Abstract
Fluoroacetate dehalogenase catalyzes the hydrolytic defluorination of fluoroacetate to produce glycolate. The enzyme is unique in that it catalyzes the cleavage of a carbon-fluorine bond of an aliphatic compound: the bond energy of the carbon-fluorine bond is among the highest found in natural products. The enzyme also acts on chloroacetate, although much less efficiently. We here determined the X-ray crystal structure of the enzyme from Burkholderia sp. strain FA1 as the first experimentally determined three-dimensional structure of fluoroacetate dehalogenase. The enzyme belongs to the α/β hydrolase superfamily and exists as a homodimer. Each subunit consists of core and cap domains. The catalytic triad, Asp104-His271-Asp128, of which Asp104 serves as the catalytic nucleophile, was found in the core domain at the domain interface. The active site was composed of Phe34, Asp104, Arg105, Arg108, Asp128, His271, and Phe272 of the core domain and Tyr147, His149, Trp150, and Tyr212 of the cap domain. An electron density peak corresponding to a chloride ion was found in the vicinity of the Nɛ1 atom of Trp150 and the Nɛ2 atom of His149, suggesting that these are the halide ion acceptors. Site-directed replacement of each of the active-site residues, except for Trp150, by Ala caused the total loss of the activity toward fluoroacetate and chloroacetate, whereas the replacement of Trp150 caused the loss of the activity only toward fluoroacetate. An interaction between Trp150 and the fluorine atom is probably an absolute requirement for the reduction of the activation energy for the cleavage of the carbon-fluorine bond.
Fluoroacetate is a naturally occurring organofluorine compound. An actinomycete, “Streptomyces cattleya” (30), and some plants in Australia and Africa (2) produce this highly toxic compound. Fluoroacetate has a carbon-fluorine bond, whose dissociation energy is among the highest found in nature (8). Despite this fact, fluoroacetate dehalogenases (FAc-DEXs) from Burkholderia sp. strain FA1 (FAc-DEX FA1) (20) and Delftia acidovorans strain B (formerly Moraxella sp. strain B; FAc-DEX H1) (16) catalyze the hydrolytic defluorination of fluoroacetate to produce glycolate. These enzymes are unique in that they catalyze the cleavage of the carbon-fluorine bond, which is much stronger than other carbon-halogen bonds.
FAc-DEXs show weak but significant sequence similarity to proteins that belong to the α/β hydrolase superfamily, such as epoxide hydrolases from Agrobacterium radiobacter AD1 (37) and humans and haloalkane dehalogenases from Sphingobium japonicum UT26 (formerly Sphingomonas paucimobilis UT26) (32) and Xanthobacter autotrophicus GJ10 (17). Although haloalkane dehalogenases of this superfamily catalyze the hydrolytic cleavage of carbon-halogen bonds, none of them catalyzes the cleavage of a carbon-fluorine bond. Dehalogenases that belong to other families, such as l-2-haloacid dehalogenase (22), dl-2-haloacid dehalogenase (28), and haloalcohol dehalogenase (42), also do not catalyze the cleavage of a carbon-fluorine bond of aliphatic organofluorine compounds.
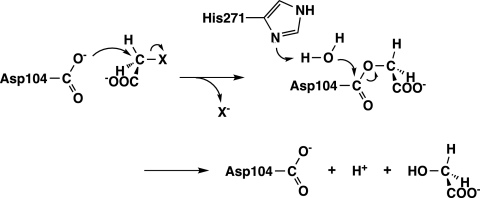
To clarify the enzymatic defluorination mechanism, we investigated the reaction mechanism of FAc-DEX H1 by site-directed mutagenesis and mass spectrometric analysis and revealed that the reaction proceeds in two steps (23). In the first step, the carboxylate group of Asp105 nucleophilically attacks the α-carbon atom of fluoroacetate to displace the fluorine atom, leading to the formation of an ester intermediate. In the second step, the ester intermediate is hydrolyzed by a water molecule activated by His272, which yields glycolate and regenerates the carboxylate group of Asp105. FAc-DEX FA1 is homologous to FAc-DEX H1, sharing 61% amino acid sequence identity, and Asp105 and His272 of FAc-DEX H1 are conserved as Asp104 and His271, respectively, in FAc-DEX FA1. Thus, the reaction catalyzed by FAc-DEX FA1 probably proceeds by the same mechanism as that catalyzed by FAc-DEX H1 (Fig. 1).
FIG. 1.
Proposed reaction mechanism of FAc-DEX FA1.
To gain a deeper insight into the catalytic mechanism of FAc-DEXs, information on the three-dimensional structure is necessary. Zhang et al. constructed a three-dimensional structure model of FAc-DEX FA1 by homology modeling by using epoxide hydrolase from mouse (Protein Data Bank [PDB] accession no. 1CR6) and haloalkane dehalogenase from X. autotrophicus GJ10 (PDB accession no. 2DHC) as templates (44). They further used this structure for computational analysis of the reaction mechanism. However, it is generally difficult to predict the precise locations and orientations of amino acid residues of proteins by homology modeling, and moreover, the degrees of sequence identity between FAc-DEX FA1 and the proteins used for the modeling are not very high (26 and 18%, respectively). Thus, an experimentally determined three-dimensional structure has been required for more solid analysis of the catalytic mechanism of FAc-DEXs.
We here report the crystal structure of FAc-DEX FA1, as the first experimentally determined three-dimensional structure of the enzyme that catalyzes the cleavage of a rigid carbon-fluorine bond of aliphatic compounds. The folding of the protein was similar to that predicted by homology modeling, but the active-site structure was significantly different. We carried out site-directed mutagenesis analysis based on the crystal structure and identified amino acid residues that are essential for defluorination.
MATERIALS AND METHODS
Materials.
Sodium fluoroacetate was purchased from Sigma-Aldrich (St. Louis, MO) and RdH (Buchs, Switzerland). Butyl-Toyopearl and DEAE-Toyopearl were purchased from Tosoh (Tokyo, Japan). Gigapite was from Toagosei Chemical Industry (Tokyo, Japan). Molecular-weight marker proteins for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Bio-Rad (Hercules, CA). Restriction enzymes and kits for genetic manipulation were purchased from Takara Shuzo (Kyoto, Japan), New England Biolabs (Beverly, MA), Sigma-Aldrich, Qiagen (Tokyo, Japan), and Stratagene (La Jolla, CA). The synthetic primers used were from either Hokkaido System Science (Sapporo, Japan) or Kurabo (Osaka, Japan). Sparse-matrix screens, Wizard I and II, were obtained from Emerald BioStructures (Bainbridge Island, WA). All other reagents were of analytical grade and were obtained from Nacalai Tesque (Kyoto, Japan), Hampton Research (Aliso Viejo, CA), or Sigma-Aldrich.
Construction of a plasmid for the overproduction of FAc-DEX FA1.
The FAc-DEX FA1 gene from Burkholderia sp. strain FA1, which assimilates fluoroacetate as a sole carbon source, was amplified by PCR using the primers PrNdeI (5′-GGGAATTCCATATGTTTGAAGGATTCGAGCGACGCC-3′) and PrBamH1 (5′-CGCGGATCCTCATGATTCTCTTCGCTCGGTCTGCTGG-3′). (The underlining indicates restriction sites.) The amplified DNA fragments were digested with restriction enzymes NdeI and BamHI and ligated with pET21a(+) digested with the same restriction enzymes. The plasmid obtained was named pET21-FA1.
Purification of the wild-type and mutant FAc-DEX FA1 enzymes.
All the purification procedures were carried out at 4°C, and 50 mM potassium phosphate buffer (pH 7.5) was used as the standard buffer unless otherwise stated. Escherichia coli BL21(DE3) cells harboring pET21-FA1 were grown aerobically in Luria-Bertani (LB) medium (1% polypeptone, 0.5% yeast extract, and 1% NaCl [pH 7.0]) containing 100 μg of ampicillin per ml at 37°C overnight. The cells harvested from a 4.8-liter LB medium culture were suspended in 80 ml of the standard buffer and disrupted by sonication. The cell debris was removed by centrifugation to obtain the crude extract. Streptomycin sulfate (1% [wt/vol]) was added to the crude extract to remove nucleic acids, and 2 M potassium phosphate (pH 7.5) was added to the resulting supernatant to a final concentration of 1 M. The solution was applied to a butyl-Toyopearl 650 M column equilibrated with 1 M potassium phosphate (pH 7.5). After the column was washed with 1 M potassium phosphate buffer (pH 7.5), the enzyme fraction was eluted with a linear gradient of 1 M to 50 mM potassium phosphate buffer (pH 7.5). The active fractions were collected and dialyzed against the standard buffer. The enzyme solution was applied to a DEAE-Toyopearl 650 M column equilibrated with the standard buffer. After the column was washed with the standard buffer, the enzyme fraction was eluted with a linear gradient of 50 to 100 mM potassium phosphate buffer (pH 7.5). The active fractions were collected and dialyzed against 5 mM potassium phosphate buffer (pH 7.5). The enzyme solution was applied to a Gigapite column equilibrated with 5 mM potassium phosphate buffer (pH 7.5). After the column was washed with 5 mM potassium phosphate buffer (pH 7.5), the flowthrough fractions were collected as the purified enzyme. The purified enzyme was dialyzed against 10 mM HEPES-NaOH (pH 7.5), concentrated to 11 mg/ml for crystallization by ultrafiltration, and stored at −80°C until use.
Mutated pET21-FA1 plasmids and E. coli BL21(DE3) were used for the overexpression of mutant FAc-DEX FA1 enzymes. E. coli BL21(DE3) cells carrying the mutated pET21-FA1 were grown aerobically in LB medium containing 100 μg of ampicillin per ml at either 18°C for 40 h or 37°C for 16 h. Expression levels of the mutant enzymes were checked by SDS-PAGE. The D128A mutant was not overproduced under these conditions. For the production of this mutant, the cells were grown aerobically in LB medium containing 100 μg of ampicillin per ml at 37°C for 3.5 h and then isopropyl-β-d-thiogalactopyranoside was added to the medium to a final concentration of 1 mM for further cultivation at 18°C for 10 h. The purification procedures for mutant enzymes were the same as that for the wild-type enzyme.
Enzyme and protein assay.
FAc-DEX FA1 was routinely assayed by determining the amount of glycolate ions produced from fluoroacetate by the method of Dagley and Rodgers (6) or by determining the amount of halide ions released from chloroacetate and bromoacetate by the method of Iwasaki et al. (12). The standard assay mixture (100 μl) contained 25 mM haloacetate, 100 mM Tris-sulfate buffer (pH 9.5), and the enzyme. After the incubation of the mixture at 30°C for 1 to 5 min, the reaction was terminated by the addition of 11.1 μl of 1.5 M sulfuric acid. One unit of the enzyme was defined as the amount of the enzyme that catalyzes the dehalogenation of 1 μmol of haloacetate per min. The protein concentration was determined by the method of Bradford (3) with bovine serum albumin as a standard.
Crystallization and data collection.
Crystals of FAc-DEX FA1 were obtained by the hanging-drop vapor diffusion method at 277 K (13). The initial screening for the crystallization conditions was carried out using commercially available sparse-matrix screens, Wizard I and II. The optimal condition required the equilibration of a mixture containing 2 μl of the purified protein solution (11 mg/ml protein, 10 mM HEPES-NaOH [pH 7.5]) and an equal volume of a reservoir solution (15 to 18% [wt/vol] polyethylene glycol 8000, 40 mM MgCl2,100 mM Tris-HCl [pH 8.5]) against 500 μl of the reservoir solution at 293 K. FAc-DEX FA1 crystals appeared within a week of incubation and grew to maximum dimensions of 0.2 by 0.1 by 0.03 mm3.
The X-ray data set for FAc-DEX FA1 to 1.5-Å resolution at 100 K was collected using a wavelength of 1.00 Å with the synchrotron radiation source at the Photon Factory NW12 station (Tsukuba, Japan). For data collection under cryogenic conditions, crystals were soaked for a few seconds in the reservoir solution containing 15% (vol/vol) glycerol. The crystals belong to monoclinic space group C2 with the following unit cell parameters: a, 85.9 Å; b, 137.6 Å; c, 52.9 Å; and β, 95.1°. Assuming two monomers per asymmetric unit, the Matthews coefficient was calculated to be 2.28 Å3 Da−1, indicating an estimated solvent content of 46.1% in the unit cell (25). A heavy-atom derivative was obtained by soaking native crystals in a solution containing 10 mM ethylmercurithiosalicylic acid sodium salt (EMTS). The X-ray data set for the Hg derivative to 1.9-Å resolution at 100 K was collected using a wavelength of 1.00 Å with BL38B1 at SPring-8 (Hyogo, Japan). All the intensity data were processed and scaled using HKL2000/SCALEPACK (36).
Structure determination and refinement.
The structure of FAc-DEX FA1 was determined by the method of single isomorphous replacement with anomalous scattering using Hg derivative data. The scaling of data sets and map calculation were performed with the CCP4 program suite (5). Phasing and density modification were carried out with programs SOLVE (40) and RESOLVE (38, 39), respectively. The map was of good quality, and the modeling of FAc-DEX FA1 was completed through several rounds of manual building with the program O (14). The structure was refined by simulated annealing and energy minimization using the program CNS (4). Water molecules were picked from the 2Fo-Fc map (where Fo and Fc are the observed and calculated structure factors, respectively) at the 1.5σ level and were analyzed for hydrogen-bonding interactions with protein residues or other water molecules. The data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Statistics | Value(s) for:
|
|
|---|---|---|
| FAc-DEX FA1 | EMTS | |
| Diffraction data | ||
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution range (Å) | 50.0-1.5 | 44.52-1.9 |
| Unique no. of reflections | 97,170 | 47,258 |
| Completeness (%) | 99.8 (99.6)a | 94.0 (94.0)a |
| Rmergeb (%) | 6.3 (35.5)a | 4.8 (17.9)a |
| MIRc data | ||
| Rdiffd (%) | 18.9 | |
| No. of sites | 2 | |
| Phasing powere | 1.15 | |
| Refinement data | ||
| Resolution limits (Å) | 20.0-1.5 | |
| Rfactor (%)f | 18.1 | |
| Rfree (%)g | 20.3 | |
| No. of solvent atoms | 332 | |
| Deviations | ||
| Bond length (Å) | 0.019 | |
| Bond angle (°) | 1.9 | |
| Mean B factorsh | ||
| Main chain atoms (Å2) | 12.4 | |
| Side chain atoms (Å2) | 13.8 | |
| Water atoms (Å2) | 18.7 | |
| Ramachandran plot (%)i | 92.7, 6.9, 0.04, 0.0 | |
The values in parentheses are for the highest-resolution shells; 1.55-1.50 Å in FAc-DEX FA1 and 1.97 to 1.90 Å in EMTS.
Rmerge = ΣhklΣi|Ihkl,i − <Ihkl>|/ΣhklΣiIhkl,i, where Ihkl,i is the ith observation of the reflection hkl and <Ihkl> is the mean intensity of the reflection hkl.
MIR, multiple isomorphous replacement.
Rdiff = Σ||FPH| − |FP||/Σ|FP|, where |FPH| and |FP| are the derivative and native structure-factor amplitudes, respectively.
Phasing power is the ratio of the root mean square of the heavy-atom scattering amplitude to the root mean square of the lack of closure error.
Rfactor = Σ||Fobs| − |Fcalc||/Σ|Fobs|.
Rfree is calculated in the same manner as Rfactor, except that the Fobs corresponds to the 10% of reflections not used in the refinement.
Mean B factor is the average value of thermal factors of all the main-chain atoms.
Percentages of residues in the most favored, additionally allowed, generously allowed, and disallowed regions, respectively, of a Ramachandran plot (21).
Graphic manipulation.
Structure diagrams were drawn with the programs Molscript (19), POVScript+ (7), and Raster3D (27).
Site-directed mutagenesis of the active-site residues.
Site-directed mutagenesis was carried out by using the QuikChange site-directed mutagenesis kit (Stratagene) to obtain mutant FAc-DEX FA1 enzymes. The synthetic mutagenic primers used are shown in Table S1 in the supplemental material. The substitution was confirmed by DNA sequencing.
Kinetic analysis of the wild-type enzyme and the W150F mutant.
Initial rates of the dehalogenation reaction were measured with various concentrations of fluoroacetate or chloroacetate as the substrate. Data were directly fitted to the Michaelis-Menten equation, and kinetic parameters were calculated using nonlinear least-squares regression with the table calculation software KaleidaGraph (Synergy Software, Reading, PA).
Inhibition experiment for the W150F mutant.
Inhibition of the enzymatic dechlorination of chloroacetate by fluoroacetate was examined for the W150F mutant to investigate whether fluoroacetate can bind to the active site of the W150F mutant. Initial velocities of the dechlorination reaction were determined by measuring the amount of chloride ions released from chloroacetate with various concentrations (10, 20, 30, and 40 mM) of chloroacetate as the substrate and a fixed concentration (0, 10, or 20 mM) of fluoroacetate as the inhibitor. The apparent inhibition constant (Ki) for fluoroacetate was obtained from plots of reciprocals of initial velocities versus fluoroacetate concentrations.
Protein structure accession number.
The atomic coordinates and structures determined in this study have been deposited in the PDB with accession number 1Y37.
RESULTS AND DISCUSSION
Properties of FAc-DEX FA1 from recombinant E. coli cells.
FAc-DEX was purified 24-fold with 61% recovery from recombinant E. coli BL21(DE3) cells harboring pET21-FA1 (Table 2). The final preparation was shown to be homogeneous by SDS-PAGE. The specific activity of the purified enzyme was 28 U/mg. The substrate specificity of FAc-DEX FA1 was examined using various haloacetates and their analogs. The relative activities for haloacetates were as follows: fluoroacetate, 100; chloroacetate, 2.6; and bromoacetate, 2.5. The following organohalogen compounds were inert: iodoacetate, dichloroacetate, trichloroacetate, 2-chloropropionate, difluoroacetate, trifluoroacetate, 1,2-dichloroethane, 2-chloroacetamide, and 2-chloroacetonitrile. Thus, FAc-DEX FA1 is specific to haloacetates and shows the highest activity toward fluoroacetate, although the dissociation energy of the carbon-fluorine bond is much higher than that of other carbon-halogen bonds.
TABLE 2.
Purification of FAc-DEX FA1 from recombinant E. coli BL21(DE3) cells harboring pET-FA1a
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1,740 | 1,520 | 1.1 | 100 | 1.0 |
| Butyl-Toyopearl | 1,200 | 160 | 7.5 | 69 | 6.6 |
| DEAE-Toyopearl | 1,140 | 49 | 23 | 66 | 20 |
| Gigapite | 1,060 | 38 | 28 | 61 | 24 |
The dehalogenase activities were determined by measuring the amount of glycolate ions produced from fluoroacetate.
FAc-DEX FA1 showed typical Michaelis-Menten kinetics toward fluoroacetate and chloroacetate as the substrates. The Km and Vmax values of FAc-DEX FA1 for fluoroacetate were 9.1 mM and 61 U/mg, respectively, and those for chloroacetate were 15 mM and 2.6 U/mg, respectively. The Vmax of the recombinant FAc-DEX FA1 was higher than that of FAc-DEX FA1 purified from Burkholderia sp. strain FA1 (11 U/mg) (20). This is probably due to the inactivation of the enzyme during purification from the original strain.
Quality of the structure of FAc-DEX FA1.
We determined the three-dimensional structure of FAc-DEX FA1 by X-ray crystallographic analysis. The final model of the native FAc-DEX FA1 comprises two subunits of 294 residues each (the C-terminal 10 residues of each subunit were not visible), 330 water molecules, 2 chloride ions, and 2 Mg ions with an Rfactor value of 18.1% at 1.5 Å resolution. Glu6 and Thr182 of subunit A and Thr182 of subunit B, the side chains of which were disordered and invisible in the electron density map, were replaced by alanine residues. The Cα atoms of subunit A were superimposed onto the corresponding atoms of subunit B with a root mean square deviation of 0.2 Å and a maximum deviation of 2.0 Å (Gly228), indicating that the two independent subunits have the same structure. The average thermal factors for the main chain atoms (N, Cα, C, and O) in the two subunits are 12.6 and 12.1 Å2. Analysis of the stereochemistry with PROCHECK (21) showed that all the main chain atoms fall in the most favorable (92.7%), additionally allowed (6.9%), and generously allowed (0.4%) regions of a Ramachandran plot.
Overall structure of FAc-DEX FA1.
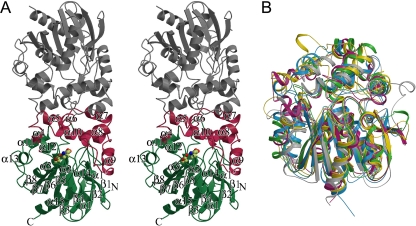
The overall structure of FAc-DEX FA1 with secondary structure assignments, as predicted with the program DSSP (15), is shown in Fig. 2A. The molecule is folded into a dimeric form with a noncrystallographic twofold axis. The surface area of the subunit interface was calculated to be 1,246 Å2. The molecular mass of the enzyme was estimated to be 79 kDa by gel filtration chromatography, and the calculated molecular mass of each subunit is 34,082 Da (20). The enzyme is thus assumed to exist as a homodimer with twofold symmetry.
FIG. 2.
Ribbon structural drawings of FAc-DEX FA1. (A) FAc-DEX FA1 dimer viewed perpendicularly to the molecular twofold axis. The core and cap domains of one subunit are represented in green and red, respectively, and the other subunit is depicted in gray. The catalytic triad, Asp104, His271, and Asp128, is shown as ball-and-stick models. (B) Superpositioning of Cα atoms of FAc-DEX FA1 (magenta) onto those of epoxide hydrolases and haloalkane dehydrogenases. The epoxide hydrolases from A. tumefaciens (PDB accession no. 1EHY) and M. musculus (PDB accession no. 1CR6) and the haloalkane dehalogenases from S. japonicum (PDB accession no. 2BFN) and X. autotrophicus (PDB accession no. 2DHC) are represented in green, yellow, cyan, and gray, respectively.
Each subunit consists of two domains, i.e., the core and cap domains. The core domain is formed by the polypeptide chains from the N terminus to Ile129 and from Thr217 to the C terminus, and the cap domain is composed of residues Ile130 to Gly216. The core domain has a typical α/β hydrolase fold with an eight-stranded β-sheet (β1 to β8) (10, 31, 35). The β-sheet (with all strands parallel except for β2), as a central core, is surrounded by four α-helices (α1′, α1, α4, and α15) on one side and five α-helices (α2, α3, α11, α12, and α13) on the other side.
The cap domain has an α-fold with seven α-helices (α4 to α10) and is inserted between strand β6 and helix α11 of the core domain. α-Helices α5 to α8 are stacked on α-helices α4, α9, and α10, forming a double-layered structure (33). The average thermal factor (29.3 Å2) of the Cα atoms in the extended loop between α-helices 8 and 9 is significantly higher than the factor of 12.4 Å2 for the cap domain. The cap domain residues of one subunit extensively interact with those of the other subunit, the surface area of the subunit interface amounting to 42.8% of the domain surface area. The active site is located at the domain interface. The subunit interface is distant from the active site and is probably not essential for the catalytic action.
Structural comparison with other proteins.
The program DALI (11) was used to search the PDB for enzymes possessing three-dimensional structures similar to that of FAc-DEX FA1. The enzymes with the highest Z scores (indicating the strength of structure similarity) are epoxide hydrolases and haloalkane dehalogenases, which belong to a large α/β hydrolase superfamily (10). The Z scores are 33.7 (PDB accession no. 1ZD5; sequence identity, 18%), 32.9 (PDB accession no. 1EHY; sequence identity, 23%), 29.0 (PDB accession no. 1CR6; sequence identity, 18%), and 27.5 (PDB accession no. 2CJP; sequence identity, 16%) for the epoxide hydrolases from Homo sapiens (9), Agrobacterium tumefaciens (33), Mus musculus (1), and Solanum tuberosum (29) and 27.5 (PDB accession no. 2BFN; sequence identity, 16%) and 24.5 (PDB accession no. 2DHC; sequence identity, 15%) for the haloalkane dehalogenases from S. japonicum UT26 (24) and X. autotrophicus GJ10 (43), respectively.
The main chain Cα atoms in FAc-DEX FA1 were superimposed onto the corresponding atoms in the α/β hydrolase fold enzymes listed above, showing that the central β-sheets of the core domains lend themselves very well to superimposition (Fig. 2B). Not only do the core domains of these enzymes have similar α/β structures, but the cap domains also have similar α structures, although superimposition is reported to work less well for the cap domains of the α/β hydrolase superfamily than for the core domains (24, 33, 34). The active sites in FAc-DEX FA1 and haloalkane dehalogenases are small compared with those in epoxide hydrolases, possibly because epoxide hydrolases accept larger substrates than FAc-DEX FA1 and haloalkane dehalogenases (17, 20, 26, 29, 32, 37).
Sequence alignment analysis was performed for epoxide hydrolases and haloalkane dehalogenases whose three-dimensional structures have been determined by X-ray crystallography (data not shown) (41). Only nine residues (Gly23, His32, Gly33, Pro35, Asp58, Gly61, Gly63, Asp104, and His271) in the core domains are conserved. Two of them (Asp104 and His271) are the residues of the catalytic triad, and three of them (His32, Gly33, and Pro35) are involved in the formation of the oxyanion hole. No conserved residue in the cap domains was observed, although the fold is conserved. The catalytic triad Asp-His-Asp/Glu plays a central role in the catalysis by epoxide hydrolases and haloalkane dehalogenases (10). The Asp-His dyad, in which Asp and His act as catalytic residues in the esterification and hydrolysis reactions, respectively, is conserved in the tertiary structure as well as the primary structure. However, the third residue (Asp or Glu) of the triad occupies one of the two conserved positions observed upon primary sequence alignment (33). Asp260 and Asp246, which are the third residues in the haloalkane dehalogenase from X. autotrophicus and the epoxide hydrolase from A. tumefaciens, respectively, occupy one of the two positions that is closer to the C terminus, whereas the haloalkane dehalogenase from S. japonicum has Glu132 as the third residue at the other position. FAc-DEX FA1 resembles the haloalkane dehalogenase from S. japonicum in the position of the third residue of the catalytic triad, Asp128. The α/β hydrolase superfamily members contain the consensus motif His-Gly-X-Pro (where X is any residue) at the active site. This motif is involved in the formation of the oxyanion hole, and X-Pro forms a sharp cis-proline turn (33). In FAc-DEX FA1, the consensus motif, His32-Gly33-Phe34-Pro35, is located between strand β3 and helix α1′ of the core domain.
Active-site structure of FAc-DEX FA1.
The active-site channel of FAc-DEX FA1 is located at the domain interface between the C-terminal side of the β-sheet in the core domain and α-helices (α5, α6, and α10) in the cap domain (Fig. 2A and 3A). The active-site residues are Phe34, Asp104, Arg105, Arg108, Asp128, His271, and Phe272 of the core domain and Tyr147, His149, Trp150, and Tyr212 of the cap domain. The channel is characterized by a number of charged or hydrophilic residues to accommodate a hydrophilic substrate, fluoroacetate. On the other hand, hydrophobic residues, apart from the catalytic triad, surround the active-site channel in the haloalkane dehalogenases from S. japonicum UT26 (24), X. autotrophicus GJ10 (43), and Rhodococcus sp. (34). The entrance of the active-site channel of FAc-DEX FA1 is encircled by the loops between β7 and α12 and β8 and α14 of the core domain, the β-turn between α5 and α6 of the cap domain, and the extended loop between α8 and α9 of the cap domain. The entrance is open, and a substrate can freely gain access to its binding site through the channel. On the other hand, the X-ray structures of haloalkane dehalogenases so far reported indicate that the entrance is almost completely closed in both native enzymes and enzymes in complexes (24, 34, 43), implying that the approach of the substrate makes the entrance open. Upon the binding of the substrate, the conformation around the substrate entrance of FAc-DEX FA1 may change to yield a closed form similar to those observed in haloalkane dehalogenases to prevent the free access of water molecules to the active site. The most probable candidate for the door of the entrance is Trp179 on the extended loop between α-helices 8 and 9, because this loop is mobile and is directed toward the solvent side, similar to those in related haloalkane dehalogenases (24, 34, 43).
FIG. 3.
Active-site structure of FAc-DEX FA1. (A) Active-site channel formed at the domain interface. The core and cap domains are shown in dark gray and light gray, respectively. The catalytic triad is shown as stick models and spheres. The channel was delineated using VOIDOO (18). (B) Stereoview of the active-site structure. The front and back of the figure represent the solvent side (entrance of the active-site channel) and the protein side (bottom of the channel), respectively. The secondary structures and side chains of the core and cap domains are depicted in green and pink, respectively. Chloride anions (Cl−) and water molecules (labeled W1 to W7) are denoted by green and blue circles, respectively. The active-site residues are represented by ball-and-stick models. Hydrogen bonds are denoted by dashed lines. The omit electron density map is contoured at the 1.0σ level. (C) Schematic drawing of the active site. Putative hydrogen bonds are denoted by dashed lines.
The active-site structure and the hydrogen-bonding scheme for the active site are shown in Fig. 3B and C, respectively. Asp104, His271, and Asp128 form a catalytic triad positioned on the C-terminal side of the β-sheet in the core domain. Asp104 is located at the sharp turn (nucleophile elbow) between strand β5 and helix α3 (35). The strand-nucleophile-helix motif referred to as the nucleophile elbow was first recognized in α/β hydrolase family proteins. Nδ1 and Nɛ2 of His271 are hydrogen bonded to the carboxylates of Asp128 and Asp104, respectively, indicating that both nitrogen atoms of His271 are protonated. The main chain NH groups of Arg105 and Phe34 in the core domain form hydrogen bonds with one of the carboxylate oxygens of Asp104 and are assumed to act as the oxyanion hole to stabilize the oxyanion formed during the hydrolysis of the ester intermediate, as commonly observed for the α/β hydrolase superfamily. These results imply that the carboxylate oxygen of Asp104, which is hydrogen bonded to Nδ2 of His271, acts as a catalytic nucleophile that attacks the α-carbon atom of the substrate.
The simulated annealing 2Fo-Fc map showed many residual electron density peaks at the active-site channel. Most of these peaks were assigned as water molecules on the basis of peak heights and distance criteria (Fig. 3B and C). One of these peaks, the atomic displacement parameter of which was 4.9 Å2, was coordinated by Trp150, His149, and four water molecules. The peak was refined as a chloride anion to give the atomic displacement parameter of 15.3 Å2 with the occupancy factor of 1.0. The peak was thus assumed to be the chloride anion originating from the crystallization buffer. The anion is within 3.0 to 3.4 Å from the Nɛ1 atom of Trp150, the Nɛ2 atom of His149, and water molecules W1 to W4 (Fig. 3B and C). The results raised the possibility that Trp150 and His149 function as halide ion acceptors in the FAc-DEX FA1 catalytic reaction.
The residues directly associated with catalysis are in the core domain, indicating that the core domain determines the reaction specificity. The active-site residues are mostly hydrophilic so as to interact with the substrate fluoroacetate. Water molecules occupying the active-site cavity form intricate hydrogen bond networks with Asp104, Arg105, Arg108, Tyr147, His149, Trp150, and Tyr212. Except for Asp104, none of these residues is conserved in the haloalkane dehalogenases from S. japonicum UT26 (24), X. autotrophicus GJ10 (43), and Rhodococcus sp. (34). The approach of the substrate to the active site of FAc-DEX FA1 probably allows most of these water molecules to be expelled from the cavity. The substrate then enters the active site, takes the place of the water molecules, and reconstructs the hydrogen bond networks with the hydrophilic residues of the active site.
Zhang et al. proposed that the main residues involved in the active site in FAc-DEX FA1 are Phe34, Tyr147, Trp148, Tyr212, Asp104, and His271, based on the three-dimensional model of the enzyme constructed by homology modeling (44). However, the structure experimentally determined by X-ray crystallography in the present study was significantly different from their model. Trp148 was not found in the active site in the crystal structure. Arg105, Arg108, Asp128, His149, Trp150, and Phe272, found in the active site by X-ray crystallography, were not identified as the active-site residues by the homology modeling study. Thus, the present crystal structure provides a new experimentally supported basis for further mechanistic analyses of FAc-DEX FA1.
Site-directed mutagenesis in the active site.
Each of the active-site residues of FAc-DEX FA1 was mutated to investigate the roles of these residues in catalysis. The activities of the mutant enzymes and the wild-type enzyme toward fluoroacetate and chloroacetate were measured by using cell extracts producing these enzymes at similar levels (Table 3). The mutant enzymes D104A, H271A, and D128A showed no activity toward both substrates, which is consistent with the interpretation that Asp104, His271, and Asp128 constitute a catalytic triad in FAc-DEX FA1. The following mutant enzymes also showed no activity toward both substrates: F34A, R105A, R108A, Y147A, H149A, W179A, Y212A, and F272A. Interestingly, the activity of the W150A mutant enzyme toward chloroacetate was not significantly different from that of the wild-type enzyme, whereas the defluorination activity toward fluoroacetate was completely lost by the W150A mutation. Trp150 was replaced also with Phe, Gln, Lys, Tyr, and Arg. None of the resulting mutants showed defluorination activity, whereas they had various degrees of dechlorination activity (Table 3). Thus, Trp150 is required for the defluorination of fluoroacetate but not for the dechlorination of chloroacetate.
TABLE 3.
Activities of the wild-type and the mutant FAc-DEX FA1 enzymes
| Enzyme | Sp act (U/mg)a for:
|
|
|---|---|---|
| Defluorinationb | Dechlorinationc | |
| Wild type | 1.04 | 0.07 |
| F34A | 0 | 0 |
| D104A | 0 | 0 |
| R105A | 0 | 0 |
| R108A | 0 | 0 |
| D128A | 0 | 0 |
| Y147A | 0 | 0 |
| H149A | 0 | 0 |
| W150A | 0 | 0.08 |
| W150F | 0 | 0.06 |
| W150Q | 0 | 0.05 |
| W150K | 0 | 0.1 |
| W150Y | 0 | 0.01 |
| W150R | 0 | 0.06 |
| W179A | 0 | 0 |
| Y212A | 0 | 0 |
| H271A | 0 | 0 |
| F272A | 0 | 0 |
Crude extracts were used for the enzyme activity assay.
The defluorination activities were determined by measuring the amount of glycolate ions produced from fluoroacetate.
The dechlorination activities were determined by measuring the amount of chloride ions released from chloroacetate.
Among the W150 mutants, W150F was selected for further analysis because Phe shows structural similarity to Trp and the effect of the W150F mutation on the active-site structure was considered to be subtle, which facilitates comparison between the wild-type and mutant enzymes. The W150F mutant enzyme was purified, and its kinetic parameters were determined. The Km and Vmax values for chloroacetate were 13 mM and 0.8 U/mg, respectively. These values were comparable to those of the wild-type enzyme (Km, 15 mM; Vmax, 2.6 U/mg). In contrast, activity toward fluoroacetate was not detected. We next examined whether fluoroacetate functions as a competitive inhibitor for the dechlorination of chloroacetate by mutant W150F to see whether fluoroacetate enters the active site. We found that the dechlorination of chloroacetate catalyzed by W150F was inhibited by fluoroacetate (data not shown). Plots of reciprocals of initial velocities versus reciprocals of chloroacetate concentrations at several fixed concentrations of fluoroacetate yielded a group of straight lines intersecting at the same point. Thus, fluoroacetate is a competitive inhibitor for the dechlorination of chloroacetate. The apparent Ki value was 7.8 mM. This value is comparable to the Km of the wild-type enzyme for fluoroacetate (9.1 mM). The results indicate that fluoroacetate gains access to the active site of the mutant enzyme W150F. Thus, the loss of activity toward fluoroacetate by the replacement of Trp150 with Phe was not due to the loss of fluoroacetate binding to the active site.
The three-dimensional structure of FAc-DEX FA1 determined in this study suggested that Trp150 at the active site binds to a chloride ion in the crystallization solution. This result implies that the residue functions as a halide ion acceptor in the dehalogenation of the substrate. The results of the site-directed mutagenesis experiments, which showed the alteration of the halogen specificity of the enzyme by the mutation of Trp150, support this view. A hydrogen-bonding interaction between Trp150 and the fluorine atom of fluoroacetate is probably an absolute requirement for the reduction of the activation energy for the cleavage of the rigid carbon-fluorine bond. This interaction may also be required for the proper orientation of fluoroacetate at the active site. This hypothesis is under investigation by computational studies of the reaction mechanism based on the structure revealed in this study. We are also analyzing the structures of enzyme-substrate complexes by using mutant enzymes, such as D104A and H271A, to reveal interactions between the enzyme and the substrate in a Michaelis complex and an ester intermediate.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-Aid for Global COE Program “Integrated Materials Science” (B-09) (to N.E. and K.J.), a Grant-in-Aid for Young Scientists (A) (14703021) from MEXT (to T.K.), and a Grant-in-Aid for Scientific Research (B) (17404021) from JSPS (to T.K.).
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Argiriadi, M. A., C. Morisseau, B. D. Hammock, and D. W. Christianson. 1999. Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase. Proc. Natl. Acad. Sci. USA 9610637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, M. L., C. M. Bothroyd, G. I. Rogers, A. Staffa, and I. D. Rae. 1987. Detection and measurement of fluoroacetate in plant extracts by 19F NMR. Phytochemistry 262293-2295. [Google Scholar]
- 3.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 5.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 502760-2763. [DOI] [PubMed] [Google Scholar]
- 6.Dagley, S., and A. Rodgers. 1953. Estimation of glycollic acid. Biochim. Biophys. Acta 12591. [DOI] [PubMed] [Google Scholar]
- 7.Fenn, T. D., D. Ringe, and G. A. Petsko. 2003. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 36944-947. [Google Scholar]
- 8.Goldman, P. 1969. The carbon-fluorine bond in compounds of biological interest. Science 1641123-1130. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, G. A., C. Morisseau, B. D. Hammock, and D. W. Christianson. 2006. Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids. Protein Sci. 1558-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heikinheimo, P., A. Goldman, C. Jeffries, and D. L. Ollis. 1999. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure 7R141-R146. [DOI] [PubMed] [Google Scholar]
- 11.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233123-138. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki, I., S. Utsumi, K. Hagino, and T. Ozawa. 1956. A new spectrophotometric method for the determination of small amounts of chloride using the mercuric thiocyanate method. Bull. Chem. Soc. Jpn. 29860-864. [Google Scholar]
- 13.Jancarik, J., and S. H. Kim. 1991. Sparse-matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 24409-411. [Google Scholar]
- 14.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 222577-2637. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki, H., N. Tone, and K. Tonumura. 1981. Plasmid-determined dehalogenation of haloacetates in Moraxella species. Agric. Biol. Chem. 4529-34. [Google Scholar]
- 17.Keuning, S., D. B. Janssen, and B. Witholt. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol. 163635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D 50178-185. [DOI] [PubMed] [Google Scholar]
- 19.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24946-950. [Google Scholar]
- 20.Kurihara, T., T. Yamauchi, S. Ichiyama, H. Takahata, and N. Esaki. 2003. Purification, characterization, and gene cloning of a novel fluoroacetate dehalogenase from Burkholderia sp. FA1. J. Mol. Catal. B 23347-355. [Google Scholar]
- 21.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 22.Liu, J. Q., T. Kurihara, A. K. M. Q. Hasan, V. Nardi-Dei, H. Koshikawa, N. Esaki, and K. Soda. 1994. Purification and characterization of thermostable and nonthermostable 2-haloacid dehalogenases with different stereospecificities from Pseudomonas sp. strain YL. Appl. Environ. Microbiol. 602389-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J. Q., T. Kurihara, S. Ichiyama, M. Miyagi, S. Tsunasawa, H. Kawasaki, K. Soda, and N. Esaki. 1998. Reaction mechanism of fluoroacetate dehalogenase from Moraxella sp. B. J. Biol. Chem. 27330897-30902. [DOI] [PubMed] [Google Scholar]
- 24.Marek, J., J. Vevodova, I. K. Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 3914082-14086. [DOI] [PubMed] [Google Scholar]
- 25.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33491-497. [DOI] [PubMed] [Google Scholar]
- 26.Meijer, J., and J. W. DePierre. 1988. Cytosolic epoxide hydrolase. Chem.-Biol. Interact. 64207-249. [DOI] [PubMed] [Google Scholar]
- 27.Merritt, E. A., and M. E. Murphy. 1994. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50869-873. [DOI] [PubMed] [Google Scholar]
- 28.Motosugi, K., N. Esaki, and K. Soda. 1982. Purification and properties of a new enzyme, dl-2-haloacid dehalogenase, from Pseudomonas sp. J. Bacteriol. 150522-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowbray, S. L., L. T. Elfstrom, K. M. Ahlgren, C. E. Andersson, and M. Widersten. 2006. X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci. 151628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, C. D., C. Schaffrath, and D. O'Hagan. 2003. Fluorinated natural products: the biosynthesis of fluoroacetate and 4-fluorothreonine in Streptomyces cattleya. Chemosphere 52455-461. [DOI] [PubMed] [Google Scholar]
- 31.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247536-540. [DOI] [PubMed] [Google Scholar]
- 32.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of a haloalkane dehalogenase of a new substrate class from a hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 633707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nardini, M., I. S. Ridder, H. J. Rozeboom, K. H. Kalk, R. Rink, D. B. Janssen, and B. W. Dijkstra. 1999. The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J. Biol. Chem. 27414579-14586. [PubMed] [Google Scholar]
- 34.Newman, J., T. S. Peat, R. Richard, L. Kan, P. E. Swanson, J. A. Affholter, I. H. Holmes, J. F. Schindler, C. J. Unkefer, and T. C. Terwilliger. 1999. Haloalkane dehalogenases: structure of a Rhodococcus enzyme. Biochemistry 3816105-16114. [DOI] [PubMed] [Google Scholar]
- 35.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, et al. 1992. The α/β hydrolase fold. Protein Eng. 5197-211. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 37.Rink, R., M. Fennema, M. Smids, U. Dehmel, and D. B. Janssen. 1997. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Biol. Chem. 27214650-14657. [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D 56965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger, T. C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 5938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Wijngaard, A. J., P. T. Reuvekamp, and D. B. Janssen. 1991. Purification and characterization of haloalcohol dehalogenase from Arthrobacter sp. strain AD2. J. Bacteriol. 173124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verschueren, K. H., F. Seljee, H. J. Rozeboom, K. H. Kalk, and B. W. Dijkstra. 1993. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363693-698. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., Z. S. Li, J. Y. Wu, M. Sun, Q. C. Zheng, and C. C. Sun. 2004. Homology modeling and S(N)2 displacement reaction of fluoroacetate dehalogenase from Burkholderia sp. FA1. Biochem. Biophys. Res. Commun. 325414-420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.