Abstract
Many human pathogens, including Corynebacterium diphtheriae, the causative agent of diphtheria, use host compounds such as heme and hemoglobin as essential iron sources. In this study, we examined the Corynebacterium hmu hemin transport region, a genetic cluster that contains the hmuTUV genes encoding a previously described ABC-type hemin transporter and three additional genes, which we have designated htaA, htaB, and htaC. The hmu gene cluster is composed of three distinct transcriptional units. The htaA gene appears to be part of an iron- and DtxR-regulated operon that includes hmuTUV, while htaB and htaC are transcribed from unique DtxR-regulated promoters. Nonpolar deletion of either htaA or the hmuTUV genes resulted in a reduced ability to use hemin as an iron source, while deletion of htaB had no effect on hemin iron utilization in C. diphtheriae. A comparison of the predicted amino acid sequences of HtaA and HtaB showed that they share some sequence similarity, and both proteins contain leader sequences and putative C-terminal transmembrane regions. Protein localization studies with C. diphtheriae showed that HtaA is associated predominantly with the cell envelope when the organism is grown in minimal medium but is secreted during growth in nutrient-rich broth. HtaB and HmuT were detected primarily in the cytoplasmic membrane fraction regardless of the growth medium. Hemin binding studies demonstrated that HtaA and HtaB are able to bind hemin, suggesting that these proteins may function as cell surface hemin receptors in C. diphtheriae.
Corynebacterium diphtheriae is a gram-positive bacterium and the etiological agent of diphtheria. Studies of C. diphtheriae have focused primarily on the structural characterization and genetic regulation of diphtheria toxin, a secreted exotoxin that is responsible for much of the morbidity and mortality associated with human infection by this pathogen (9). Expression of diphtheria toxin is repressed by iron, and transcription of the structural gene, tox, is regulated by the diphtheria toxin repressor protein, DtxR, in association with iron (4, 36). DtxR is a global iron-dependent repressor in C. diphtheriae that controls the expression of at least 50 genes at more than 20 different promoters (7, 14, 15, 29, 34, 38). While iron-dependent regulation of bacterial virulence factors has been well studied, it is also known that acquisition of iron from the extracellular environment is often critical for microbial pathogens to cause disease (5, 6). Bacteria have developed a variety of mechanisms for uptake and utilization of iron. These mechanisms include high-affinity siderophore uptake systems (48) and binding protein-dependent transporters that facilitate the acquisition of iron from various host sources, including transferrin, lactoferrin, and heme, which is bound by various host proteins, such as hemoglobin (27, 35, 42). Siderophore transport systems are ubiquitous in bacteria, and it has been known for many years that C. diphtheriae secretes a siderophore and contains a siderophore-specific uptake system (10, 30). The structure of the C. diphtheriae siderophore has not been determined; however, a gene required for siderophore biosynthesis and genes encoding an ABC-type siderophore transporter were recently identified in C. diphtheriae and were shown to be regulated by DtxR and iron (15).
Hemin is the oxidized form of heme that is found in extracellular environments, and it is the form of heme transported by bacterial uptake systems. Hemin iron transport and utilization systems have been identified in numerous bacterial species and were initially described in gram-negative pathogens, where it was shown that an outer membrane receptor and a periplasmic binding protein-dependent ABC-type transporter are required for hemin uptake (13, 18, 41). Certain gram-negative species also obtain hemin iron through the use of hemophores, which are low-molecular-weight secreted hemin binding proteins that are able to extract hemin from hemoglobin and then transfer the hemin to receptors in the bacterial outer membrane (20).
Hemin uptake systems have recently been described in gram-positive bacteria, including Staphylococcus aureus (24, 46), Streptococcus pyogenes (3, 19), and Corynebacterium species (11, 35). S. aureus binds hemin or hemoglobin to its cell envelope through various surface-anchored proteins, termed iron-regulated surface determinants (Isd), which contain sortase recognition signals at their C termini that are essential for the covalent linkage of these proteins to the peptidoglycan (24, 46). It is believed that hemin is transported through the cell envelope via a cascade mechanism in which hemin is transferred between various Isd receptors (22, 50). In S. pyogenes, hemin is proposed to bind initially to surface-exposed proteins, designated Shp and Shr, which appear to be anchored to the membrane by a putative transmembrane region in their C termini (21, 49). In both S. aureus and S. pyogenes it is thought that hemin is transferred from the surface-anchored proteins to a substrate binding component that is associated with a heme-dependent ABC transporter that facilitates the passage of hemin into the cytosol.
In Corynebacterium ulcerans, the transport of hemin involves use of the ABC-type transporter encoded by the hmuTUV genes, which share sequence similarity to genes encoding hemin uptake systems in other organisms, including C. diphtheriae (11, 35). The hmuTUV genes in C. diphtheriae were shown to complement an hmuTUV mutation in C. ulcerans, and it was further demonstrated that the HmuT protein in C. diphtheriae is a hemin binding lipoprotein that is tethered to the cytoplasmic membrane by an N-terminal lipid moiety (11). An insertion mutation in hmuT had no effect on hemin iron utilization in C. diphtheriae; however, it is suspected that the medium conditions used in the study were not optimal for detection of a hemin uptake deficiency (16, 35). The completion of the genome sequence of a clinical isolate of C. diphtheriae (8) resulted in identification of htaA, an iron- and DtxR-regulated gene that is located immediately upstream of hmuT (14). No promoter activity was detected in the htaA-hmuT intergenic region, and it was suspected that htaA and the hmuTUV genes may constitute a DtxR-regulated operon (14, 35). Two other genes, designated htaB and htaC, were also identified in this hemin transport gene cluster. The function of the predicted products of htaA, htaB, and htaC has not been determined; however, the linkage of these genes to the hmuTUV operon suggests that they have a possible function in hemin uptake or hemin iron utilization. The use of heme as an iron source in C. diphtheriae also involves the hmuO gene, which encodes a heme oxygenase that catalyzes the degradation of intracellular heme and the subsequent release of the heme-associated iron (16, 31, 47). The hmuO gene in C. diphtheriae is not linked to the hemin transport gene cluster, but transcription of hmuO is regulated by DtxR and iron, as well as by heme (32, 33).
In this study, we extended our characterization of the hemin transport locus, designated hmu, to examine the expression and cellular localization of specific hmu gene products. We show here that the hmuTUV and htaA genes are involved in the use of hemin as an iron source in C. diphtheriae. Protein localization studies revealed that HtaA, HmuT, and HtaB are associated predominantly with the cytoplasmic membrane when bacteria are grown in minimal medium, which suggests that these proteins may function to transport hemin. We also demonstrate that HtaA and HtaB are surface-exposed hemin binding proteins that may function as hemin receptors in C. diphtheriae.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli and C. diphtheriae strains used in this study are listed in Table 1. Luria-Bertani (LB) medium was used for culturing E. coli, and heart infusion broth (Difco, Detroit, MI) containing 0.2% Tween 80 (HIBTW medium) was used for routine growth of C. diphtheriae strains. Bacterial stocks were maintained in 20% glycerol at −80°C. Antibiotics were added to LB medium at concentrations of 34 μg/ml for chloramphenicol, 50 μg/ml for kanamycin, and 100 μg/ml for ampicillin for E. coli cultures and to HIBTW medium at concentrations of 2 μg/ml for chloramphenicol and 50 μg/ml for kanamycin for C. diphtheriae cultures. HIBTW medium was made a low-iron medium by addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) at a concentration of 12 μg/ml (unless indicated otherwise). Modified PGT (mPGT) medium is a semidefined low-iron medium that has been described previously (44). Antibiotics, EDDA, Tween 80, hemin (bovine), and hemoglobin (human) were obtained from Sigma Chemical Co. (St. Louis, MO).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or use | Reference or source |
|---|---|---|
| C. diphtheriae strains | ||
| 1737 | Wild type, Gravis biotype, tox+ | 28 |
| 1737htaAΔ | htaA deletion mutant of 1737 | This study |
| 1737TUVΔ | hmuTUV deletion mutant of 1737 | This study |
| 1737htaBΔ | htaB deletion mutant of 1737 | This study |
| 1737hmuΔ | Deletion mutation in hmu locus of 1737 | This study |
| C7(−) | Wild type, tox | 2 |
| C7dtxR | dtxR(R47H) mutant of C7(−) | This study |
| E. coli strains | ||
| BL21(DE3) | Protein expression | Novagen |
| DH5α | Cloning strain | Invitrogen |
| S17-1 | RP4 mobilization functions | 45 |
| TOP10 | Cloning of PCR fragments | Invitrogen |
| Plasmids | ||
| pCR-Blunt-II-TOPO | Cloning of PCR fragments, Kanr | Invitrogen |
| pCM502 | Promoter probe, Cmr | 32 |
| phtaB-Z | htaB promoter-lacZ fusion in pCM502 | This study |
| pET24a(+) | Expression vector (His fusion), Kanr | Novagen |
| pET28a(+) | Expression vector (His fusion), Kanr | Novagen |
| pGEX-6P-1 | Expression vector (GST fusion) Ampr | GE Healthcare |
| pKN2.6Z | C. diphtheriae shuttle vector, Knr | 11 |
| pKhtaA | pKN2.6Z carrying the htaA gene | This study |
| pCM2.6 | C. diphtheriae shuttle vector, Cmr | 36 |
| pCD842 | pCM2.6 carrying the hmuTUV genes | 11 |
| pCC1 | Copy control vector for htaA cloning | Epicentre |
Mutant construction.
A previously described allelic replacement technique (45) was used to construct nonpolar deletion mutations in the C. diphtheriae 1737 htaA, htaB, and hmuTUV genes, as well as in the complete hmu gene cluster (htaC through htaB). Mutant construction utilized PCR to clone DNA fragments located upstream and downstream of the region targeted for deletion. The deleted regions in the htaA, htaB, and hmuTUV mutants are predicted to encode peptides consisting of approximately 20 amino acids that contain in-frame fusions of residues derived from the N and C termini of the original proteins. Primers used for construction of the mutants are listed in Table 2. PCR was used to confirm the mutations in all of the deletion mutants (not shown). An R47H point mutation was introduced into the C7 dtxR gene using a previously described procedure (15).
TABLE 2.
Primers used in this study
| Primer | Amplified region | Approx fragment size (bp) | Sequencea |
|---|---|---|---|
| Deletion mutants | |||
| htaAUP4 | Upstream of htaA | 1,000 | 5′-GCGGATATCTCGAGTGCTTTGCCAGG-3′ |
| htaAUP2-2 | 5′-GCGGATCCTTGAATTATCACGGTGC-3′ | ||
| htaADN1-2 | Downstream of htaA | 500 | 5′-GCGGGATCCATTTCCAATCCAGAAATTG-3′ |
| htaADN2-2 | 5′-GCGAAGCTTGATATCACACACCACGGACATAC-3′ | ||
| UPB1 | Upstream of htaB | 900 | 5′-GCGGTCGACGGTTGAGATAAATAACC-3′ |
| UPB2 | 5′-GCGAAGCTTGCAGCCTGAGTCGTTGG-3′ | ||
| DNB1-1 | Downstream of htaB | 700 | 5′-GCGGTCGACTCTAGGTTTAGATTGCG-3′ |
| DNB2 | 5′-GCGGATATCCGAACTCGGTGCAGACG-3′ | ||
| UPC1 | Upstream of htaC | 1,100 | 5′-GCGGTCGACCAAACGATCAAGCCACC-3′ |
| UPC2-1 | 5′-GCGAAGCTTGATATCAGCTGATTTTTGCCATAGCG-3′ | ||
| hm1-1 | Upstream of hmuT | 750 | 5′-GCGATATCCAATGGAACCTTGATGG-3′ |
| hm1-2 | 5′-GCGGATCCACAAACACGAAGCAAAGA-3′ | ||
| hm1-3 | Downstream of hmuV | 650 | 5′-GCGGATCCCAAAACCAAGATGATTTTG-3′ |
| hm1-4 | 5′-GCAAGCTTGATATCCAATTGCGAATAAAAGG-3′ | ||
| Promoter fusions | |||
| htaB-PO1 | htaB promoter | 360 | 5′-GCGGTCGACGCGAGGTCGTACTACGC-3′ |
| htaB-PO2 | 5′-GCGGGATCCTCAGCGACAGCTTGAGG-3′ | ||
| Protein expression | |||
| 24FHtA-His | htaA for pET24(a)+ without | 1,600 | 5′-GCGCATATGGCCCACCACCACCACCACCACGCTGATAGCAATCAATGC-3′ |
| RHtA-TM | transmembrane region | 5′-GCGAAGCTTCTAGGGATCAATACCGGAATC-3′ | |
| GEXFHta-GST | htaA for pGEX-6P-1 without | 1,600 | 5′-GCGGATATCAGCTGATAGCAATCAATGC-3′ |
| RXhoHta-TM | transmembrane region | 5′-GCGCTCGAGCTAGGGATCAATACCGGAATC-3′ | |
| 24FHtB-His | htaB for pET24(a)+ | 5′-GCGCATATGGCCCACCACCACCACCACCACGCTGAAGAACCGGCAGCC-3′ | |
| 28FHtB | htaB for pET28(a)+ without | 780 | 5′-GCGCATATGGCTGAAGAACCGGCAGCC-3′ |
| RHtB-TM | transmembrane region | 5′-GCGAAGCTTTTAGTTAAAACCAGAAGTAGA-3′ |
Restriction sites are underlined, stop codons are indicated by bold type, and inserted bases are double underlined.
Hemoglobin iron utilization assays.
The hemoglobin iron utilization assay has been described previously (16). Briefly, C. diphtheriae strains were grown overnight (20 to 22 h at 37°C) in HIBTW medium and then inoculated to an optical density at 600 nm (OD600) of 0.2 into fresh HIBTW medium that contained 12 μg/ml of the iron chelator EDDA. Strains were grown for several hours at 37°C until log phase, at which time bacteria were recovered by centrifugation, resuspended in mPGT medium, and then inoculated at an OD600 of 0.03 into fresh mPGT medium that contained various supplements, as indicated below. After 20 to 22 h of growth at 37°C, the OD600s of the cultures were determined.
Experiments that examined the effect of secreted or purified HtaA on hemoglobin iron utilization used HtaA that was either purified from E. coli or obtained in native form from C. diphtheriae culture supernatants. Culture supernatants were prepared as follows. C. diphtheriae strain 1737hmuΔ containing either the cloned htaA gene on pKhtaA or the vector pKN2.6Z was grown to late log phase in low-iron HIBTW medium, and culture supernatants were concentrated by ammonium sulfate precipitation. One milliliter of a culture supernatant was concentrated to 50 μl and then dialyzed in phosphate-buffered saline (PBS) to remove the ammonium sulfate. The concentrated and dialyzed supernatant (50 μl) was added to 1 ml of mPGT medium in the hemoglobin utilization assay described above. Purified glutathione S-transferase (GST)-HtaA and HtaA were used similarly in the hemoglobin utilization assay, and approximately 1 μg of purified protein was added to 1 ml of mPGT culture medium.
Plasmid construction.
Plasmids used in this study are listed in Table 1. PCR-derived DNA fragments were initially cloned into the pCR-Blunt II-TOPO vector (Invitrogen), and genomic DNA derived from C. diphtheriae strain 1737 was used as a template for all PCRs (unless otherwise indicated). The promoter probe vector pCM502 (32) was used for construction of all lacZ promoter fusions; this vector contains a promoterless lacZ gene and replicates at a low copy number in C. diphtheriae. The htaB promoter-lacZ fusion construct, phtaB-Z, contains a 360-bp PCR-derived DNA product that includes the 3′ end of the hmuV gene and the 5′ coding region of htaB. To construct plasmid pKhtaA, PCR was used to generate an approximately 2-kb DNA fragment that contains the complete coding region and promoter sequence for htaA. Since the cloned htaA gene was unstable at a high copy number in E. coli, the htaA gene was initially cloned into the copy control vector pCC1 (Epicentre, Madison, WI). The insert was subsequently ligated into the EcoRV site in vector pKN2.6Z (11) and transformed into C. ulcerans 712 (40). The DNA sequence of the cloned htaA gene on plasmid pKhtaA was found to be identical to that reported for the htaA gene from the C. diphtheriae strain NCTC13129 genome (8).
The vector pET24a+ (Novagen, Madison, WI) was used for expression of six-histidine (His)-tagged HtaA and HtaB proteins in E. coli. Internal regions of the htaA and htaB genes that lack the N-terminal leader sequences (residues 1 to 30 for htaA and residues 1 to 24 for htaB) and C-terminal transmembrane and charged tail regions (residues 565 to 591 for htaA and residues 293 to 325 for htaB) were amplified by PCR and subsequently ligated into the NdeI-HindIII sites in pET24a+. The pET28a+ vector, which contains a thrombin protease recognition site, was also used for expression of htaB. A thrombin cleavage and capture kit (Novagen) was used to cleave the N-terminal His tag from the HtaB protein according to the manufacturer's protocol. The purified HtaB protein without the His tag was used in the hemin binding studies. The pGEX-6P-1 vector (GE Healthcare) was used for expression of a GST-HtaA fusion protein that contains an N-terminal GST tag. The same region of htaA that was cloned into the pET24a+ vector was ligated into pGEX-6P-1; however, EcoRV and XhoI sites were introduced into the PCR primers used to amplify the htaA gene.
LacZ assays.
C. diphtheriae strains containing lacZ fusion constructs were grown overnight in HIBTW medium and then inoculated at an OD600 of 0.2 into fresh HIBTW medium and grown to log phase. Log-phase cultures were used to inoculate fresh HIBTW medium to an OD600 of 0.1 and grown overnight in the presence of various supplements, as indicated below. LacZ activity was determined for overnight cultures as previously described (37).
Protein expression and antibody production.
E. coli strain BL21(DE3) carrying the cloned htaA or htaB gene on the pET24a+ expression vector was grown in 100 ml of LB medium at 37°C to mid-log phase, at which time 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cultures were allowed to grow for an additional 2 to 3 h before they were harvested. The cultures were washed in 10 ml of 20 mM Tris-HCl (pH 7.5), and each pellet was stored at −20°C overnight. The pellet was thawed and resuspended in buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole (pH 8.0) at 4°C, and the bacteria were lysed with a French pressure cell, which was followed by removal of cell debris by centrifugation at 10,000 × g at 4°C. The soluble fraction containing the HtaB protein was collected and purified using Ni-nitrilotriacetic acid resin (Qiagen) by a batch affinity method according to the manufacturer's instructions. It was observed that the HtaA-His protein formed inclusion bodies and could not be purified with the Ni-nitrilotriacetic acid affinity resin. However, we observed that HtaA could be significantly enriched after solubilization in 8 M urea, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then excision of the HtaA protein band from the acrylamide gel. The gel-purified HtaA and affinity-purified HtaB proteins were used for production of polyclonal antibodies in guinea pigs by standard methods (Cocalico Biologicals Inc.,).
The procedure used for expression of HtaA with the pGEX-6P-1 vector was similar to the procedure described above for the pET expression system. The GST-HtaA protein was found predominantly in the soluble fraction after lysis of the bacteria, and the fusion protein was purified using GST resin (GE Healthcare) by a batch method according to the manufacturer's instructions. Purified HtaA protein was obtained after removal of the GST tag by cleavage with PreScission protease (GE Healthcare), which was followed by purification steps performed according to the manufacturer's protocol.
SDS-PAGE and Western blot analysis.
Proteins were separated by SDS-PAGE (17) and stained with Bio-Safe Coomassie blue (Bio-Rad) by following the manufacturer's instructions. Western blot procedures were performed as described previously (12), and anti-HtaA and anti-HtaB antibodies were used at dilutions of 1:10,000 and 1:20,000, respectively. Antibodies raised against HmuT (35), DtxR (39), and diphtheria toxin (a gift from R. K. Holmes) were used at dilutions of 1:40,000, 1:5,000, and 1:10,000, respectively. Binding of the primary antibodies to immobilized proteins was detected by using appropriate alkaline phosphatase-labeled secondary antibodies. Alkaline phosphatase activity was detected by using established procedures (35).
Protein localization studies.
To determine whether proteins were secreted or cell associated, 1-ml cultures were grown to mid-log phase at 37°C in HIBTW medium containing 12 μg/ml EDDA (low-iron medium). Cultures were centrifuged briefly to pellet the cells, and the supernatant was recovered and passed through a 0.2-μm filter to remove bacteria. Proteins in the filtered supernatant were precipitated with 10% trichloroacetic acid, washed with ethanol, dried, and then resuspended in Tris-EDTA buffer (pH 7.4). One half of the sample was analyzed after SDS-PAGE and Coomassie blue staining, and the other half of the sample was used for Western analysis. The resulting cell pellet was lysed and then resuspended in SDS loading buffer, and samples were analyzed as described above.
A previously described procedure was used to determine the cellular location of cell-associated proteins (11), with the following modifications. Briefly, 200-ml cultures of various C. diphtheriae strains were grown to late log phase in HIBTW medium at 37°C. Cells were harvested by centrifugation, washed once with PBS (pH 7.4) (Invitrogen), and then resuspended in 10 ml of PBS, which was followed by lysozyme treatment and lysis of the bacteria using a French pressure cell. Cell debris was removed by centrifugation, and the soluble fraction, which contained both soluble and membrane proteins, was centrifuged at 100,000 × g to pellet the cytoplasmic membrane. The supernatant, which contained the soluble intracellular proteins, was recovered and stored at 4°C, while the membrane fraction was solubilized in 0.1% Triton X-100. C. diphtheriae cultures grown in 50 ml of mPGT medium were prepared similarly.
Proteinase K experiments.
C. diphtheriae 1737htaAΔ/pKhtaA was grown as described above for the hemoglobin utilization assay, except that 0.5 μM FeSO4 was added to mPGT medium (low-iron conditions) in place of hemoglobin. Cultures were harvested after overnight growth and centrifuged at 7,000 × g for 10 min. The cells were resuspended in PBS, which was followed by addition of proteinase K at a final concentration of 50 μg/ml to the cell suspension. The reaction mixture was incubated for 30 min at 37°C, at which time the cells were pelleted and then washed two times with PBS. The bacteria were then treated with lysozyme followed by 0.75% Sarkosyl to lyse the bacteria. Whole-cell protein preparations were boiled under reducing conditions prior to analysis by SDS-PAGE and Western blotting. Control samples were not treated with proteinase K. Plasmid pKhtaA was used to obtain better detection of the HtaA protein.
Hemin binding analysis.
Purified GST-HtaA and HtaB, from which the leader peptide and the transmembrane region were deleted (HtaB also lacked the six-His tag), were analyzed to determine their hemin binding properties by UV-visible spectroscopy using a Beckman DU 640 spectrophotometer. Proteins at a concentration of 3.5 μM in PBS buffer containing glycerol were assessed to determine their abilities to bind hemin in the concentration range from 0.5 to 20 μM. Proteins were incubated in the presence of hemin for at least 15 min before spectrophotometric analysis. UV-visible absorption scans of HtaA and HtaB were done using wavelengths between 280 and 600 nm. Absorbance spectra for all protein-hemin samples were zeroed against a reference cuvette that contained the same concentration of hemin in PBS buffer in the absence of protein.
Detection of heme-dependent peroxidase activity.
The chromogenic compound 3,3′,5,5′-tetramethylbenzidine (TMBZ) turns blue in the presence of heme-dependent peroxidase activity and was used to detect hemin-protein complexes as previously described (43). Prior to separation of proteins by SDS-PAGE, samples were incubated for 30 min at room temperature in the presence or absence of hemin. Hemin was prepared in 0.1 N NaOH and was used at a concentration of 0.625 μM for HtaA detection or at a concentration of 2.5 μM for studies with HtaB. Samples were also incubated in the absence of hemin with 0.1 N NaOH. Proteins were not boiled or exposed to reducing agents prior to SDS-PAGE. Lithium dodecyl sulfate (LDS) was used in place of SDS in the HtaB studies as described previously (43). HtaA in culture supernatants was concentrated by precipitation with 75% ammonium sulfate followed by dialysis in 50 mM Tris (pH 8.0).
Computer analysis.
Amino acid sequence similarity searches were done using the BLAST program (1) at the National Center for Biotechnology Information and also using the BLAST server provided at the online site for the Sanger Institute (http://www.sanger.ac.uk/Projects/C_diphtheriae). The annotated genome sequence of C. diphtheriae strain NCTC13129 (8) is accessible in the EMBL/GenBank database under accession number BX248353.
RESULTS
Analysis of the C. diphtheriae hmu hemin transport gene cluster.
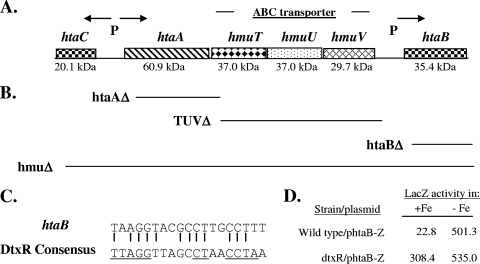
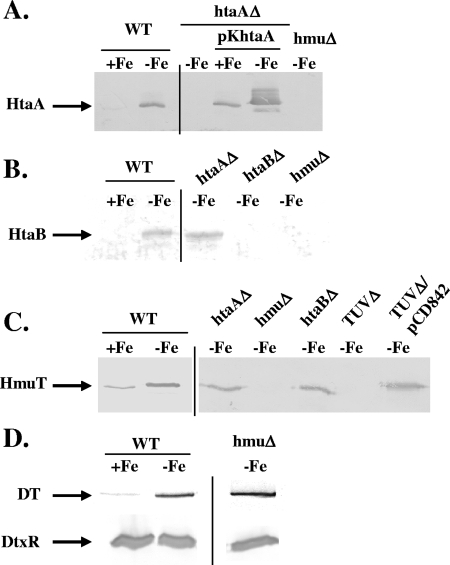
Previous studies of C. diphtheriae identified three genes, hmuT, hmuU, and hmuV, which encode components of an ABC-type hemin transporter (11). The HmuT protein was shown to be a membrane-anchored hemin binding lipoprotein, while HmuU and HmuV were predicted to be the permease and ATPase components, respectively. In a subsequent study (14), we showed that the hmuTUV genes were part of a larger gene cluster that included the htaA, htaB, and htaC genes (Fig. 1A), and we reported that htaA and htaC were transcribed from divergent DtxR- and iron-regulated promoters (14). In this study, we identified a putative DtxR binding site upstream of htaB that aligns with 12 of the 19 residues in the consensus DtxR binding sequence and matches 9 of the 11 most highly conserved bases (Fig. 1A and C). Analysis of an htaB-lacZ transcriptional fusion construct (phtaB-Z) in the wild type and in a dtxR mutant of C. diphtheriae showed that expression from the htaB promoter is regulated by iron and DtxR (Fig. 1D). Transcription from the htaB promoter was not affected by the divalent metals Mn and Zn, and the presence of hemoglobin resulted in a slight decrease in expression, since hemin serves as an iron source in C. diphtheriae (data not shown). Western analysis using antibodies raised against HtaA, HtaB, and HmuT demonstrated that the production of each of these proteins is repressed by iron (Fig. 2A to C), which is consistent with the study described above and with previous findings (14). Diphtheria toxin, a well-characterized iron-regulated protein, and DtxR, which is constitutively expressed, were included as controls in this study (Fig. 2D).
FIG. 1.
(A) Genetic map of the hmu locus in C. diphtheriae. The predicted sizes of the various gene products are indicated below the map. P indicates the presence of a DtxR-regulated promoter, and the arrows indicate the direction of transcription. (B) Regions deleted in the various C. diphtheriae 1737 mutants constructed. The deleted regions are aligned with the genetic map shown in panel A. (C) Alignment of the 19-bp putative DtxR binding site upstream of the C. diphtheriae htaB gene with the consensus DtxR binding site. The underlined sequences are the most highly conserved residues (14). (D) Assessment of the promoter activity (LacZ activity) of an htaB-lacZ transcriptional fusion construct, phtaB-Z, in high-iron HIBTW medium (+Fe) and in iron-depleted conditions (HIBTW medium containing 12 μg/ml EDDA) (−Fe). The values are the means of three experiments. Each result varied less than 15% from the mean. The difference between high- and low-iron conditions for the dtxR mutant was statistically significant (P < 0.05), and this suggests that the point mutant (R47H) maintains some low-level Fe-dependent repressor activity. See Materials and Methods for experimental details.
FIG. 2.
Western blot analysis of proteins produced by C. diphtheriae wild-type strain 1737 (WT) and various 1737 deletion mutants. Strains were grown in 1 ml of iron-replete HIBTW medium (+Fe) or iron-depleted medium (HIBTW medium containing 12 μg/ml EDDA) (−Fe), and proteins present either in culture supernatants (HtaA and diphtheria toxin [DT] in panels A and D, respectively) or in whole-cell extracts (HtaB, HmuT, and DtxR in panels B, C, and D, respectively) were detected by Western blot analysis using antiserum raised against the proteins indicated on the left. Samples were normalized using OD600 before SDS gels were loaded. htaAΔ, 1737htaAΔ; TUVΔ, 1737TUVΔ; hmuΔ, 1737hmuΔ; htaBΔ, 1737htaBΔ.
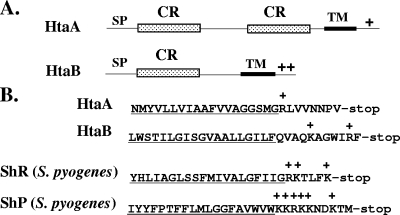
BLAST analysis of the predicted amino sequence of HtaA reveals that it has homology (ranging from 20% to 60% identity) with proteins in several bacterial species that are related to C. diphtheriae, including C. ulcerans, Corynebacterium jeikeium, Corynebacterium glutamicum, Propionibacterium acnes, and Streptomyces coelicolor. HtaA also contains approximately 150 to 200 residues in its N-terminal region that share almost 50% sequence similarity to a segment in its C terminus. These two conserved regions are approximately 35% similar to a region in HtaB (Fig. 3A). HtaA and HtaB both contain predicted signal peptides and a putative transmembrane region in their C termini, which is followed by one (HtaA) or two (HtaB) positively charged residues (Fig. 3A and B). The htaC gene is predicted to encode a putative membrane protein that shares limited amino acid sequence similarity with proteins with unknown functions.
FIG. 3.
(A) Comparison of various structural characteristics of the HtaA and HtaB proteins. HtaA and HtaB contain a region of sequence similarity consisting of approximately 200 amino acids designated the conserved region (CR). SP, signal peptide; TM, transmembrane region; +, positively charged residues. (B) Comparison of the amino acid sequences in the C-terminal tail region for HtaA, HtaB, and the surface-anchored hemin binding proteins ShR and ShP from S. pyogenes. The putative membrane-spanning region is underlined, and positively charged residues are indicated by plus signs.
Mutations in htaA and hmuTUV result in a reduced ability to utilize hemoglobin as an iron source in C. diphtheriae.
A function for HtaA in C. diphtheriae has not been determined previously, although the close proximity of htaA to genes involved in hemin transport suggests that HtaA may have a role in hemin uptake or hemin iron utilization. In previous studies (11, 35), the C. diphtheriae hmuTUV genes were shown to complement mutations in the hmuTUV genes in C. ulcerans; however, an insertion mutation in C. diphtheriae hmuT did not result in a defect in hemin iron use (35). Since the previous report, we have developed a more sensitive method for measuring hemin iron utilization in Corynebacterium (16). To determine the functions of proteins encoded in the hmu gene cluster in C. diphtheriae, we constructed various nonpolar deletions in this region (Fig. 1B). The various mutations in the hmu gene cluster were confirmed by PCR (data not shown) and Western analysis (Fig. 2A to C). C. diphtheriae strain 1737 was used to construct the various hmu mutants and to assess hemoglobin iron utilization. The 1737 strain is a clinical isolate from the recent Russian diphtheria epidemic (28) and is very closely related to the strain used for analysis of the genome sequence (8).
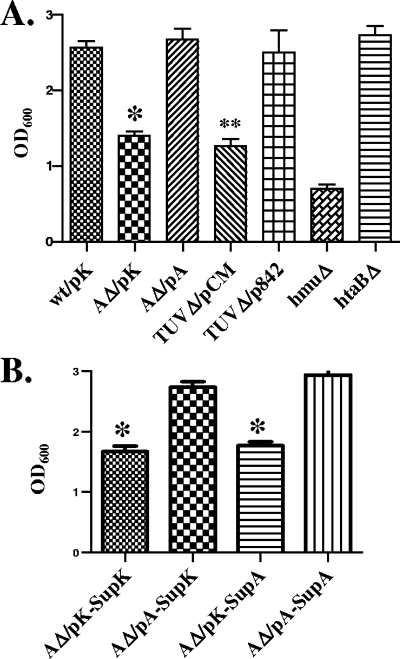
The ability to use hemoglobin as an iron source was analyzed using low-iron mPGT medium in the presence of 25 μg/ml hemoglobin after 22 to 24 h of growth at 37°C. All strains grew to similar OD600s when 1 μM FeSO4 was added to mPGT medium (the OD600s were between 3.0 to 3.5), and the growth of all strains was fully inhibited in low-iron mPGT medium in the absence of an iron source (low-iron mPGT medium contains 10 μM EDDA). Deletion of the C. diphtheriae 1737 htaA gene, the hmuTUV genes, or the complete hmu gene cluster resulted in significantly diminished growth compared to the growth of the wild-type strain when hemoglobin was supplied as the sole iron source (Fig. 4A). Deletion of the htaB gene had no effect on the use of hemoglobin as an iron source in these assays (Fig. 4A). The presence of the cloned htaA and hmuTUV genes restored the ability to utilize hemoglobin as an iron source in the htaA and hmuTUV mutants, respectively (Fig. 4A). These findings suggest that the products of htaA and hmuTUV are involved in hemin transport or in hemin iron utilization in C. diphtheriae. However, the results also suggest that additional genes outside the hmu locus encode products that have the ability to transport hemin, since hemoglobin was still able to stimulate growth of all of the deletion strains. All of the mutants described with mutations in the C. diphtheriae hmu locus that showed reduced growth when hemoglobin was added as the sole iron source exhibited similar reductions in growth compared to the wild-type strain when hemin was used as the only iron source (data not shown).
FIG. 4.
Utilization of hemoglobin as an iron source by C. diphtheriae wild-type strain 1737 and mutant strains. (A) OD600s of cultures grown for 20 to 22 h in mPGT medium containing 10 μM EDDA and 25 μg/ml hemoglobin (human). The plasmids carried by the various strains included pKhtaA containing the cloned htaA gene (pA), vector pKN2.6Z (pK), vector pCM2.6 (pCM), and pCD842 containing the cloned hmuTUV genes (p842). wt, wild-type strain 1737; AΔ, 1737htaAΔ; TUVΔ, 1737TUVΔ; hmuΔ, 1737hmuΔ; htaBΔ, 1737htaBΔ. (B) Results of experiments performed like the experiments described for panel A, except that cultures of 1737htaAΔ were supplemented with 50 μl of concentrated culture supernatant from 1737hmuΔ that contained either the vector pKN2.6Z (SupK) or the cloned htaA gene on pKhtaA (SupA). The presence of HtaA in culture supernatants in 1737hmuΔ/pKhtaA was confirmed by SDS-PAGE analysis (not shown). The results are the averages and standard deviations of three independent experiments. The values for 1737htaAΔ/pKN2.6Z are significantly different from the values for 1737/pKN2.6Z and 1737htaAΔ/pKhtaA (*, P < 0.05), and the values for 1737TUVΔ/pCM2.6 are significantly different from the values for 1737/pKN2.6Z and 1737TUVΔ/pCD842 (**, P < 0.05).
Cellular localization of the C. diphtheriae hmu gene products.
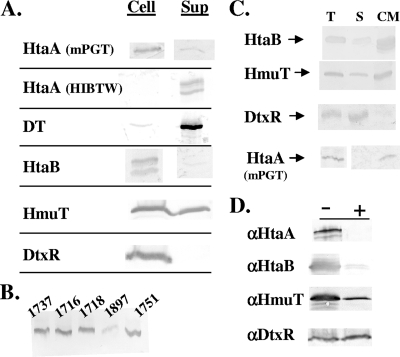
To better understand the function of the hmu gene products in hemin iron utilization, studies were done to identify the cellular locations of the various proteins encoded in this gene cluster. Studies were initially done to determine whether various proteins are cell associated or secreted into the culture medium. As shown in Fig. 5A, HtaA was predominantly cell associated when C. diphtheriae was grown in low-iron mPGT minimal medium, although low levels of HtaA were detected in the culture supernatant. Surprisingly, HtaA was found almost exclusively in the supernatant fraction when bacteria were grown in low-iron HIBTW medium, and HtaA exhibited a secretion profile almost identical to that of diphtheria toxin, a well-known secreted protein in C. diphtheriae (Fig. 5A). Examination of four additional C. diphtheriae strains, strains 1716, 1718, 1897, and 1751 (28), showed that they also secreted HtaA during growth in HIBTW medium (Fig. 5B). The HtaB and HmuT proteins were both found to be predominantly cell associated, although smaller quantities were observed in the supernatant fraction (Fig. 5A). DtxR, an intracellular protein used as a control to monitor cell lysis, was found exclusively in the cellular fraction, which indicates that the presence of HtaA in the supernatant was not the result of leakage or lysis of the bacteria during growth in HIBTW medium. The localization of diphtheria toxin, HtaB, and HmuT after growth in mPGT medium was similar to that observed after growth in HIBTW medium (data not shown).
FIG. 5.
(A) Western blot analysis to measure secretion or cell association of proteins expressed from C. diphtheriae wild-type strain 1737. One-milliliter cultures were grown for 20 to 22 h at 37°C in iron-depleted HIBTW medium or in mPGT medium (only results for HtaA are shown for growth in mPGT medium). Polyclonal antiserum (specific to the proteins indicated on the left) was used for detection of proteins associated with the cell pellet (Cell) and the supernatant fraction (Sup). Doublet bands for HtaA and HtaB indicate breakdown products, which exhibited some variability between protein preparations. See Materials and Methods for a description of the method used for sample preparation. DT, diphtheria toxin. (B) Western blot detection of HtaA in culture medium from various C. diphtheriae clinical isolates after growth in iron-depleted HIBTW medium. (C) Western blot analysis to identify the localization of proteins after cell fractionation of C. diphtheriae wild-type strain 1737. Cultures were grown in HIBTW medium or in mPGT medium (only results for HtaA are shown for mPGT medium), and polyclonal antiserum (specific to proteins indicated on the left) was used for detection of total cellular proteins. T, membrane and cytosolic fractions; S, soluble cytosolic proteins; CM, cytoplasmic membrane proteins. The total cellular fraction for HtaA was collected from the whole-cell lysate when the cells were harvested. Samples were loaded using equivalent protein levels. (D) Western blotting to detect protein levels after proteinase K treatment of C. diphtheriae 1737htaΔ/pKhtaA grown in low-iron mPGT medium. The antisera used for detection are indicated on the left (αHtaA, anti-HtaA; αHtaB, anti-HtaB; αHmuT, anti-HmuT; αDtxR, anti-DtxR), and the presence (+) or absence (−) of proteinase K (50 μg/ml) is indicated at the top. See Materials and Methods for a description of experimental details.
The results of cellular fractionation studies and Western analysis indicate that HtaA is associated primarily with the cytoplasmic membrane when C. diphtheriae 1737 is grown in mPGT medium (Fig. 5C). HtaA and diphtheria toxin were not detected in any cellular fraction when bacteria were grown in HIBTW medium, since these proteins are secreted into the supernatant in this medium (Fig. 5A). HtaB and HmuT are localized primarily to the cytoplasmic membrane, while the transcriptional regulatory protein, DtxR, is found exclusively in the soluble cytosolic fraction (Fig. 5C).
Secreted HtaA is not involved in hemin iron utilization.
While it is not clear what function, if any, the secreted HtaA protein has in hemin iron utilization in C. diphtheriae, addition of HtaA (either purified or concentrated from culture supernatants) to cultures of 1737htaAΔ did not enhance the growth of this htaA mutant in the presence of hemoglobin (Fig. 4B). These results suggest that the secreted HtaA protein is not involved in the use of hemin iron by C. diphtheriae but that the dominant form of HtaA involved in the uptake of hemin is most likely bound to the cytoplasmic membrane.
Surface exposure of hmu gene products.
To determine whether HtaA, HtaB, or HmuT is exposed on the cell surface and therefore may be accessible to bind hemin or possibly hemoglobin, C. diphtheriae was grown in low-iron mPGT medium, which was followed by treatment of the intact bacteria with proteinase K. This protease is predicted to cleave proteins that are exposed outside the cell wall. As shown in Fig. 5D, HtaA and HtaB were fully digested by proteinase K, suggesting that these proteins are exposed on the surface of the bacteria. The levels of HmuT were reduced after protease treatment, but this protein was not fully digested like HtaA and HtaB. This finding suggests that HmuT is not as accessible to proteinase K as the other two proteins and may be partially protected by the cell wall; a similar observation was reported previously for the lipoprotein component of the ABC-type hemin transporter in S. aureus (24). DtxR, which is an intracellular protein, was not digested by proteinase K (Fig. 5D).
HtaA and HtaB are hemin binding proteins.
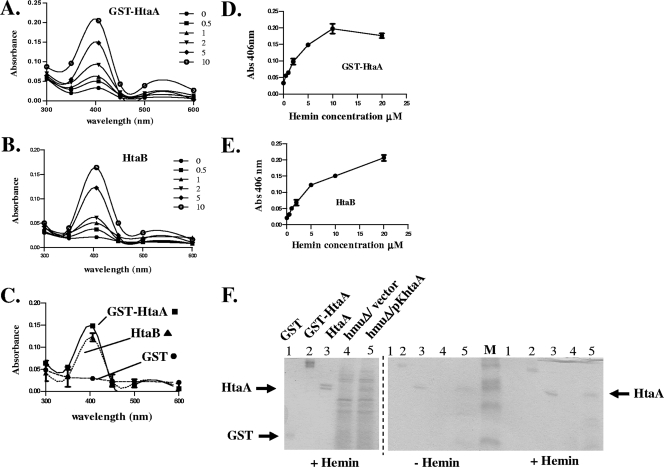
UV-visible spectroscopy was used to analyze the hemin binding properties of the purified GST-HtaA and HtaB proteins. The GST-HtaA and HtaB proteins at a concentration of 3.5 μM were incubated at room temperature for at least 15 min with hemin at concentrations ranging from 0.5 μM to 10 μM. UV-visible absorption scans of the protein-hemin samples indicated that both GST-HtaA and HtaB showed strong absorption peaks at 406 nm (Fig. 6A and B), which is an absorption maximum consistent with the binding of hemin (24). Absorption measurements for HtaA in the absence of the GST tag could not be obtained due to proteolytic degradation of HtaA, which occurred after cleavage of the GST tag and the subsequent dialysis that was needed to remove the GST-protease cleavage buffer. The GST-protease cleavage buffer was found to interfere with absorbance measurements for the HtaA-hemin complex. When the GST protein alone was incubated with 5 μM hemin, it showed no absorption peak in the 400-nm region (Fig. 6C), indicating that it is does not bind hemin, as previously reported (24). Measurements of absorption at 406 nm for GST-HtaA and HtaB with increasing hemin concentrations were used to determine dissociation constants of 1.9 ± 0.4 μM for GST-HtaA and 4.9 ± 0.7 μM for HtaB (Fig. 6D and E).
FIG. 6.
HtaA and HtaB are hemin binding proteins. (A and B) UV-visible spectroscopy was used to examine GST-HtaA (A) or HtaB (B) at a concentration of 3.5 μM in the presence of various hemin concentrations. Absorption scans were done at 300 to 600 nm. (C) Absorption scans for GST-HtaA, HtaB, and GST in the presence of 5 μM hemin. (D and E) Absorption at 406 nm for GST-HtaA (D) and HtaB (E) at various hemin concentrations. Different concentrations of hemin (0 to 20 μM) were incubated with protein (3.5 μM) for at least 15 min at 20 to 25°C prior to measurement of absorbance. The values are the means and standard errors of three independent experiments. (F) Protein preparations were incubated with 0.625 μM hemin (+ Hemin) or with control buffer (− Hemin) prior to separation by SDS-PAGE and staining either with Coomassie blue (left panel) or with TMBZ to detect heme-dependent peroxidase activity as described in Materials and Methods (right panel). Lane 1, purified GST protein; lane 2, purified GST-HtaA fusion; lane 3, purified HtaA; lanes 4 and 5, ammonium sulfate-precipitated supernatant of 1737hmuΔ (hmuΔ) carrying either the vector (lane 4) or the cloned htaA gene on plasmid pKhtaA (lane 5); lane M, molecular weight marker. The + Hemin gel in the right panel (TMBZ stained) is a duplicate of the left panel (Coomassie blue stained).
The chromogenic substrate TMBZ, which has been used previously to detect heme-dependent peroxidase activity that is associated with hemin binding proteins (43), was used to analyze the ability of the native form of the HtaA protein to bind hemin. TMBZ staining of secreted proteins from C. diphtheriae 1737hmuΔ carrying plasmid pKhtaA grown in low-iron HIBTW medium showed that heme-dependent peroxidase activity was uniquely associated with the HtaA protein (Fig. 6F, + Hemin gel, lane 5). Peroxidase activity was also associated with HtaA that was present in culture supernatants of wild-type strain 1737 in the absence of any plasmid (not shown). No proteins were stained with TMBZ in supernatants from 1737hmuΔ carrying only the plasmid vector (Fig. 6F, + Hemin gel, lane 4). It was noted that HtaA exhibited hemin binding even in the absence of prebinding with hemin (Fig. 6F, − Hemin gel), which suggests that HtaA produced in C. diphtheriae is bound to hemin (Fig. 6F, − Hemin gel, lane 5). Purified HtaA (without the GST tag) and the GST-HtaA fusion protein (lacking the signal peptide and the putative C-terminal membrane-spanning region) exhibited heme-dependent peroxidase activity in the presence of TMBZ (Fig. 6F, + Hemin gel, lanes 2 and 3). Compared to HtaA, the purified HtaB protein showed very weak, nonspecific, heme-dependent peroxidase activity in the presence of TMBZ (data not shown). This observation suggests that the HtaB-hemin complex is not stable under the conditions used to detect TMBZ-dependent peroxidase activity.
DISCUSSION
While hemin transport systems were first described in gram-negative bacteria (13, 41), the Corynebacterium HmuTUV system was the first hemin transporter to be identified in a gram-positive species (11). Over the last several years, hemin transport systems have been described in several additional gram-positive organisms, including the important human pathogens S. aureus (24) and S. pyogenes (3, 19). While both gram-negative and gram-positive bacteria employ ABC-type transporters and an associated substrate binding protein to move hemin through the cytoplasmic membrane, these organisms utilize distinctly different components to bind hemin or hemoglobin to the cell surface. While gram-negative bacteria bind hemin or hemoglobin to large outer membrane proteins, gram-positive bacteria, which do not have distinct outer membranes, bind hemin or hemoglobin to proteins anchored to the cell envelope. These surface receptors either are covalently linked to the cell wall peptidoglycan via sortases, as observed with the Isd proteins in S. aureus (24, 46, 50), or are tethered to the cytoplasmic membrane through a transmembrane C-terminal tail region, as proposed for the Shp and Shr proteins in S. pyogenes (3, 19, 21, 49) and for HtaA and HtaB in C. diphtheriae.
Both HtaA and HtaB contain N-terminal signal sequences and have putative transmembrane regions in their C termini, structural features that are also present in the S. pyogenes heme binding proteins Shr and Shp (3, 19). Although HtaA and HtaB share structural and possibly functional similarities with these S. pyogenes proteins, BLAST analysis shows that HtaA and HtaB have no significant sequence similarity to either Shp or Shr. A comparison of the C-terminal regions of these four proteins reveals that they all contain a putative membrane-spanning region that is followed by a positively charged tail sequence, which in S. pyogenes is proposed to be involved in anchoring the proteins to the cytoplasmic membrane (Fig. 3B) (3, 19). HtaA contains only a single charge in this tail region, and membrane prediction models suggest that the HtaA transmembrane segment is relatively weak compared to the membrane-spanning regions in HtaB, Shp, and Shr (http://www.ch.embnet.org/software/TMPRED_form.html).
In this study, the hmu hemin transport system in C. diphtheriae was analyzed and shown to include genes whose products are involved in the utilization of hemin and hemoglobin as iron sources. Deletion of htaA, hmuTUV, and the complete hmu locus resulted in a reduction in hemin iron use; however, each of the mutants maintained the ability to use hemin iron for growth, which indicates that additional uptake and/or utilization systems for hemin iron acquisition are active in C. diphtheriae. The presence of more than one hemin iron utilization and uptake system has been described or proposed for several bacteria, including the gram-negative organisms Vibrio cholerae (26) and Yersinia enterocolitica (41) and the gram-positive organisms S. pyogenes (3) and S. aureus (24). For many of these organisms, it was reported that mutations in the hemin-specific ABC transporters did not eliminate the ability of these species to use hemin as an iron source, which suggested that alternative transporters were active. Analysis of the genome of C. diphtheriae has revealed several ABC-type iron or siderophore transporters (8), all of which show some sequence similarity to previously identified hemin transporters. However, the products encoded by the hmuTUV genes show the highest levels of sequence similarity to previously described hemin uptake systems (11). Genes that are predicted to encode proteins with sequence similarity to HtaA (DIP0522 and ChtA) and HtaB (ChtB) are present in the C. diphtheriae genome, and all of these factors contain putative N-terminal secretion signals, as well as C-terminal transmembrane regions (8, 14). It is possible that DIP0522 and/or ChtA may contribute to the hemin iron uptake activity observed in an htaA mutant, and similarly, the product of chtB may complement an htaB mutant. Further studies are needed to determine whether DIP0522, ChtA, ChtB, HtaB, or any of the other related iron uptake systems encoded in the C. diphtheriae genome is involved in hemin iron utilization.
BLAST analysis (1; http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) has revealed that bacterial species related to C. diphtheriae, including C. ulcerans, C. glutamicum, C. jeikeium, and Streptomyces coelicolor, all contain genes whose predicted products have significant sequence similarity to HtaA, and all of these putative proteins contain transmembrane regions in their C termini followed by positively charged residues, an observation that indicates that there is a common anchoring mechanism in this group of related proteins. However, some differences in the anchoring of these proteins to the cell envelope have been observed. A recent study of Propionibacterium acnes, an organism related to Corynebacterium, identified a putative hemin iron transport protein (PA-21693) that has sequence similarity to HtaA but contains a sortase anchoring motif in its C terminus, suggesting that PA-21693 is covalently anchored to the cell wall through the action of a sortase (23). It is not clear why certain hemin or hemoglobin binding proteins in gram-positive bacteria are covalently anchored to the cell wall by sortases, while others, such as HtaA and Shp, are tethered to the surface by a single membrane-spanning region. A covalent attachment to the cell wall appears to be more secure than binding to the cell by a single transmembrane region. All of the gram-positive species that are known to contain surface-anchored hemin binding proteins produce sortases and numerous sortase substrates, indicating that a lack of sortases is not the reason for the differences in the anchoring of these proteins. It is possible that differences in how these proteins are attached to the cell envelope may be attributed to the different environments that these organisms inhabit and/or to additional functions that may be associated with these surface proteins, such as adhesins.
An unexpected finding in this study was the observation that HtaA is secreted during growth in nutrient-rich HIBTW medium but is predominantly cell associated during growth in mPGT medium, a semidefined minimal medium that is commonly used to culture C. diphtheriae strains (44). The presence of a C-terminal transmembrane region in HtaA predicts that the protein is associated with the cytoplasmic membrane, which is observed for HtaB when C. diphtheriae is grown in either HIBTW or mPGT medium. However, analysis of the C-terminal amino acid sequence of HtaA indicates that HtaA has a relatively weak transmembrane region compared to the membrane-spanning region in HtaB (http://www.ch.embnet.org/software/TMPRED_form.html). Although a less-than-optimal membrane-spanning sequence may indicate a weaker association with the membrane, it would not account for the differences in localization observed between organisms grown in HIBTW and mPGT media. The reason for the difference in localization of HtaA between organisms grown in the two media is not known, although differences in membrane composition, permeability, cellular metabolism, or the presence of proteases may contribute to the observed differences in the location of HtaA. While it is not known if HtaA is secreted or membrane bound in vivo, studies reported here showed that when the HtaA protein was added to mPGT culture medium, there was no stimulation of growth of the C. diphtheriae 1737 htaA deletion mutant when hemoglobin was the sole iron source (Fig. 4B). This finding suggests that the secreted form of HtaA may not be involved in hemin iron uptake.
In a previous report (35), we were unable to demonstrate a hemin iron utilization deficiency in a C. diphtheriae hmuT vector integration mutant (RT5), but we observed that a similarly constructed hmuT mutant of C. ulcerans was defective in hemin iron use. This previous study was performed using nutrient-rich HIBTW medium, whereas mPGT medium was used as the growth medium in the studies described in this report. The C. diphtheriae RT5 mutant does exhibit a hemin iron utilization defect similar to that observed with the hmuTUV deletion mutant when cells are grown in mPGT medium (M. P. Schmitt, unpublished observation). The reason for the lack of a phenotype for the C. diphtheriae hmuT mutant in HIBTW medium is not known; however, as noted above, C. diphtheriae hmu mutants are able to use hemin iron even in mPGT medium, which suggests that there are alternate hemin uptake or utilization systems. It is possible that these systems (or others) may have more robust activity during growth in HIBTW medium than during growth in mPGT medium and can fully substitute for the hmu system during growth in HIBTW medium. Differences in metabolism or in heme or iron requirements may also contribute to the observed difference in hemin iron use between the two media.
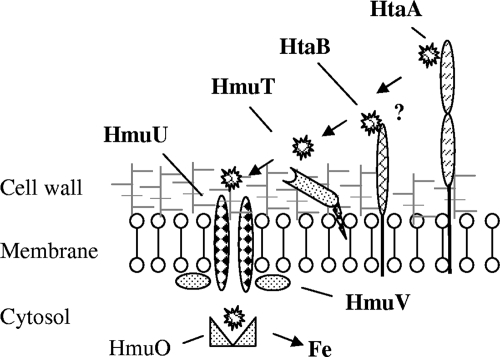
Work in our laboratory has demonstrated that hemin and hemoglobin are equally capable of supplying iron for growth of C. diphtheriae strains (16, 31, 35). While it is not known how C. diphtheriae acquires the hemin moiety from hemoglobin, a recent study has suggested that one mechanism by which hemin iron is obtained for use by bacteria is through the spontaneous release of hemin from hemoglobin (25). It is also possible that secreted or surface-exposed proteases in C. diphtheriae contribute to the breakdown of hemoglobin and subsequent release of hemin. In the model shown in Fig. 7, we propose that hemin initially binds to HtaA at one or both of the conserved domain regions. Hemin binding sites have not been identified for HtaA, and future studies should determine if these conserved regions are involved in hemin binding or transport. It is predicted that hemin bound to HtaA is transferred to HtaB, which may function in an intermediate step in the movement of hemin from HtaA to HmuT. This mechanism of action for HtaB would be similar to the function proposed for some of the Isd proteins from S. aureus, which are involved in the movement of hemin through the cell wall (22, 46, 50). HmuT is proposed to deliver hemin to the HmuU permease, a component of the ABC transporter (HmuU and HmuV), which facilitates the uptake of hemin into the cytosol, where it is thought to be degraded by the heme oxygenase enzyme HmuO, releasing iron for cellular metabolism.
FIG. 7.
Model for hemin transport in C. diphtheriae. It is proposed that hemin initially binds to HtaA, a surface-anchored hemin binding protein that is associated with the cytoplasmic membrane through a C-terminal transmembrane region. Hemin would then be transferred from HtaA to membrane-anchored HtaB (?), or, alternatively, hemin would be passaged directly to the HmuT lipoprotein. Hemin that is bound to HmuT would be transferred to HmuU, which is a membrane-bound permease component of an ABC transporter, and in conjunction with HmuV, an ATPase, would facilitate the movement of hemin into the bacterial cytosol. HmuO is proposed to degrade the intracellular heme and release the iron for cellular metabolism.
Acknowledgments
We thank Lori Keating and Karen Meysick for helpful comments on the manuscript.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Altschul, S. F., G. Warren, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bardsdale, W. L., and A. M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, C. S., G. E. Montanez, C. R. Woods, R. M. Vincent, and Z. Eichenbaum. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 711042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, J. M., O. N. Manish, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 875968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V. 2005. Bacterial iron transport related to virulence. Contrib. Microbiol. 12210-233. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by gram-positive bacterial pathogens. Microbes Infect. 41149-1156. [DOI] [PubMed] [Google Scholar]
- 7.Brune, I., H. Werner, A. T. Huser, J. Kalinowski, A. Puhler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 316516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier, R. J. 2001. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 391793-1803. [DOI] [PubMed] [Google Scholar]
- 10.Cryz, S. J., L. M. Russell, and R. K. Holmes. 1983. Regulation of toxinogenesis in Corynebacterium diphtheriae: mutations in the bacterial genome that alter the effects of iron on toxin production. J. Bacteriol. 154245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from hemin and hemoglobin are homologous to ABC hemin transporters. Mol. Microbiol. 3668-84. [DOI] [PubMed] [Google Scholar]
- 12.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Henderson, D. P., and S. M. Payne. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7461-469. [DOI] [PubMed] [Google Scholar]
- 14.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 1856826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkle, C. A., and M. P. Schmitt. 2005. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 187422-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkle, C. A., and M. P. Schmitt. 2007. Comparative analysis of hmuO function and expression in Corynebacterium species. J. Bacteriol. 1893650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C. B. 1995. Quelling the red menace: hemin capture by bacteria. Mol. Microbiol. 18383-390. [DOI] [PubMed] [Google Scholar]
- 19.Lei, B., L. M. Smoot, H. M. Menning, J. M. Voyich, S. V. Kala, F. R. Deleo, S. D. Reid, and J. M. Musser. 2002. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect. Immun. 704494-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letoffe, S., J. M. Ghigo, and C. Wandersman. 1994. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl. Acad. Sci. USA 919876-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, M., and B. Lei. 2005. Heme transfer from streptococcal cell surface protein Shp to HtsA of transporter HtsABC. Infect. Immun. 735086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, M., W. N. Tanaka, H. Zhu, G. Xie, D. M. Dooley, and B. Lei. 2008. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 2836668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodes, M. J., H. Secrist, D. R. Benson, S. Jen, K. D. Shanebeck, J. Guderian, J. F. Maisonneuve, A. Bhatia, D. Persing, S. Patrick, and Y. A. Skeiky. 2006. Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology 1523667-3681. [DOI] [PubMed] [Google Scholar]
- 24.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299906-909. [DOI] [PubMed] [Google Scholar]
- 25.Mocny, J. C., J. S. Olson, and T. D. Connell. 2007. Passively released heme from hemoglobin and myoglobin is a potential source of nutrient iron for Bordetella bronchiseptica. Infect. Immun. 754857-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Occhino, D. A., E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 291493-1507. [DOI] [PubMed] [Google Scholar]
- 27.Otto, B. R., V.-V. Vught, A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18217-233. [DOI] [PubMed] [Google Scholar]
- 28.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, I. K. Wachsmuth, and J. D. Wenger. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 1741064-1072. [DOI] [PubMed] [Google Scholar]
- 29.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 1844846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, L. M., S. J. Cryz, and R. K. Holmes. 1984. Genetic and biochemical evidence for a siderophore-dependent iron transport system in Corynebacterium diphtheriae. Infect. Immun. 45143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenase and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 654634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 1815330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt, M. P. 2004. Corynebacterium diphtheriae, p. 344-359. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 35.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 1831476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 591899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 593903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor and iron. J. Bacteriol. 1761141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt, M. P., E. M. Twiddy, and R. K. Holmes. 1992. Purification and characterization of the diphtheria toxin repressor. Proc. Natl. Acad. Sci. USA 897576-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serwold-Davis, T. M., N. B. Groman, and M. Rabin. 1987. Transformation of Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium glutamicum, and Escherichia coli with the C. diphtheriae plasmid pNG2. Proc. Natl. Acad. Sci. USA 844964-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojiljkovic, I., and K. Hantke. 1994. Transport of hemin across the cytoplasmic membrane through a hemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13719-732. [DOI] [PubMed] [Google Scholar]
- 42.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21281-295. [DOI] [PubMed] [Google Scholar]
- 43.Stugard, C. E., P. A. Daskaleros, and S. M. Payne. 1989. A 101-kilodalton heme-binding protein associated with Congo red binding and virulence of Shigella flexneri and enteroinvasive Escherichia coli strains. Infect. Immun. 573534-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tai, S.-P. S., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9267-273. [DOI] [PubMed] [Google Scholar]
- 45.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]
- 46.Torres, V. J., G. Pishchany, M. Humayun, O. Schneewind, and E. P. Skaar. 2006. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 1888421-8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273837-841. [DOI] [PubMed] [Google Scholar]
- 48.Winkelmann, G. 2002. Microbial siderophore-mediated transport. Biochem. Soc Trans. 30691-696. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, H., M. Liu, and B. Lei. 2008. The surface protein Shr of Streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein Shp. BMC Microbiol. 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, H. G. Xie, M. Liu, J. S. Olson, M. Fabian, D. M. Dooley, and B. Lei. 2008. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants (Isd) system of Staphylococcus aureus. J. Biol. Chem. 28318450-18460. [DOI] [PMC free article] [PubMed] [Google Scholar]