Abstract
The ability of Enterococcus faecalis to form robust biofilms on host tissues and on abiotic surfaces such as catheters likely plays a major role in the pathogenesis of opportunistic antibiotic-resistant E. faecalis infections and in the transfer of antibiotic resistance genes. We have carried out a comprehensive analysis of genetic determinants of biofilm formation in the core genome of E. faecalis. Here we describe 68 genetic loci predicted to be involved in biofilm formation that were identified by recombinase in vivo expression technology (RIVET); most of these genes have not been studied previously. Differential expression of a number of these determinants during biofilm growth was confirmed by quantitative reverse transcription-PCR, and genetic complementation studies verified a role in biofilm formation for several candidate genes. Of particular interest was genetic locus EF1809, predicted to encode a regulatory protein of the GntR family. We isolated 14 independent nonsibling clones containing the putative promoter region for this gene in the RIVET screen; EF1809 also showed the largest increase in expression during biofilm growth of any of the genes tested. Since an in-frame deletion of EF1809 resulted in a severe biofilm defect that could be complemented by the cloned wild-type gene, we have designated EF1809 ebrA (enterococcal biofilm regulator). Most of the novel genetic loci identified in our studies are highly conserved in gram-positive bacterial pathogens and may thus constitute a pool of uncharacterized genes involved in biofilm formation that may be useful targets for drug discovery.
Nosocomial infections by multiply antibiotic-resistant opportunistic pathogens such as Enterococcus faecalis have a major impact on morbidity, mortality, and health care costs (22, 37, 46, 52). E. faecalis is a normal resident of the human intestinal tract and can also be cultured from a variety of plants, animals, insects, and other environmental sources. Although E. faecalis is generally nonpathogenic in healthy humans, it is an extremely hardy and adaptable organism capable of survival and even robust growth under a variety of conditions that would be lethal for many phylogenetically related bacteria, such as pathogenic streptococci (19). Enterococci isolated from clinical sources almost invariably carry a cornucopia of mobile genetic elements, including plasmids, transposons, and genomic islands, that encode gene products that mediate horizontal transfer of high-level antibiotic resistance and virulence (11, 12, 54). Intra- and interspecies horizontal transfer of these elements has a high degree of medical significance, since it accelerates the evolution of increased bacterial virulence and resistance to antimicrobial agents. Both the core genome and the mobile elements of enterococci carry a variety of sensing systems that mediate cell-cell communication and detection of environmental signals; such systems are involved in the remarkable ability of these bacteria to adapt and proliferate in a variety of different ecological niches (2, 15, 16, 24).
Genetic analyses of enterococci have focused primarily on genetic determinants that are found more frequently in clinical isolates than in intestinal strains from healthy individuals (3, 13, 14, 39). These studies have provided important insights into the evolution of enterococcal virulence and resistance by horizontal transfer of mobile elements in the hospital environment. To date, however, there is no single virulence determinant associated with a mobile element that has been demonstrated to be essential for all enterococcal infections. Indeed, it could be argued that the E. faecalis core genome encodes a considerable capacity for adaptation to survival and growth under a variety of conditions and that this inherent adaptability provides an excellent evolutionary scaffold for the emergence of new clones via acquisition of mobile elements that enhance competitive fitness in immunocompromised patients subjected to extensive antibiotic treatment. We hypothesized that the core genome of E. faecalis includes a conserved minimal set of genetic determinants essential for biofilm formation and that disruption of any of these determinants would impair the ability of E. faecalis to cause infections that involve a biofilm component. While several loci that affect enterococcal biofilm formation have been identified (23, 24, 27, 42, 55), there has not been a comprehensive interrogation of the entire core genome for such genes. Many previously identified enterococcal biofilm determinants encode surface proteins involved in adhesion and thus represent a minority of the required functions for biofilm growth.
We set out to test our hypothesis by a systematic search for essential biofilm determinants in the genome of E. faecalis OG1RF, a recently sequenced laboratory strain that lacks plasmids and several other mobile DNA elements that encode antibiotic resistance and virulence genes that were identified previously in the genome of clinical isolate V583 (6). Because any single approach to this goal has limitations, we simultaneously employed two complementary methods, transposon mutagenesis (32) and recombinase-in vivo expression technology (RIVET) (10, 36), to identify chromosomally encoded biofilm determinants. Here we describe the results obtained from the RIVET screen and from follow-up gene expression analysis and functional complementation studies; the latter experiments confirmed a direct role in biofilm formation for several genes identified in the RIVET screen. For this screen, we employed submerged cellulose coupons, previously shown to support the development of robust biofilms with extensive three-dimensional structures (17); we felt that this system had the potential to identify biofilm determinants that were not identified in the 96-well polystyrene microtiter dish assay used in the transposon screen. Bioinformatic analyses revealed that a large number of novel potential biofilm determinants identified in our cumulative studies are highly conserved in the core genomes of many important bacterial pathogens. This work highlights the advantages of using multiple simultaneous approaches to functional genomic analysis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
E. faecalis OG1RF planktonic cells were grown in brain heart infusion broth (BHI; Difco) at 37°C unless otherwise noted. Mutants harboring nonpolar in-frame deletions of selected E. faecalis open reading frames (ORFs) were constructed by using a previously described allelic exchange system (30). Each deletion removed the entire predicted ORF except for the first and last three codons. For the sequences and genome locations of all of the primers used to create the deletions described in this paper, see Table S3 in the supplemental material. These primers were used to amplify the up- and downstream regions; the PCR products obtained were fused, inserted into pCJK47, and employed for allelic exchange as previously described (30). Complementation analysis used the genes immediately downstream from the predicted promoters identified in RIVET clones (along with their cognate ribosome binding site [RBS]), cloned, and expressed from plasmid pMSP3535 (8). This plasmid allows the expression of cloned sequences from a nisin-inducible promoter (8). These strains were grown in the presence of nisin at 25 ng/ml and erythromycin at 10 μg/ml. All plasmid and chromosomal constructs were confirmed by sequencing. Biofilms were grown on submerged sterilized cellulose coupons (17) (SpectraPor RC; Spectrum Laboratories) at either 30 or 37°C for the specified amounts of time in tryptic soy broth without glucose (TSB-gluc; Difco) in 3 ml in a plastic six-well dish (Costar 3516) unless otherwise noted. Coupons containing biofilms were washed three times in phosphate-buffered saline, sonicated two times (VirSonic 475; VirTis) at a setting of 2, and vortexed for 30 s twice in a plastic tube containing 500 μl of fresh phosphate-buffered saline in order to harvest biofilm cells.
Construction of a resolvase reporter fusion vector, pJMA61.
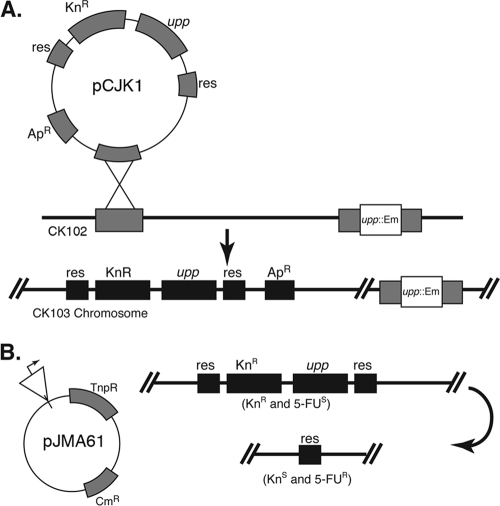
A SalI fragment bearing a transcriptional terminator (to prevent transcriptional readthrough from upstream sequences), a unique BglII site (to insert genomic DNA), and the gene that encodes TnpR resolvase preceded by a modified RBS (to enable efficient translation in E. faecalis) was excised from staphylococcal vector pES95 (36) and cloned into broad-host-range shuttle vector pAM401 (58) to produce resolvase reporter fusion vector pJMA61 (Fig. 1).
FIG. 1.
RIVET plasmid and strains. (A) RIVET host strain CK103 was created via Campbell insertion of pCJK1 containing the res cassettes flanking selectable and counterselectable genes into the CK102 chromosome. (B) An OG1RF genomic library was created in the resolvase fusion reporter vector pJMA61. This vector contains a promoterless copy of the resolvase gene, tnpR, adjacent to the cloning site. When tnpR transcription is activated, the resulting resolvase protein catalyzes site-specific recombination between the duplicated res sites (depicted on the right side), leaving a heritable genetic change in the host strain that can be screened for at a later time. The phenotype of the host strain before and after TnpR-mediated excision is indicated in parentheses.
Construction of an E. faecalis RIVET host strain, CK103.
A 2.2-kb BamHI fragment containing the Ω-Km2 cassette (48) was cloned into the BamHI site of pRR51 (49) to insert the kanamycin resistance determinant between two copies of the res sequence, creating pRRΩ. Subsequently, pRRΩ was digested at the unique PacI site (external to the res-Kan-res cassette) and PacI-digested E. faecalis genomic DNA was inserted. One recombinant clone (pRRΩ12b) bearing a 0.8-kb PacI fragment of genomic DNA, including the entire EF2078 ORF, annotated as a hypothetical protein, plus portions of the upstream and downstream ORFs, was found to efficiently integrate into the EF2078 locus of the OG1RF chromosome via Campbell-type insertion (pRRΩ12b cannot replicate in E. faecalis), and the resulting integrants did not display any noticeable defects in planktonic or biofilm growth. To introduce a counterselectable marker into pRRΩ12b, the P-upp cassette from pCJK2 (31) was amplified and inserted into the unique SphI site with primer-encoded SphI restriction sites, placing the counterselectable marker adjacent to the kanamycin resistance determinant between the res sites, thereby creating pCJK1. The resulting plasmid was introduced into E. faecalis CK102 (31) by electroporation with selection for kanamycin resistance, generating strain CK103. CK102 carries the upp1::erm allele, rendering the cells resistant to the toxic base analog 5-fluorouracil (5FU) (31). Recombinant strain CK103 was verified by PCR to contain pCJK1 integrated into the EF2078 locus and found to be phenotypically sensitive to 5FU, as expected due to the production of functional upp gene product from the integrated P-upp cassette.
RIVET screen for promoters induced during biofilm formation.
Genomic DNA from E. faecalis OG1RF was partially digested with Sau3AI. Fragments of 0.5 to 2.5 kb were extracted from an agarose gel and cloned into BglII-digested pJMA61 (H. S. Gold, C. T. Eliopoulos, G. M. Eliopoulos, R. C. Moellering, Jr., and D. T. Beattie, 36th Intersci. Conf. Antimicrob. Agents Chemother., 1996) to create a library of E. faecalis genomic DNA upstream of the tnpR reporter. pJMA61 confers resistance to chloramphenicol. Escherichia coli clones carrying the library were pooled, and plasmid DNA was isolated and introduced into CK103 by electroporation. Transformants were pooled and frozen at −80°C in aliquots with 30% glycerol. For RIVET screening, thawed aliquots were cultured for 8 h at 30°C in 3 ml TSB-gluc with an initial inoculation of 1:50 in the presence of 2,000 μg/ml kanamycin and 20 μg/ml chloramphenicol. After 8 h, the cells were collected by centrifugation and fresh medium containing 2,000 μg/ml kanamycin and 20 μg/ml chloramphenicol was added. This culture was incubated for 8 h at 30°C. The remaining cells were washed three times in TSB-gluc containing no antibiotics and then grown as biofilms as described above at 30°C for 2 h, 3 days, or 5 days in TSB-gluc with 10 μg/ml chloramphenicol. For the 3-day and 5-day screens, the liquid medium was removed and replaced every 24 h. The cellulose coupons containing the RIVET screen biofilms were removed from the wells after growth, and cells were harvested as described above. The resulting suspensions were diluted serially and plated on a medium containing 20 μg/ml chloramphenicol and 75 μg/ml 5FU. The resulting colonies were picked and patched to BHI agar plates containing either 1,000 μg/ml kanamycin and 10 μg/ml chloramphenicol or 75 μg/ml 5FU and 10 μg/ml chloramphenicol. Those colonies that did not grow on kanamycin were inoculated in 10 ml BHI liquid medium supplemented with 10 μg/ml chloramphenicol and grown overnight for plasmid isolation with a kit (Qiagen). The inserts in the plasmids were then sequenced by using primers that would produce sequence from both strands of the insert (forward, 5′ AGC GTC GAC TCT AGA GAT CCA G 3′; reverse, 5′ TAC CCG TGC GTA ACC AAA AAG TCG 3′).
Bioinformatic analysis.
The resulting sequences were compiled and batch BLASTed against the omniome at http://www.jcvi.org/, as well as imported into a sequence analysis program, Sequencher 4.8 (http://www.genecodes.com), and allowed to “assemble automatically” into a contig carrying the E. faecalis V583 genome ORFs. Once the E. faecalis OG1RF genome was made available, the sequence was added to this contig file to verify the results in OG1RF as well. These results were compared with those from the BLAST analysis, and the ORF downstream of the cloned chromosomal insert fragment containing the putative promoter was identified.
Quantitative reverse transcription (qRT)-PCR.
Biofilms were grown as described above to obtain cells for RNA extraction. Planktonic cells were isolated from the same vessel in which biofilms were grown and at the same time points. RNA Protect reagent (Qiagen) was used to stabilize the RNA until extraction. The RNA was extracted from biofilm and planktonic cells with the RNeasy kit (Qiagen) by following the suggested protocol for bacterial RNA with the following modifications. Prior to using the kit, the cell walls were subjected to enzymatic degradation at 37°C for 10 min with 50 mg/ml lysozyme (Sigma) and 1,000 U/ml mutanolysin (Sigma) in 10 mM Tris made with RNase-free water. The resulting total RNA was DNase treated with the Turbo DNA-Free kit (Ambion) by following the “rigorous” protocol. The DNase-treated RNA was then checked for DNA contamination by PCR with previously published 16S (56) and gyrB (7) primers. The RNA was reverse transcribed to cDNA by using the Superscript III First Strand synthesis system for RT-PCR kit (Invitrogen). PCR primers were designed for genes of interest using the published genome sequence and primer 3 (51). qRT-PCR was undertaken by following the instructions from Bio-Rad with iQ SYBR green Supermix and using a Bio-Rad iQ5 thermocycler. Data were analyzed with the software supplied with the iQ5 thermocycler.
Biofilm phenotype assay.
Biofilms for the biofilm phenotype assay were grown as described above, and biofilm cells were harvested by sonication. The resulting harvested cells were serially diluted and plated to an appropriate medium. For complementation experiments, both the liquid and agar media contained erythromycin at 10 μg/ml. The markerless null mutants were grown with no antibiotics. All mutants were compared to the isogenic wild-type E. faecalis control strain.
RESULTS
Construction of a resolvase reporter system for E. faecalis.
RIVET screens are designed to identify promoters that are active under a condition of interest, such as during animal infections, and that drive very low or undetectable levels of transcription during growth in laboratory medium. The use of recombination as a reporter for gene expression has been described by Camilli and coworkers (10, 36). The system relies on the ability of the site-specific TnpR recombinase of Tnγδ to excise DNA that is flanked by its target sequences (res), generating a heritable change in genotype. If res sites flank a counterselectable marker, expression of resolvase results in loss of the intervening marker and therefore a selectable phenotype (Fig. 1). While this approach has been well developed for use with gram-negative bacteria, several modifications were required for use with E. faecalis. Our modifications included the introduction of appropriate selectable and counterselectable markers into a cassette flanked by res sequences, integration of this cassette into the chromosome of an E. faecalis reporter strain, and construction of a resolvase fusion vector that encodes tnpR with an RBS modified for efficient recognition in E. faecalis (Fig. 1). When a pJMA61 derivative containing a cloned genomic fragment that encodes an active promoter is propagated in CK103, expression of resolvase and excision of the res-Kan-upp-res cassette from the chromosome occur. This permanent genotypic change results in a selectable phenotype: the cells become resistant to the toxic base analog 5FU (31), enabling selection to be used to identify clones that have undergone resolution.
RIVET for genetic analysis of biofilm formation.
To identify genes specifically activated during biofilm growth, we constructed a library of E. faecalis genomic DNA in resolvase fusion reporter vector pJMA61, introduced the resulting library into RIVET host strain CK103, and cultured the library in liquid medium containing high levels of kanamycin and normal levels of chloramphenicol; this served to eliminate most of the clones containing promoters expressed during planktonic growth. We then inoculated the surviving bacteria into cultures containing submerged cellulose membranes as a surface for biofilm growth. After various time periods, the membranes were removed and rinsed to dislodge loosely attached bacteria and the biofilm cells were removed by sonication and plated on agar medium containing 5FU. Colonies appearing on these plates were tested for kanamycin sensitivity, and the inserts in the plasmid DNA from approximately 300 Kans 5FUr clones were sequenced and analyzed as described in Materials and Methods to identify the cloned genomic regions. We expected that expression of genes important in biofilm development would occur in a temporal fashion and that some important genes might be transiently expressed. Genes that were either transiently expressed or expressed at a relatively low level would still undergo a permanent and heritable genetic change, and the clones containing these promoters should still be present in libraries harvested at any time point following the activation of their expression; thus, the time point at which a given clone was isolated does not provide a precise indication of when the relevant promoter was activated or of the duration of its activation. However, in cells harvested from biofilms grown for prolonged periods, the representation of the transiently expressed promoters in the resulting library of RIVET clones might be very low. To increase the representation of such promoters in our library, we isolated clones from biofilms grown for various lengths of time chosen arbitrarily. Approximately 30% of the clones analyzed were from 72-h biofilms, 50% were from 120-h biofilms, and 20% were from 2-h biofilms. It should also be noted that clones containing a genomic fragment with a biofilm active promoter and a transcription termination signal would not allow for readthrough into the TnpR recombinase and would not be represented in the results. Most of the clones contained one or more ORFs (or partial ORFs) and associated 5′ untranslated regions oriented such that transcription from a promoter within the untranslated region would read into the flanking vector sequences and result in tnpR expression. Exceptions to this pattern will be discussed below.
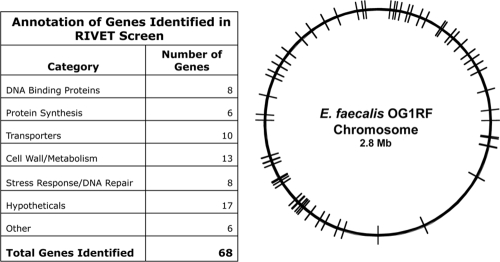
From the screen described above, we were able to identify 68 unique cloned regions containing putative biofilm-induced promoters (see Table S1 in the supplemental material). As illustrated in Fig. 2, the adjacent genes whose expression is driven by these promoters are distributed around the entire genome and encode proteins with a variety of predicted functions; for a complete listing of these genes, see the supplemental material. The genes in this group include those previously implicated in biofilm formation (1, 4, 5, 7, 9, 20, 25, 26, 28, 29, 33-35, 39-44, 47, 53, 57), as well as numerous genes not known to affect the formation of biofilms and in many cases never analyzed functionally. The previously reported biofilm genes of the enterococci are not represented in this group. This is not surprising, given that most previously identified determinants likely mediate adherence and would therefore need to be expressed in planktonic cells to allow the initiation of surface growth. Such genes would have been selected against in our screen during the planktonic growth of the inocula used for biofilm formation. Several of the loci were represented in multiple unique clones, suggesting that the screen was approaching saturation, at least at the later time points under the sampling conditions we used. Table 1 shows the 18 genes that were identified in nonsibling clones more than twice. The most frequently identified locus contained EF1809, a putative transcription regulator of the GntR family (21), was isolated in 14 unique clones. Further analysis of EF1809 is described below.
FIG. 2.
Genome distribution of determinants identified in a RIVET screen. The list on the left indicates the number of putative biofilm-related genes identified by RIVET in various categories based on the TIGR (The Institute for Genomic Research) gene annotation for the genome of E. faecalis V583 (45) and on the recently published genome sequence of OG1RF (6). In the circular map on the right, the location of each gene in the chromosome is indicated by a line. For a complete compilation of all 68 genetic determinants identified in this study, see Table S1 in the supplemental material; further analysis of selected genes is presented in other sections of this paper.
TABLE 1.
Genes identified in nonsibling clones more than twice
| TIGRa identifier | Annotation | No. of unique clones |
|---|---|---|
| EF038 | Glutamate 5-kinase | 5 |
| EF059 | UDP-N-acetylglucosamine pyrophosphorylase | 8 |
| EF365 | Conserved hypothetical protein | 11 |
| EF417 | Conserved hypothetical protein | 5 |
| EF457 | PTSb system, IIB component | 4 |
| EF798 | Hypothetical protein | 9 |
| EF977 | N utilization substance protein B | 4 |
| EF1081 | Conserved hypothetical protein | 3 |
| EF1591 | Transcriptional regulator, AraC family | 3 |
| EF1727 | EbsA protein | 3 |
| EF1755 | Phosphate ABC transporter, ATP binding protein | 3 |
| EF1809 | Transcriptional regulator, GntR family | 14 |
| EF1626 | Alcohol dehydrogenase, zinc containing | 5 |
| EF1918 | Conserved hypothetical protein | 8 |
| EF1978 | DNA-3-methyladenine glycosylase | 5 |
| EF2207 | DNA binding protein, Fis family | 6 |
| EF2570 | Aldehyde oxidoreductase, putative | 5 |
| EF2744 | Peptidase, M42 family | 5 |
TIGR, The Institute for Genomic Research.
PTS, phosphotransferase.
Confirmation of RIVET screen results.
Several genes were chosen for initial follow-up confirmation studies based on several criteria. EF3282 was chosen because homologues of this gene have been implicated in biofilm formation in other organisms. EF798 and EF984 were chosen because they are physically linked to genes identified in the complementary biofilm transposon screen, and EF1809 and EF2207 were chosen for further examination because they were identified in the RIVET screen more than three times. These genes were identified from many different regions of the genome and likely encoded diverse biological functions.
qRT-PCR was used to confirm that selected cloned genomic fragments identified in the RIVET screen actually contained promoters that were differentially activated during biofilm growth relative to planktonic growth. For this analysis, RNA was harvested from E. faecalis OG1RF cells at 24 h of planktonic or biofilm growth under the same conditions employed in the RIVET screen. The 24-h time point was chosen for these conformational studies based on biofilm kinetic data that showed that 24-h biofilms are indistinguishable, in terms of bacterial population levels, from 72- and 120-h biofilms (unpublished data). The results of this experiment are shown in Fig. 3. Expression levels were checked for four different loci, EF1809, EF2207, EF3282, and EF984. In all cases, there was a higher level of expression seen in the 24-h biofilm sample than in the 24-h planktonic sample. In the case of EF1809, the biofilm expression level was 12.5 times the planktonic expression level. The high level of expression of this locus in biofilms is consistent with the frequent isolation of EF1809-containing clones in the RIVET screen. The fact that differential expression was observed for all four randomly selected clones suggests that the isolation of E. faecalis loci in the RIVET screen is predictive of increased expression of those loci during biofilm growth.
FIG. 3.
Confirmation of differential expression of RIVET-identified genes during biofilm growth. The transcription of four genes identified in the RIVET screen was examined by qRT-PCR analysis of RNA extracted from either planktonic cells or biofilm cells after 24 h of growth. All of the graphs show the n-fold expression change on the y axis normalized to that of the reference gene, gyrB (7). This gene was shown to be expressed at the same level by biofilm and planktonic cells of E. faecalis strain OG1RF (data not shown). The level of expression of the gene of interest relative to gyrB in the planktonic sample was set at 1 in all cases and is shown in gray on the x axis along with the biofilm sample shown in black. (A) Expression of EF1809. (B) Expression of EF3282. (C) Expression of EF2207. (D) Expression of EF984. Error bars represent 1 standard deviation. The asterisk indicates a P value of 0.001.
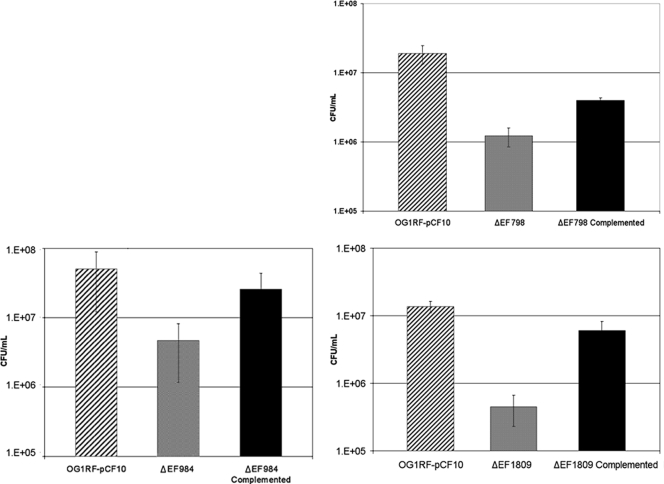
We have begun to assess the functional role of the RIVET-identified loci in biofilm formation by constructing in-frame markerless deletions in selected genes and examining the mutants for defects in biofilm formation by using a CFU-based accumulation assay. Mutant and wild-type cells were grown as biofilms for 24 h under the same conditions used for the RIVET screen. As shown in Fig. 4, null mutations in three RIVET-identified genes, EF1809, EF798, and EF984, all resulted in major reductions in the CFU counts of 24-h biofilms relative to those of the wild type. In all of the cases tested, these defects were complemented by nisin-induced expression of the cloned wild-type gene. Thus, for the genes that have been tested to date, the RIVET screen has successfully identified genetic determinants that are specifically upregulated in biofilm growth and that play a significant role in biofilm formation.
FIG. 4.
Biofilm defects of E. faecalis strains carrying null mutations in RIVET-identified genes. Isogenic strains carrying nonpolar null mutations in either EF1809, EF798, or EF984 were examined for biofilm formation on submerged cellulose membranes. The populations of mutant cells in biofilms after 24 h were compared with those obtained with either the wild-type strain or the null strain expressing the cloned wild-type gene in trans as described in Materials and Methods. This assay was repeated on five separate occasions. In planktonic growth, neither the null strains nor the complemented strains showed a defect compared to the wild type (data not shown). The strain designations are indicated on the x axis, and numbers of CFU per milliliter are shown on the y axis. Error bars represent 1 standard deviation. P values: EF1809, 0.01; EF798, 0.01; EF984, 0.01.
RIVET screen results are complementary to those of a transposon screen for biofilm determinants in E. faecalis.
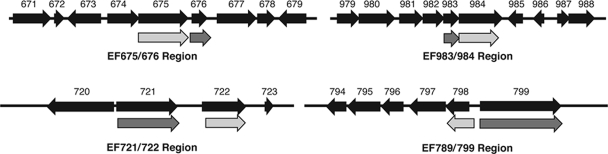
We recently described a new random transposon mutagenesis system for use in gram-positive organisms and demonstrated its utility for investigating biofilm formation in E. faecalis (32). Comparison of the data sets from the two screens was noteworthy in that each screen identified unique genes, but in several cases these genes were closely linked and possibly in shared regulatory networks as well. Figure 5 shows maps of pairs of adjacent genes where one gene was identified in the transposon screen and the other was identified in the RIVET screen. The annotation of the predicted proteins and the linkage and organization of these pairs of genes indicate that at least some of these pairs may represent a regulator and its target and suggest experimental approaches to examine these relationships further. Interactions between these gene pairs are supported in some cases by their molecular organization and by the fact that in two of the cases tested thus far, inactivation of one of the putatively linked genes (EF 799, identified by transposon mutagenesis [32], and EF984, identified in the RIVET screen [Fig. 5]) produced phenotypic defects in biofilm formation similar to those observed following disruption of the other predicted linked genes (EF798 [Fig. 5] and EF983 [32]). Further analysis is required to verify the predicted regulatory interactions, but the data suggest that the physical linkages could reflect functional linkage.
FIG. 5.
RIVET and transposon screens for biofilm determinants identify unique but linked genes. Linear maps of four regions (black arrows) where the RIVET screen and transposon screens identified adjacent genes are depicted at the top. Annotation of the genes is shown at the bottom; RIVET-identified genes are in light gray, and transposon-identified genes are in dark gray.
Comprehensive genomic screening reveals multiple novel regulatory genes related to E. faecalis biofilm formation and extensive conservation of putative novel biofilm determinants in gram-positive pathogens.
An important conclusion from our cumulative studies is that the E. faecalis core genome contains many predicted transcription factors, heretofore not investigated, that appear to be important in biofilm formation. Table 2 lists seven predicted DNA binding proteins implicated in biofilm formation based on our screens. The E. faecalis hrcA (EF1306) gene and other, neighboring genes are highly similar to a conserved stress response operon that also plays a role in biofilm development in the streptococci (33). The other seven putative regulators (or orthologs in other organisms) listed in Table 2 had not been analyzed prior to our studies. This list of predicted transcription factors is probably an underestimate since our complete collection of candidate biofilm genes (see Table S1 in the supplemental material) includes many possible regulatory proteins (such as EF798; Fig. 5) whose functions and biochemical activities are not known. Interestingly, a majority of previously uncharacterized genes identified in our studies are highly conserved among a large group of low-G+C gram-positive bacteria. This suggests the possibility that a large number of conserved biofilm determinants remain to be identified and functionally analyzed in many important pathogenic bacteria (see Table S1 and Fig. S1 in the supplemental material).
TABLE 2.
Predicted transcription factors
| Gene designator | Family annotation | Screen | Confirmed biofilm phenotype |
|---|---|---|---|
| EF117 | GntR | RIVET | NDa |
| EF676 | ArgR | Transposon | Yes |
| EF873 | Cro/Cl | RIVET | ND |
| EF983 | ArgR | Transposon | Yes |
| EF1306 | HrcA | Transposon | Yes |
| EF1591 | AraC | RIVET | ND |
| EF1809 | GntR | RIVET | Yes |
| EF2203 | TetR | RIVET | ND |
ND, not determined.
DISCUSSION
The work presented in this paper demonstrates the utility of a RIVET screen for the identification of novel genetic determinants of biofilm formation in the gram-positive nosocomial pathogen E. faecalis. In this screen, 68 candidate biofilm genes were identified; these genetic determinants include those annotated to have functions previously associated with biofilm formation (stress response and polysaccharide metabolism), as well as many that are completely novel. We identified 17 conserved hypothetical genes and 7 genes predicted to encode DNA binding proteins not previously linked to biofilm formation. As noted above, the present results may be relevant to elucidation of the genetic basis of biofilm formation in other organisms (see Table S1 and Fig. S1 in the supplemental material). This RIVET screen and a recent transposon mutagenesis study also undertaken by our group (32) both focused on genetic determinants in the core genome of E. faecalis. While the former study required the use of a microtiter plate biofilm assay, the RIVET study enabled us to use a submerged-coupon system previously shown to support the robust formation of E. faecalis biofilms with distinctive three-dimensional architecture and abundant production of extracellular matrix (18). We suspect that this may account for the fact that many more predicted biofilm determinants were identified in the RIVET screen. While it is certainly likely that additional determinants could be identified by changing the conditions for biofilm growth used in the screens, the cumulative set of genes already identified may represent a large percentage of the genes involved in biofilm formation by E. faecalis. This work thus constitutes the first comprehensive investigation of determinants of biofilm formation in the core genome of E. faecalis or closely related organisms.
Comprehensive validation of the RIVET screen would require confirmation of differential expression during biofilm growth and biofilm defects of nonpolar mutants for every genetic locus identified. This was beyond the scope of the present study, but it was important to sample a few representative loci to justify longer-term follow-up studies. In an attempt to provide an unbiased sample of predicted biofilm loci, we examined a partially overlapping subset of candidate genes widely distributed around the genome and having diverse predicted functions. The results presented in Fig. 3 and 4 suggest that isolation of a particular locus in the RIVET screen is predictive of the involvement of that locus in biofilm formation. Current and future studies will extend these analyses to genes predicted to function in the same pathway or process as the genes selected initially and to other loci. EF1809, which encodes a predicted DNA binding protein of the GntR family (21), was most frequently identified in nonsibling clones by the RIVET screen. It has also been confirmed to be differentially expressed via qRT-PCR (Fig. 3) and shown to be necessary for the ability of E. faecalis to form a wild-type biofilm (Fig. 4). The GntR family of transcriptional regulators consists of an N-terminal helix-turn-helix region and a C-terminal effector binding domain (50). The GntR family includes the subcategories MocR, YtrR, FadR, AraR, HutC, PlmA, DevA, and DasR (2, 50). Based on sequence similarity, it appears that EF1809 is a member of the HutC/FarR subfamily (2). EF1809 and the other members of this group contain a C-terminal UbiC transcription regulator-associated domain as their effector binding domain which has been shown experimentally in other family members to modulate the activity of some of these transcription factors in response to small molecules (2). The EF1809-null mutant showed 50-fold-reduced biofilm formation compared to that of the wild-type strain. From these results, we conclude that EF1809 is a novel regulator involved in biofilm formation and propose to designate this gene ebrA (enterococcal biofilm regulator).
In addition to the 68 genomic regions described above and in Table S1 in the supplemental material, we isolated eight clones (see Table S2 in the supplemental material) containing genomic regions where (i) the entire insert was within an ORF, (ii) the predicted transcription of all of the ORFs within and flanking the insert was in the opposite direction from that of the recombinase reporter gene in the recombinant plasmid, or (iii) both i and ii were true. It is possible that these clones are artifacts or that they encode small regulatory RNAs or peptides whose expression is increased during biofilm development. This suggests that the RIVET screen has the potential to identify significant small, non-protein-coding genetic determinants that might be missed in other screens due to transient expression, to the small size of the genetic locus, or to the confounding effects of other, overlapping determinants in the same region. We are currently examining selected clones from this group to assess their significance.
Null mutants with biofilm defects (e.g., Fig. 4) are likely to be reduced in virulence in experimental animal models of infections that involve biofilm formation such as endocarditis (38), and such studies are being undertaken. If this prediction holds, and if the conserved genes in other pathogens have similar functions, these determinants might make useful targets for the development of antimicrobial drugs that could block infections without directly killing the bacterial agent. Such compounds would also be useful tools in more detailed basic studies of biofilm development. The large number of novel genes identified in our studies constitutes a significant pool of target candidates for such studies.
Supplementary Material
Acknowledgments
We thank Dawn Manias and Vy Nguyen for assistance with and advice on these experiments and Tim Leonard for help in preparing the figures. Computational resources and support were provided by the Center for Biomedical Research Informatics at the University of Minnesota.
During a portion of this work, C.J.K. was supported by NRSA fellowship F32-AI56684 and K.S.B. was supported by the Dennis Watson fellowship from the Department of Microbiology. This research was supported by NIH grant AI058134 to G.M.D.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, G. G., S. Moreau-Marquis, B. A. Stanton, and G. A. O'Toole. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 761423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and V. Anantharaman. 2003. HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold. FEMS Microbiol. Lett. 22217-23. [DOI] [PubMed] [Google Scholar]
- 3.Baldassarri, L., R. Creti, C. R. Arciola, L. Montanaro, M. Venditti, and R. Di Rosa. 2004. Analysis of virulence factors in cases of enterococcal endocarditis. Clin. Microbiol. Infect. 101006-1008. [DOI] [PubMed] [Google Scholar]
- 4.Becker, P., W. Hufnagle, G. Peters, and M. Herrmann. 2001. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 672958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benkert, B., N. Quack, K. Schreiber, L. Jaensch, D. Jahn, and M. Schobert. 2008. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 1543053-3060. [DOI] [PubMed] [Google Scholar]
- 6.Bourgogne, A., D. Garsin, X. Qin, K. Singh, J. Sillanpaa, S. Yerrapragada, Y. Ding, S. Dugan-Rocha, C. Buhay, H. Shen, G. Chen, G. Williams, D. Muzny, A. Maadani, K. Fox, J. Gioia, L. Chen, Y. Shang, C. Arias, S. Nallapareddy, M. Zhao, V. Prakash, S. Chowdhury, H. Jiang, R. Gibbs, B. Murray, S. Highlander, and G. Weinstock. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgogne, A., K. V. Singh, K. A. Fox, K. J. Pflughoeft, B. E. Murray, and D. A. Garsin. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 1896490-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44183-190. [DOI] [PubMed] [Google Scholar]
- 9.Bush, A., and B. K. Rubin. 2003. Macrolides as biological response modifiers in cystic fibrosis and bronchiectasis. Semin. Respir. Crit. Care Med. 24737-748. [DOI] [PubMed] [Google Scholar]
- 10.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 912634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 251377-1388. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B. 2007. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58205-227. [DOI] [PubMed] [Google Scholar]
- 13.Creti, R., M. Imperi, L. Bertuccini, F. Fabretti, G. Orefici, R. Di Rosa, and L. Baldassarri. 2004. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 5313-20. [DOI] [PubMed] [Google Scholar]
- 14.Donelli, G., and E. Guaglianone. 2004. Emerging role of Enterococcus spp. in catheter-related infections: biofilm formation and novel mechanisms of antibiotic resistance. J. Vasc. Access 53-9. [DOI] [PubMed] [Google Scholar]
- 15.Dunny, G. M. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signaling, gene transfer, complexity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3621185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny, G. M., and B. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51527-564. [DOI] [PubMed] [Google Scholar]
- 17.Erlandsen, S. L., C. J. Kristich, and G. M. Dunny. 2004. Ultrastructure of Enterococcus faecalis biofilms. Biofilms 1131-137. [Google Scholar]
- 18.Erlandsen, S. L., C. J. Kristich, G. M. Dunny, and C. L. Wells. 2004. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem. Cytochem. 521427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Facklam, R., M. da Gloria, S. Carvalho, and L. M. Teixeira. 2002. History, taxonomy, biochemical characteristics and antibiotic susceptibility testing of enterococci, p. 1-54. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology and antibiotic resistance. ASM Press, Washington, DC.
- 20.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51675-690. [DOI] [PubMed] [Google Scholar]
- 21.Fujita, Y., and T. Fujita. 1987. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc. Natl. Acad. Sci. USA 844524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furtado, G. H. C., R. E. Mendes, A. C. Campos Pignatari, S. B. Wey, and E. A. S. Medeiros. 2006. Risk factors for vancomycin-resistant Enterococcus faecalis bacteremia in hospitalized patients: an analysis of two case-control studies. Am. J. Infect. Control 34447-451. [DOI] [PubMed] [Google Scholar]
- 23.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 1865629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock, L. E., and M. Perego. 2004. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J. Bacteriol. 1867951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, T.-P., E. B. Somers, and A. C. L. Wong. 2006. Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 1883116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 1472517-2528. [DOI] [PubMed] [Google Scholar]
- 27.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189420-430. [DOI] [PubMed] [Google Scholar]
- 28.Kemp, K. D., K. V. Singh, S. R. Nallapareddy, and B. E. Murray. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 755399-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, M. H., W.-C. Choi, H. O. Kang, J. S. Lee, B. S. Kang, K.-J. Kim, Z. S. Derewenda, T.-K. Oh, C. H. Lee, and J.-K. Lee. 2005. The molecular structure and catalytic mechanism of a quorum-quenching N-acyl-l-homoserine lactone hydrolase. Proc. Natl. Acad. Sci. USA 10217606-17611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristich, C. J., J. R. Chandler, and G. M. Dunny. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristich, C. J., D. A. Manias, and G. M. Dunny. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 715837-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristich, C. J., V. T. Nguyen, T. Le, A. M. T. Barnes, S. Grindle, and G. M. Dunny. 2008. Development and use of an efficient system for Random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl. Environ. Microbiol. 743377-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemos, J. A. C., Y.-Y. M. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 1836074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 1856241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27967-976. [DOI] [PubMed] [Google Scholar]
- 37.Marothi, Y., H. Agnihotri, and D. Dubey. 2005. Enterococcal resistance—an overview. Indian J. Med. Microbiol. 23214-219. [PubMed] [Google Scholar]
- 38.McCormick, J. K., H. Hirt, C. M. Waters, T. J. Tripp, G. M. Dunny, and P. M. Schlievert. 2001. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect. Immun. 693305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 723658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohamed, J. A., F. Teng, S. R. Nallapareddy, and B. E. Murray. 2006. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type I and fibronectin. J. Infect. Dis. 193231-240. [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee, P. K., S. Mohamed, J. Chandra, D. Kuhn, S. Liu, O. S. Antar, R. Munyon, A. P. Mitchell, D. Andes, M. R. Chance, M. Rouabhia, and M. A. Ghannoum. 2006. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect. Immun. 743804-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nallapareddy, S., K. Singh, J. Sillanpaa, D. A. Garsin, M. Hook, S. Erlandsen, and B. E. Murray. 2006. Endocarditis and biofilm associated pili of Enterococcus faecalis. J. Clin. Investig. 1162799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. von Holy, and V. S. Brozel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 682770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 45.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 46.Pearman, J. W. 2006. 2004 Lowbury Lecture: the Western Australian experience with vancomycin-resistant enterococci—from disaster to ongoing control. J. Hosp. Infect. 6314-26. [DOI] [PubMed] [Google Scholar]
- 47.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1811203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 1732617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed, R. R., and N. D. F. Grindley. 1981. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell 25721-728. [DOI] [PubMed] [Google Scholar]
- 50.Rigali, S., M. Schlicht, P. Hoskisson, H. Nothaft, M. Merzbacher, B. Joris, and F. Titgemeyer. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 323418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 52.Sharma, R., L. Sharma, and B. Kapoor. 2005. Antibacterial resistance: current problems and possible solutions. Indian J. Med. Sci. 59120-129. [PubMed] [Google Scholar]
- 53.Sheikh, J., S. Hicks, M. Dall'Agnol, A. D. Phillips, and J. P. Nataro. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41983-997. [DOI] [PubMed] [Google Scholar]
- 54.Sung, J. M.-L., and J. A. Lindsay. 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 512189-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tendolkar, P. M., A. S. Baghdayan, and N. Shankar. 2006. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 1882063-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trampuz, A., K. E. Piper, J. M. Steckelberg, and R. Patel. 2006. Effect of gamma irradiation on viability and DNA of Staphylococcus epidermidis and Escherichia coli. J. Med. Microbiol. 551271-1275. [DOI] [PubMed] [Google Scholar]
- 57.Ueda, A., and T. K. Wood. 2008. Potassium and sodium transporters of Pseudomonas aeruginosa regulate virulence to barley. Appl. Microbiol. Biotechnol. 79843-858. [DOI] [PubMed] [Google Scholar]
- 58.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.