on SMG medium (Fig. 4A, B, and C). For confirmation, we used aminotriazole (AT) minimal medium supplemented with all amino acids except histidine to confirm that rshNsp complements the growth defect in CF1693 but not in CF1652 (Fig. 4A, B, and D). AT is a histidine analog that induces histidine starvation in E. coli. On AT-containing minimal medium supplemented with all amino acids except histidine, only strains that produce enough ppGpp to activate histidine biosynthesis can grow (20).
FIG. 4.
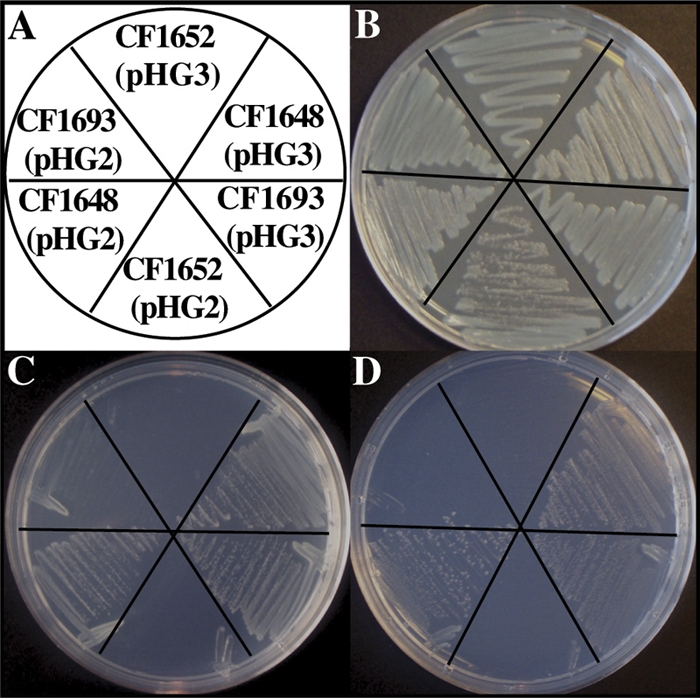
Functional analysis of RshNsp in E. coli mutants. (A) Complementation analysis. E. coli relA mutant CF1652, E. coli relA spoT double mutant CF1693, and E. coli wild-type strain CF1648 were used. Each strain is with rsh (pHG3) and without rsh (pHG2, empty vector). (B) Growth of the same strains as in panel A on rich undefined LB agar medium. (C) Restoration of growth of the E. coli relA spoT double mutant transformed with pHG3 on M9-glucose medium supplemented with the amino acids serine, methionine, and glycine at 1 mM each (SMG medium). (D) Restoration of growth of the E. coli relA spoT double mutant transformed with pHG3 on AT medium.
Mutation in rsh leads to soluble polysaccharide production and promotes cell aggregation.
The mucoid phenotype of Rr 2-17 did not change with the presence of the pRK290 vector control or pHG4. In contrast, the hypomucoid phenotype of Hx 699 was complemented by pHG4 but not pRK290. In Hx 699(pHG4) the mucoid phenotype was restored to one equivalent to those of Rr 2-17(pRK290) and Rr 2-17(pHG4) (data not shown). Consistent with the hypomucoid phenotype, measurement of total soluble polysaccharide showed that Hx 699(pRK290) produces about 30% of the soluble polysaccharide produced by the trans-complemented Hx 699(pHG4) and Rr 2-17 containing pRK290 or pHG4 (Fig. 5A).
FIG. 5.
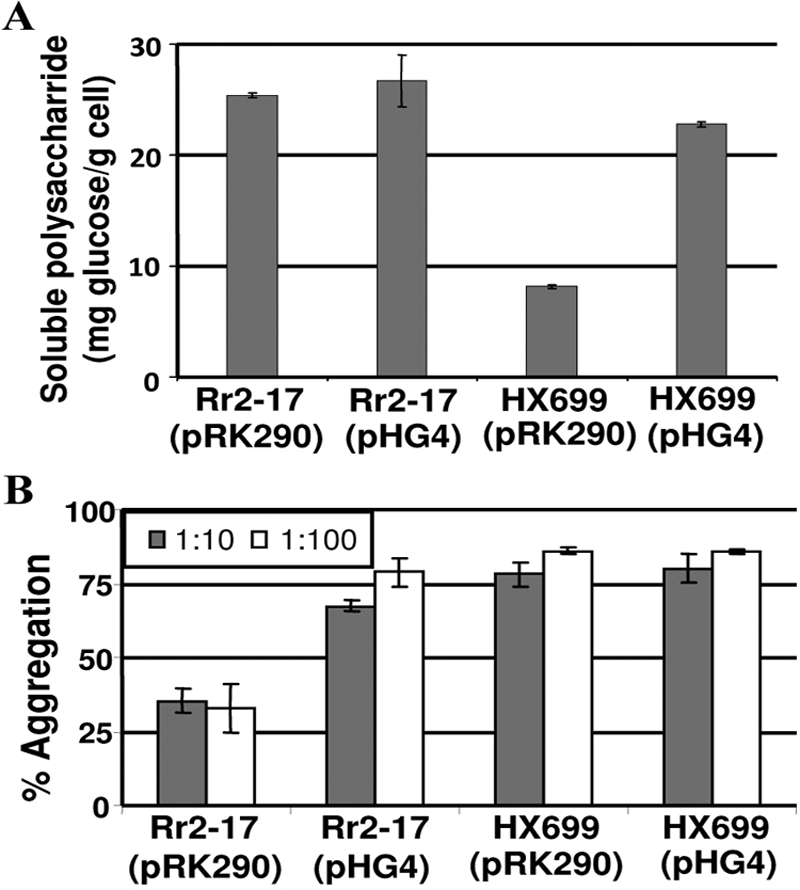
Quantification of soluble polysaccharides and cell aggregation by Rr 2-17 and Hx 699. (A) Quantification of soluble polysaccharides from Rr 2-17 and Hx 699 with and without rsh. Each strain was grown for 3 days on PD agar medium. Cells were harvested in sterile water, and total soluble glucose was determined. Rr 2-17(pHG4), Rr 2-17 containing extra copies of rsh; Hx 699(pHG4), Hx 699 containing rsh. The data are means from three replications, and bars represent standard deviations. (B) Quantification of cell aggregation. The dilution of the overnight cultures at the beginning of the experiments is indicated. Cultures were grown in each experiment for 7 days on TYE medium.
We also tested the four constructed strains for cell aggregation. The cell aggregates were minimal for Rr 2-17(pRK290). Cell aggregates that became pronounced as the length of the experiment increased to 7 days were evident in cultures of Rr 2-17(pHG4) and of Hx 699 with pHG4 or pRK290 (Fig. 5B).
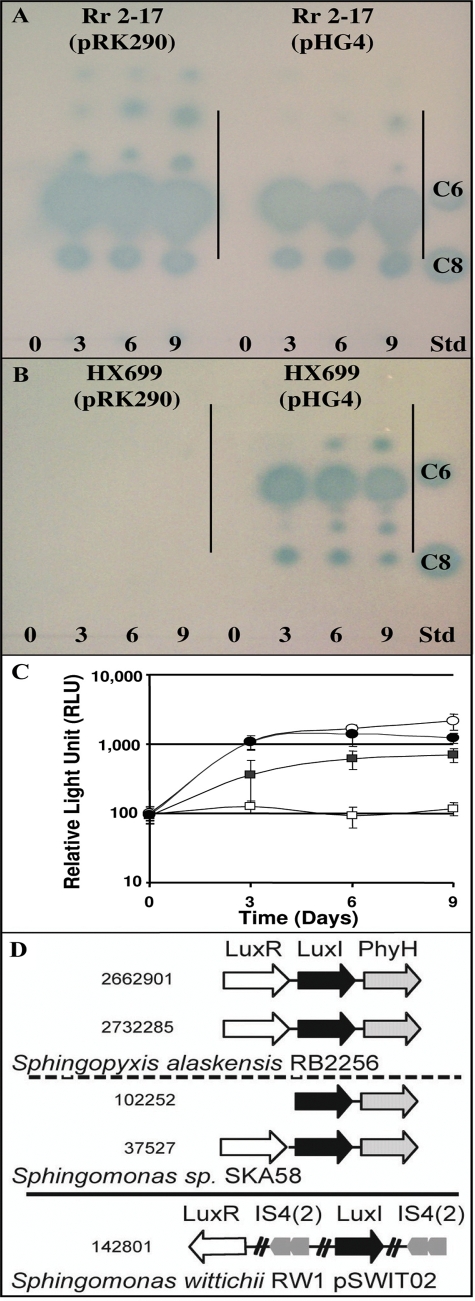
AHL signal accumulation in wild-type and mutant strains with and without multiple copies of rshNsp and genomic context of LuxI homologs in genomes of the Sphingomonadales.
When grown on PD agar medium, AHL signal accumulation in Rr 2-17(pRK290) was not altered after 9 days of growth (Fig. 6A). The number of AHLs and their detection intensities from extracts of Rr 2-17(pHG4) were less than those of the same host with pRK290 (Fig. 6A). The number of AHL signals produced by Rr 2-17 on rich agar medium appears to be between five and seven, using four times the amount of extract used in the TLC shown in Fig. 1B (Fig. 6A). Multicopy rshNsp (pHG4) partially complemented AHL production by Hx 699(pHG4) after growth for 9 days. Hx 699(pHG4) accumulated near-wild-type numbers of AHLs but at slightly reduced intensities (Fig. 6B) and at reduced levels when total AHLs were measured (Fig. 6C), whereas Hx 699(pRK290) failed to accumulate any detectable AHLs (Fig. 6B). This complementation pattern suggests that the AHL QS system in Rr 2-17 is regulated by rshNsp and that our rsh clone complements the rsh mutation in Hx 699.
FIG. 6.
(A to C) Analysis of QS molecules from Rr 2-17 and Hx 699 grown on rich PD agar (A and B) and estimation of total accumulated AHL signals detectable with a LuxR-dependent biosensor (C). (A) AHL signal accumulation in Rr 2-17(pRK290) and Rr 2-17 with extra copies of rsh on pHG4 at 0, 3, 6, and 9 days. The culture extract applied per lane is equal to 40 μl of culture supernatant. AHL standards of C6-HL (100 pmol) and C8-HL (33 pmol) are included in the standard lane (Std). The origin of loading is indicated. (B) AHL signal accumulation in Hx 699(pRK290) and complemented Hx 699(pHG4) at 3, 6, and 9 days. (C) Quantification of AHL signal accumulation in aEtOAc extracts of Rr 2-17(pRK290) (open circles), Rr 2-17(pHG4) (solid circles), Hx 699(pRK290) (open squares), and Hx 699(pHG4) (solid squares) at 0, 3, 6, and 9 days. The same samples were used in the TLC assays (A and B). The extract assayed was equivalent to 400 μl of culture supernatant. The experiment was repeated three times, and the mean values and standard errors are shown. (D) Alignment of gene context/neighborhoods around luxI homologs in the sequenced genomes of the Sphingomonadales order. Genes that have conserved neighborhoods are shown by boxed arrows, and the direction of the arrow indicates the transcriptional direction. Names of representative encoded protein are given, as are the species in which it is present and the approximate nucleotide location number of each LuxI (solid black arrows). Broken cross lines in the S. wittichii RW1(pSWIT02) LuxI gene context indicate intervening regions contain additional genes.
To acquire functional insights into LuxI homologs in the Sphingomonadales by means of contextual information, we searched for LuxI homologs in the sequenced bacterial genomes as a data set (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi, accessed on 19 September 2008). The S. alaskensis RB2256 (accession number NC_008048) and Sphingomonas sp. strain SKA58 (accession number NZ_AAQG01000009) genomes each contained two LuxI homologs, and plasmid pSWIT02 of the S. wittichii RW1 (accession number NC_009508) genome contained one LuxI homolog. Three of the LuxI homologs, two from S. alaskensis RB2256 and one from Sphingomonas sp. strain SKA58, showed a partner LuxR homolog directly adjacent and transcribed in the same direction, each encompassing a complete QS regulatory circuit (Fig. 6D) (6). The single LuxI homolog of S. wittichii RW1 contained in pSWIT02 was physically distant from its potential partner LuxR homolog (Fig. 6D). This LuxI homolog is flanked by insertion sequences of the IS4 family of transposases encompassing an approximately 18-kb fragment in pSWIT02. This gene context separates the LuxI homolog from its predicted receptor (LuxR homolog) in pSWIT02 of S. wittichii and contrasts with the gene context found around the LuxI homologs in the S. alaskensis RB2256 and Sphingomonas sp. strain SKA58 genomes (Fig. 6D).
DISCUSSION
In this work, we describe a grapevine nopaline tumor isolate (Rr 2-17) as a cluster III Novosphingobium sp. strain (Fig. 1A). This further expands the environmental distribution of the Sphingomonas genus. The full-length 16S rRNA gene sequence of Rr 2-17 was elucidated, and the nearest phylogenetic neighbor is 98% identical to that of Novosphingobium sp. strain TUT562 (Fig. 1A). Novosphingobium sp. strain TUT562 was proposed as having a distinct taxonomic position as a single genospecies with the genus Novosphingobium and was described as Novosphingobium naphthalenivorans (43). Further phenotypic and chemotaxonomic work with Rr 2-17 is necessary to validate its status within the Novosphingobium genospecies.
Plant-associated bacteria that produce AHL signals have been known for over a decade (7, 48, 51, 57). However, an analysis of species associated with the diseased tissue of a crown gall tumor has not been reported. This is the first report of a nonagrobacterial species that produces AHL isolated from a crown gall tumor (Fig. 1). Since the TraR-based biosensor NTL4(pZLR4) can detect the AHLs produced by Rr 2-17, it is tempting to speculate that in nature nonagrobacterial tumor-colonizing strains can donate signals and influence pTi replication and conjugation in plant tumors and thus influence virulence of A. tumefaciens and A. vitis. The inability of Rr 2-17 to utilize nopaline secreted from nopaline crown gall tumor as a sole source of carbon appears to indicate that its purpose in putatively increasing virulence of A. vitis and indirectly causing crown gall tumor is to alter the nutrient composition in the microbial population and perhaps lessen its competition. However, it is also possible that intermediates in nopaline catabolism could be utilized by Rr 2-17. The AHL signals produced by Rr 2-17 are in the process of being structurally characterized; however, Rr 2-17 appears to produce multiple short-chain AHL signals, and their synthesis varies depending upon growth in liquid or agar medium (Fig. 1B and 6 and data not shown). The identification of LuxI orthologs in sequenced members of the Sphingomonadales (Fig. 6D) is strong evidence that a QS system based on AHL signals may be common to certain species of this order. We are currently identifying the luxI homologs of Rr 2-17. Further analysis aided by the sequencing of the Rr 2-17 genome will be useful in gathering information about genes involved in QS signal production and perception.
Our results show that a mutation in a single gene, rsh, that encodes a member of the RelA/SpoT-like family of proteins affects Rr 2-17 in different ways, which suggests its role in the higher gene regulatory circuit. The full-length nucleotide sequence of rshNsp revealed that it shares significant percent sequence identity with rsh genes of other members of the Sphingomonadaceae family as well as other plant-associated alphaproteobacteria, including A. tumefaciens and R. etli (Fig. 3; see Table S2 in the supplemental material). Bioinformatic analyses of the conserved domains in rshNsp and known rsh and spoT genes show significant conserved percent amino acid identity with the conserved motifs I, II, and IV of the entire superfamily of metal-dependent phosphohydrolases (Fig. 4C). These results, the results of our additional work with rshNsp complementing the E. coli relA spoT double mutation (Fig. 4), and the fact that only a single rsh gene was found in the five sequenced genomes of the Sphingomonadales order (21) support that our rshNsp codes for a dual-function enzyme displaying both (p)ppGpp synthetase and hydrolase activities (1, 2, 8, 38). A possible explanation for the inability of rshNsp to complement the growth of CF1652 on SMG and AT media may be excess net hydrolase activity relative to net synthase activity, leading to insufficient (p)ppGpp to induce the stringent response (Fig. 4). These observations are consistent with earlier work on the rsh gene of R. etli (4). Further experimental work is necessary to confirm our hypothesis of the bifunctionality of rshNsp.
Phylogenetic analysis placed rshNsp in the rsh group of Novosphingobium, thus providing strong support for the identity of Rr 2-17, in addition to our 16S rRNA analysis (Fig. 3D and 1A). Based on the phylogenetic distribution of rsh, relA, and spoT in this study, we proposed an alternative model for the evolution of (p)ppGpp metabolism proteins. The ancestral rsh gene diverged into more specialized rsh genes, namely, spoT-like in alphaproteobacteria or rel in actinobacteria, while still retaining its dual function. However in beta- and gammaproteobacteria, lateral gene transfer of an additional rsh (rel-like) from an ancestor of actinobacterial origin to the existing spoT-like rsh gene imposed a selection pressure on both rsh genes to coexist, thus causing differentiation to an even more specific protein (high hydrolase/low synthase [SpoT] and high synthase/no hydrolase [RelA]).
In our work, rshNsp was found to be involved in the regulation of the cell density-dependent QS regulon (Fig. 6). The stringent response is induced during the transition from a nutrient-sufficient/low-stress environment to low-nutrient/high-stress conditions and occurs simultaneously with the increase in cell density, as exemplified during the transition from the exponential to the stationary growth phase in a culture. By setting nutrient availability as a determinant for QS, it is possible to prevent unnecessary utilization of nutrients for the synthesis of the QS signal. It is worth noting that the AHL profile in Hx 699(pHG4) is noticeably less intense than that in the wild-type strain. The presence of a stop codon upstream of the R6Kγori origin of replication in the transposon which is in frame with the rsh open reading frame (data not shown) and the transposon insertion downstream of the hydrolase domain (Fig. 3B) might lead to the expression of a truncated RshNsp protein which still retains hydrolase activity but lacks regulatory activity. Such a scenario would lead to a nonoptimal (p)ppGpp level in Hx 699(pHG4) to achieve a wild-type AHL profile. Other workers have identified an effect of relA/spoT-like genes on QS signals produced by other alpha- and gammaproteobacteria. In the plant-symbiotic bacterium R. etli, the expression of the luxI homologs, cinI and raiI, and the accumulated levels of the corresponding AHL signals were reduced in a relA mutant (35). In the opportunistic pathogen P. aeruginosa, overexpression of relA led to premature expression of the las and rhl QS circuitry (16, 50). Interestingly, the AHL degradation system of the plant pathogen A. tumefaciens has been shown to be regulated by the alarmone (p)ppGpp (63). In a study of rhizosphere isolates, a member of the Sphingopyxis genus that produces and degrades AHL was identified, and this feature has been found with only three additional strains, P. aeruginosa, A. tumefaciens, and Variovorax paradoxus (24, 25, 30, 48). It will be interesting to determine if Rr 2-17 is capable of degrading AHL.
In our study, the absence of rsh resulted in the hypomucoid phenotype and a reduction of total polysaccharides (Fig. 2B and 5A), and exogenously or endogenously produced AHLs supplied to Hx 699 failed to rescue this phenotype, suggesting that it is not controlled solely through the QS circuitry. Members of the Sphingomonadaceae and other bacteria isolated from terrestrial sources, such as soil, produce large quantities of extracellular polymeric substances composed largely of polysaccharides and smaller amounts of protein and DNA that form a matrix in which the cells are embedded (12, 18). The regulation of this phenotype by rsh is consistent with the fact that a polymeric matrix provides a stable protective milieu for bacterial growth and survival, particularly under stress environments, and to facilitate transmission by acting as a source for the dissemination of microorganisms (15, 22). In contrast, the results of our cell aggregation assay show that only in Rr 2-17(pRK290) did planktonic cells dominate the culture after 7 days of growth, suggesting that the disruption of rshNsp shifts the balance to favor cell aggregates (Fig. 5B).
The results of this work contribute to a growing body of evidence that links bacterial lifestyles to the nutritional/stress status of the environment and host (6, 8, 9, 26, 27). Adaptation is essential for surviving periods of stress and nutrient depletion but is also important in the interaction of bacteria with plant hosts during colonization in symbiotic and pathogenic interactions (2, 38). The rsh genes of alphaproteobacteria, which are involved in the regulation of the (p)ppGpp alarmone, have been implicated in the regulation of pathways in pathogenic and symbiotic bacteria. The list of genera can now be extended to include the grapevine tumor-associated commensal bacteria such as Novosphingobium sp. strain Rr 2-17 and its rshNsp gene. There is a growing amount of information suggesting that ppGpp made by chloroplasts enhances the adaptive ability of plants under stress (2), and it is interesting to speculate that Rr 2-17, by producing (p)ppGpp, donates this signal to the plant crown gall tumor cells, thereby playing an adaptive role in the stress of the diseased plant. Conversely, under microbial stress, a plant host may donate (p)ppGpp to colonizing Rr 2-17. Additional work is aimed at cloning and characterization of the QS circuitry in Rr 2-17 and elucidating its association with the stringent response.
Supplementary Material
Acknowledgments
We thank Michael Cashel, National Institute of Health, Bethesda, MD, for providing E. coli strains and plasmids and particularly useful discussions. We thank Stephen K. Farrand, Department of Microbiology, University of Illinois at Urbana-Champaign, Urbana, IL, for providing bacterial biosensor strain NTL4(pZLR4), and we thank the anonymous reviewers of this work for their insightful comments and suggestions. We also thank Dawn Carter for technical support throughout this work and Michael Cashel for critically reviewing an earlier version of the paper.
Ernő Szegedi was the recipient of Hungarian National Scientific Foundation (OTKA) grant no. K-68053. Han Ming Gan and Michael Savka were supported by the College of Science, Department of Biological Sciences, Rochester Institute of Technology, Rochester, NY, and Han Ming Gan was also supported by a Merck/AAAS grant for undergraduate research.
Footnotes
Published ahead of print on 6 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal dependent phosphohydrolases. Trends Biochem. Sci. 23469-472. [DOI] [PubMed] [Google Scholar]
- 2.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 1445-54. [DOI] [PubMed] [Google Scholar]
- 3.Burr, T. J., C. Bazzi, S. Süle, and L. Otten. 1998. Crown gall of grape: biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis. 821288-1297. [DOI] [PubMed] [Google Scholar]
- 4.Calderon-Flores, A., G. Du Pont, A. Huerta-Saquero, H. Merchant-Larios, L. Servin-Gonzalez, and S. Duran. 2005. The stringent response is required for amino acid and nitrate utilization, Nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J. Bacteriol. 1875075-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Case, R. J., M. Labbate, and S. Kejelleberg. 2008. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2345-349. [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., D., J. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 7.Cha, C., P. Gao, Y.-C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 111119-1129. [DOI] [PubMed] [Google Scholar]
- 8.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1795854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4160-165. [DOI] [PubMed] [Google Scholar]
- 10.Chilton, M.-D., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detectable in crown gall tumors. Proc. Natl. Acad. Sci. USA 713672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Angelo-Picard, C., D. Faure, I. Penot, and Y. Dessaux. 2005. Diversity of N-acyl homoserine lactone-producing and degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 71796-1808. [DOI] [PubMed] [Google Scholar]
- 12.Davey, M. E., and G. G. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Re, B., A. Busetto, G. Vignola, B. Sgorbati, and D. L. Palenzona. 1998. Autoaggregation and adhesion ability in a Bifidobacterium suis strain. Lett. Appl. Microbiol. 27307-310. [PubMed] [Google Scholar]
- 14.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 777347-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and, D. G., Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 725638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 18.Flemming, H. C., R. T. Neu, and D. J. Wozniak. 2007. The EPS matrix: the house of biofilm cells. J. Bacteriol. 1897945-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg. 1994. Methods for general and molecular bacteriology, p. 518. American Society for Microbiology, Washington, DC.
- 20.Gropp, M., Y. Strausz, M. Gross, and G. Glaser. 2001. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 183570-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, R. S., and A. Mok. 2007. Phylogenomics and signature proteins for the alpha Proteobacteria and its main groups. BMC Microbiol. 7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 295-108. [DOI] [PubMed] [Google Scholar]
- 23.Hogg, T., U. Mechold, H. Malke, M. Cashel, and R. Hilgenfeld. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p) ppGpp metabolism during the stringent response. Cell 11757-68. [DOI] [PubMed] [Google Scholar]
- 24.Hu, J. Y., Y. Fan, Y. H. Lin, H. B. Zhang, S. L. Ong, N. Dong, J. L. Xu, W. J. Ng, and L. H. Zhang. 2003. Microbial diversity and prevalence of virulent pathogens in biofilm development in a water reclamation system. Res. Microbiol. 154623-629. [DOI] [PubMed] [Google Scholar]
- 25.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 695941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain, V., M. Kumar, and D. Chatterji. 2006. ppGpp: stringent response and survival. J. Microbiol. 441-10. [PubMed] [Google Scholar]
- 27.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 161260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseleau-Petit, D., D. Vinella, and R. D'Ari. 1999. Metabolic alarms and cell division in Escherichia coli. J. Bacteriol. 1819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer, G. F., J. C. Baker, and B. N. Ames. 1988. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and pppGpp as components of an adaptive response. J. Bacteriol. 1702344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 1826921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1433703-3711. [DOI] [PubMed] [Google Scholar]
- 32.Meade, H. M., and E. R. Signer. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 742076-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p) ppGpp synthetases/hydrolases (the Rel, RelA, and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3585-600. [PubMed] [Google Scholar]
- 34.Mocali, S., E. Bertelli, F. Di Cello, A. Mengoni, A. Sfalanga, F. Viliani, A. Caciotti, S. Tegli, G. Surico, and R. Fani. 2003. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res. Microbiol. 154105-114. [DOI] [PubMed] [Google Scholar]
- 35.Moris, M., K. Braeken, E. Schoeters, C. Verreth, S. Beullens, J. Vanderleyden, and J. Michiels. 2005. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J. Bacteriol. 1875460-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otten, L., T. Burr, and E. Szegedi. 2008. Agrobacterium: a disease-causing bacterium, p. 1-46. In T. Tzfira and V. Citovsky (ed.), Agrobacterium: from biology to biotechnology. Springer, New York, NY.
- 37.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs) J. Gen. Appl. Microbiol. 491-19. [DOI] [PubMed] [Google Scholar]
- 38.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 6235-51. [DOI] [PubMed] [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 40.Scott, R. A., J. Weil, P. T. Le, P. Williams, R. G. Fray, S. Beck von Bodman, and M. S. Savka. 2006. Long- and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil. Mol. Plant-Microbe Interact. 19227-239. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 946036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, S., and A. Hiraishi. 2007. Novosphingobium naphthalenivorans sp. nov., a naphthalene-degrading bacterium isolated from polychlorinated-dioxin-contaminated environments. J. Gen. Appl. Microbiol. 53221-228. [DOI] [PubMed] [Google Scholar]
- 44.Swofford, D. L. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA.
- 45.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of the phylogenetic and chemotaxonomic analyses. Int. J. Syst. Bacteriol. 511405-1417. [DOI] [PubMed] [Google Scholar]
- 46.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 47.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 10111030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uroz, S., C. d'Angelo, A. Carlier, M. Elasri, C. Sicot, A. Petit, P. Oger, D. Faure, and Y. Dessaux. 2003. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 1491981-1989. [DOI] [PubMed] [Google Scholar]
- 49.Uzan, M., and A. Danchin. 1976. A rapid test for the relA mutation in E. coli. Biochem. Biophys. Res. Commun. 69751-758. [DOI] [PubMed] [Google Scholar]
- 50.Van Delden, C., R. Comte, and M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1835376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Bodman, S. B., J. M. Wiley, and S. P. Diggle. 2008. Cell-cell communication in bacteria: united we stand. J. Bacteriol. 1904377-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, J., N. Gardiol, T. Burr, G. P. C. Salmond, and M. Welch. 2007. RelA-dependent (p)ppGpp production controls exoenzyme synthesis in Erwinia carotovora subsp. atroseptica. J. Bacteriol. 1897643-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weisburg, G. W., S. M. Barns, D. A. Pelletier, and D. A. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 171697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 431115-1127. [DOI] [PubMed] [Google Scholar]
- 55.White, C. E., and S. C. Winans. 2007. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos. Trans. R. Soc. London B 3621135-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, D. C., S. D. Sutton, and D. B. Ringelberg. 1996. The genus Sphingomonas; physiology and ecology. Curr. Opin. Biotechnol. 7301-306. [DOI] [PubMed] [Google Scholar]
- 57.Williams, P. 2007. Quorum sensing, communication and cross-kingdom signaling in the bacterial world. Microbiology 1533923-3938. [DOI] [PubMed] [Google Scholar]
- 58.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl-homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163185-192. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spot null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]
- 60.Yabuuchi, E., Y. Kosako, N. Fujiwara, T. Nake, I. Matsunaga, H. Ogura, and K. Kobayashi. 2002. Emendation of the genus Sphingomonas Yabuuchi et al. 1990. and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int. J. Syst. Evol. Microbiol. 521485-1496. [DOI] [PubMed] [Google Scholar]
- 61.Yabuuchi, E., I. Yano, H. Oyaizu, Y. Hashimoto, T. Ezaki, and H. Yamamoto. 1990. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol. Immunol. 3499-229. [DOI] [PubMed] [Google Scholar]
- 62.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13amp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, H. B., C. Wang, and L. H. Zhang. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p) ppGpp. Mol. Microbiol. 521389-1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.