Abstract
Clostridial spore germination requires degradation of the spore's peptidoglycan (PG) cortex by cortex-lytic enzymes (CLEs), and two Clostridium perfringens CLEs, SleC and SleM, degrade cortex PG in vitro. We now find that only SleC is essential for cortex hydrolysis and viability of C. perfringens spores. C. perfringens sleC spores did not germinate completely with nutrients, KCl, or a 1:1 chelate of Ca2+ and dipicolinic acid (Ca-DPA), and the colony-forming efficiency of sleC spores was 103-fold lower than that of wild-type spores. However, sleC spores incubated with various germinants released most of their DPA, although slower than wild-type or sleM spores, and DPA release from sleC sleM spores was very slow. In contrast, germination and viability of sleM spores were similar to that of wild-type spores, although sleC sleM spores had 105-fold-lower viability. These results allow the following conclusions about C. perfringens spore germination: (i) SleC is essential for cortex hydrolysis; (ii) although SleM can degrade cortex PG in vitro, this enzyme is not essential; (iii) action of SleC alone or with SleM can accelerate DPA release; and (iv) Ca-DPA does not trigger spore germination by activation of CLEs.
Clostridium perfringens is a gram-positive, spore-forming anaerobic bacterium and a significant cause of histotoxic and gastrointestinal diseases in humans and animals (19, 20). C. perfringens isolates are classified into five types, A through E, based on their ability to produce alpha-, beta-, epsilon- and iota-toxin (20). A small percentage (<5%) of type A isolates produce the C. perfringens enterotoxin and cause type A food poisoning and non-food-borne gastrointestinal diseases (19). C. perfringens isolates also can form metabolically dormant spores that are resistant to many stress factors and can survive for long periods in the environment. Under favorable conditions these surviving spores can germinate, outgrow, return to vegetative growth, and then release toxins and cause disease (20).
Bacterial spores initiate germination when they sense compounds termed germinants, including nutrients, a 1:1 chelate of Ca2+ and pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) (Ca-DPA) and cationic surfactants (27, 30, 42). In spores of Bacillus subtilis and related species, nutrient germinants are sensed by receptors located in the spore's inner membrane, with subsequent initiation of biophysical events including release of monovalent cations (H+, Na+, and K+), and the spore core's large depot of DPA present as a 1:1 chelate with divalent cations, predominantly Ca2+ (42). Hydrolysis of the spore's peptidoglycan (PG) cortex is a later event in germination, and in B. subtilis is triggered at least in part by Ca-DPA release (29, 42). Cortex PG hydrolysis is essential for completion of spore germination, since this removes a physical constraint allowing the core to expand and take up water to the level found in vegetative cells (33). This full hydration now allows enzyme activity in the spore core leading to initiation of energy metabolism, macromolecular synthesis and spore outgrowth (9, 42).
In Bacillus spores the PG cortex comprises ≥80% of spore PG, with the remainder in the nascent germ cell wall that becomes the cell wall of the outgrowing spore (21). Cortex PG generally has three structural differences from germ cell wall and vegetative cell PG, as studied best in B. subtilis (2, 34, 35). (i) Although about one-fourth of cortex N-acetylmuramic acid (NAM) residues are substituted with short peptides, essentially all NAM residues in germ cell wall and vegetative cell PG carry these short peptides. As a consequence cortex PG is less highly cross-linked than germ cell wall or vegetative PG. (ii) About one-fourth of NAM residues in cortex PG carry a single l-alanine residue, a modification not found in germ cell wall or vegetative cell PG, although this modification is not present in C. perfringens cortex PG (26). (iii) Approximately every second muramic acid residue in cortex PG has been converted to muramic-δ-lactam, a modification again not found in germ cell wall or vegetative cell PG (47, 48). Muramic-δ-lactam appears to be the recognition element for cortex-lytic enzymes (CLEs) that hydrolyze the cortex during spore germination but do not act on germ cell wall PG (17, 42).
In B. subtilis and Bacillus megaterium there are two redundant CLEs that degrade cortex PG during germination, SleB and CwlJ, as well as a third enzyme SleL (YaaH) that by itself is not sufficient for cortex hydrolysis during germination, although it may contribute to the hydrolysis in some way (9, 16, 17, 42). SleB is synthesized in the developing forespore and is located primarily at the inner edge of the cortex in a mature form possessing lytic transglycosylase activity (4, 8), but the mechanism for regulation of SleB activity is unclear. In contrast to SleB, CwlJ is synthesized in the mother cell and is located primarily at the outer edge of the spore cortex (3, 8, 13). While CwlJ appears to be synthesized in an active form, the specificity of this enzyme has not been determined. However, CwlJ is activated during germination by Ca-DPA, either released from the spore core or supplied exogenously (17, 42).
Work with C. perfringens strain S40 has identified two CLEs, SleC and SleM, as potentially involved in cortex PG hydrolysis during spore germination. The sleC and sleM genes are present in the genomes of all sequenced C. perfringens strains (23, 43), and homologues are also present in other Clostridium species (5, 24, 39). SleC is synthesized in the mother cell compartment of the sporulating cell as a precursor with four domains, and the N-terminal preregion and the C-terminal pro-region are removed during sporulation. SleC exists in the spore as the inactive zymogen, pro-SleC, consisting of an N-terminal pro-sequence and the mature active enzyme, and pro-SleC is converted to the active enzyme early in spore germination through removal of the pro-region by germination-specific proteases (22, 25, 45). However, it is not clear what regulates germination-specific protease activity. Active SleC is a bifunctional enzyme with lytic transglycosylase and N-acetylmuramoyl-l-alanine amidase activity on cross-linked peptide moieties in the cortex (15), and the purified enzyme is active on the cortex in decoated spores (15). SleM is also synthesized in the mother cell compartment during sporulation but as the mature enzyme and has N-acetylmuramidase activity. SleM appears to degrade only SleC-modified cortex PG and has no activity on decoated spores (7). Both SleC and SleM appear to be located on the outer edge of the spore cortex and are removed when spores are decoated (7).
While significant knowledge has been obtained on the properties of SleC and SleM in vitro, the actual role of these enzymes during C. perfringens spore germination has not been established. Consequently, in the current study we have constructed sleC, sleM, and sleC sleM mutant strains of C. perfringens and used these strains to determine the roles of SleC and SleM during spore germination.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The C. perfringens strains and the plasmids used in the present study are described in Table S1 in the supplemental material.
Construction of gusA fusion plasmids and β-glucuronidase assay.
DNA fragments (300 to 400 bp) upstream of sleC and sleM from C. perfringens SM101, which included the 52- and 74-bp intergenic regions between sleC and cspB and between sleM and CPR1312, respectively, that most likely contain these genes' promoters, as shown for C. perfringens strain S40 (18), were PCR amplified using the primer pairs CPP399/CPP388 and CPP396/CPP398. The forward and reverse primers (see Table S2 in the supplemental material) had SalI and PstI cleavage sites, respectively. These PCR fragments were digested with SalI and PstI and cloned between SalI and PstI sites in pMRS127 to create sleC- and sleM-gusA fusions, giving plasmids pDP86 and pDP87 (see Table S1 in the supplemental material). These plasmids were introduced by electroporation (10) into C. perfringens SM101, and erythromycin-resistant (50 μg/ml) transformants were selected. Transformants carrying plasmids with the sleC-gusA and sleM-gusA fusions were grown vegetatively in TGY medium (3% Trypticase soy, 2% glucose, 1% yeast extract, 0.1% l-cysteine) (14) and in Duncan-Strong (DS) (11) sporulation medium, and assayed for β-glucuronidase (GUS) activity as described previously (49). GUS specific activity was expressed in Miller units that were calculated as described previously (36).
Construction of a C. perfringens sleC deletion mutant.
To isolate a derivative of C. perfringens SM101 with a deletion of sleC, a ΔsleC suicide vector was constructed as follows. A 2,235-bp DNA fragment carrying 114 bp from the N-terminal coding region and 2,121 bp upstream of sleC was PCR amplified by using the primer pair CPP357/CPP365 (forward and reverse primers [see Table S2 in the supplemental material] had KpnI and BamHI cleavage sites at the 5′ ends, respectively). A 2,087-bp DNA fragment carrying 242 bp from the C-terminal coding region and 1,845 bp downstream of sleC was PCR amplified using the primer pair CPP359/CPP364 (forward and reverse primers [see Table S2 in the supplemental material] had PstI and XhoI cleavage sites at the 5′ ends, respectively). These PCR fragments were cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA), yielding plasmids pDP60 and pDP62. An ∼2.2-kb KpnI-BamHI fragment from pDP60 was cloned in pDP25 (see Table S1 in the supplemental material) giving plasmid pDP61, and an ∼2.1-kb PstI-XhoI fragment from pDP62 was cloned into pDP61 (see Table S1 in the supplemental material) giving pDP63. Finally, an ∼5.5-kb KpnI-XhoI fragment from pDP63 was cloned into pMRS104 (see Table S1 in the supplemental material), yielding plasmid pDP64, which cannot replicate in C. perfringens. Plasmid pDP64 was introduced into C. perfringens SM101 by electroporation, and a chloramphenicol-resistant (Cmr; 20 μg/ml) sleC mutant was isolated as described previously (38). The identity of the sleC strain DPS107 was confirmed by PCR and Southern blot analyses (data not shown).
Construction of a C. perfringens sleM deletion mutant.
To isolate a derivative of C. perfringens SM101 with a deletion of sleM, a ΔsleM suicide vector was constructed as follows. A 1,006-bp DNA fragment carrying 61 bp from the N-terminal coding region and 945 bp upstream of sleM was PCR amplified using the primer pair CPP401/CPP403 (forward and reverse primers [see Table S2 in the supplemental material] had KpnI and SpeI cleavage sites at the 5′ ends, respectively). A 1,436-bp DNA fragment carrying 225-bp from the C-terminal coding region and 1,211 bp downstream of sleM was PCR amplified using the primer pair CPP404/CPP400 (forward and reverse primers [see Table S2 in the supplemental material] had PstI and XhoI cleavage sites at the 5′ ends, respectively). These PCR fragments were cloned into pCR-XL-TOPO, yielding plasmids pDP91 and pDP92. An ∼1.0-kb KpnI-SpeI fragment from pDP91 was cloned in pDP25 (see Table S1 in the supplemental material), yielding plasmid pDP93, and an ∼1.4-kb PstI-XhoI fragment from pDP92 was cloned into pDP93 (see Table S1 in the supplemental material), yielding plasmid pDP94. Finally, an ∼3.7-kb KpnI-XhoI fragment from pDP94 was cloned into pMRS104 (see Table S1 in the supplemental material), yielding pDP95, which cannot replicate in C. perfringens. Plasmid pDP95 was introduced into C. perfringens SM101 by electroporation, and a Cmr sleM mutant was isolated as described previously (38). The identity of the sleM strain DPS109 was confirmed by PCR and Southern blot analyses (data not shown).
Construction of a sleC sleM double mutant.
To isolate a derivative of C. perfringens SM101 with deletions of both sleC and sleM, a ΔsleC suicide vector encoding tetracycline resistance (2 μg/ml) was constructed as follows. A 3.2-kb BamHI-PstI fragment carrying tetM from pDP35 was cloned into pDP64 (see Table S1 in the supplemental material), yielding plasmid pDP65. Next, a ∼1.1-kb SmaI fragment carrying ermB from pJIR599 was cloned into pDP65 (see Table S1 in the supplemental material), yielding pDP66, which cannot replicate in C. perfringens. Plasmid pDP66 was introduced into C. perfringens DPS109 by electroporation, and a Cmr tetracycline-resistant sleC sleM mutant was isolated as described previously (38). The identity of the sleC sleM strain DPS110 was confirmed by PCR and Southern blot analyses (data not shown).
Construction of a ΔsleC strain complemented with sleC.
To construct a sleC strain complemented with wild-type sleC, a suicide-complementing plasmid targeted to the plc locus was constructed as follows. A 1,704-bp PCR fragment carrying 1,590 bp upstream and 114 bp of the N-terminal coding region of plc was PCR amplified using the primer pair CPP507/CPP511 (forward and reverse primers [see Table S2 in the supplemental material] had SacI and KpnI cleavage sites at the 5′ ends, respectively). A 1,264-bp PCR fragment carrying 309 bp from the C-terminal coding region and 955 bp downstream of plc was PCR amplified using the primer pair CPP516/CPP510 (forward and reverse primers [see Table S2 in the supplemental material] had SalI and SphI cleavage sites at the 5′ ends, respectively). These PCR fragments were cloned into pCR-XL-TOPO (Invitrogen), yielding the plasmids pDP126 and pDP127 (see Table S1 in the supplemental material). An ∼1.7-kb SacI-KpnI fragment from pDP126 was cloned into pMRS104 (see Table S1 in the supplemental material) giving plasmid pDP128, and an ∼1.3-kb SalI-SphI fragment from pDP127 was cloned into pDP128 (Table S1), yielding plasmid pDP129. Next, an ∼1.9-kb PCR fragment (including 437 bp upstream of sleC and the sleC coding region) was PCR amplified with Phusion High-Fidelity DNA polymerase (New England Biolabs, Inc.) with the primer pair CPP482/CPP488 (forward and reverse primers [see Table S2 in the supplemental material] had KpnI and SalI cleavage sites at the 5′ ends, respectively), and cloned into Zero-Blunt-TOPO (Table S1) giving pDP115. As shown by assays of GUS activity (Fig. 1B), the 437-bp region upstream of sleC contained a strong promoter, as expected (18). Finally, an ∼1.9-kb KpnI-SalI fragment from pDP115 was cloned into pDP129, yielding pDP138 (see Table S1 in the supplemental material), which cannot replicate in C. perfringens. Plasmid pDP138 was introduced into the C. perfringens ΔsleC strain DPS107 by electroporation (10), and erythromycin-resistant Cmr transformants were selected. The presence of plasmid pDP138 in DPS107(pDP138) was confirmed by PCR and Southern blot analyses (data not shown).
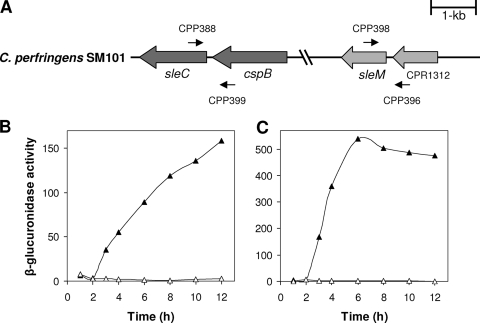
FIG. 1.
(A to C) Arrangement and expression of sle genes in C. perfringens SM101. (A) The arrangement of sleC and sleM in C. perfringens SM101 is shown, and the locations of the primers used to amplify the upstream regions of each gene are indicated. The sleC and sleM promoters were predicted to be within the intergenic regions between sleC and cspB and between sleM and CPR1312, respectively (18). (B and C) GUS specific activities from sleC-gusA (B) and sleM-gusA (C) fusions in C. perfringens SM101 grown in TGY vegetative (▵) and DS sporulation (▴) media were determined as described in Materials and Methods. The data represent averages of three independent experiments, and time zero denotes the time of inoculation of cells into either TGY or DS medium.
Spore preparation and purification.
Starter cultures (10 ml) of C. perfringens isolates were prepared by overnight growth at 37°C in fluid thioglycolate broth (Difco) as described previously (14). C. perfringens sporulating cultures were prepared by inoculating 0.2 ml of an fluid thioglycolate starter culture into 10 ml of DS sporulation medium (11), followed by incubation for 24 h at 37°C, and the presence of spores was confirmed by phase-contrast microscopy. Large amounts of spores were prepared by scaling up the latter procedure, as described previously (30). Spore preparations were cleaned by repeated centrifugation and washing with sterile distilled water until spore suspensions were >99% free of sporulating cells, cell debris, and germinated spores and then suspended in distilled water at a final optical density at 600 nm (OD600) of ∼6 and stored at −20°C.
Decoating treatment.
Spores at an OD600 of 20 were decoated in 1 ml of 50 mM Tris-HCl (pH 8.0)-8 M urea-1% (wt/vol) sodium dodecyl sulfate-50 mM dithiothreitol for 90 min at 37°C, and the spores were washed three times with 150 mM NaCl and twice with water (32). This extraction procedure did not kill the spores, as determined on brain heart infusion (BHI) agar plates (see below) containing 1 μg of lysozyme/ml.
Assessment of the colony-forming efficiency of spores.
Untreated and decoated spores at an OD600 of 1.0 were heat activated at 80°C for 10 min, aliquots of various dilutions were plated on BHI agar with or without lysozyme (1 μg/ml) in the plates, and the plates were incubated at 37°C anaerobically for 24 h. The colonies were counted, and the results were expressed as CFU/ml/OD600.
Spore germination.
Spore suspensions were heat activated at 80°C for 10 min, cooled in water at an ambient temperature for 5 min, and incubated at 40°C for 10 min as described previously (30). Germination of spores with an OD600 of 1 with BHI broth, Ca-DPA (50 mM CaCl2, 50 mM DPA adjusted to pH 8.0 with Tris-HCl), or KCl (100 mM KCl-25 mM sodium phosphate buffer [pH 7.0]) (30) was routinely measured by monitoring the OD600 of spore cultures (Smartspec 3000 spectrophotometer; Bio-Rad Laboratories, Hercules, CA), which falls ∼60% upon complete germination of wild-type spores. The levels of germination were also confirmed by phase-contrast microscopy. All reported values are averages of two experiments performed on at least two independent spore preparations, and individual values varied by <15% from the average values shown.
DPA release.
DPA release from spores during nutrient and nonnutrient germination was measured by heat activating (80°C, 10 min) a spore suspension (OD600 of 1.5), cooling, and incubation in BHI broth or in 25 mM sodium phosphate buffer (pH 7.0) alone or with 100 mM KCl (pH 7.0) or in 25 mM Tris-HCl (pH 8.0) alone or with 50 mM Ca-DPA adjusted to pH 8.0 with Tris-HCl at 40°C for 1 or 18 h. A 1-ml aliquot was centrifuged in a microcentrifuge (13,200 rpm, 2 min), and the spore pellet was washed four times with 1 ml of distilled water. Control experiments were done for each experiment and reveal that losses of spores due to these multiple centrifugations were <10% of the initial amount, and appropriate corrections for such losses were made accordingly. The residual spore DPA content was determined by boiling the samples for 60 min, cooling them on ice for 5 min, centrifugation at 13,200 rpm in a microcentrifuge for 5 min, and measuring the OD270 of the supernatant fluid as described previously (6, 41). The DPA content of the initial dormant spores was measured by boiling an aliquot (1 ml) for 60 min, centrifugation at 13,200 rpm in a microcentrifuge for 5 min, and measuring the OD270 of the supernatant fluid as described previously (6, 30). In B. subtilis spores, ≥85% of the material absorbing at 270 nm is DPA (1, 6). To confirm that the material from C. perfringens spores that absorbed at 270 nm was indeed DPA, total DPA and DPA released upon germination from C. perfringens spores were also measured by a colorimetric assay in parallel with control assays with pure DPA (37). Comparison of the measurements by OD270 and the colorimetric assay indicated that ∼90% of the material absorbing at 270 nm was indeed DPA (data not shown).
For measuring DPA release during dodecylamine germination, spores (OD600 of 1.5) were incubated at 60°C with 1 mM dodecylamine in 25 mM Tris-HCl (pH 7.4). Aliquots (1 ml) of germinating cultures were centrifuged for 3 min at 13,200 rpm in a microcentrifuge, and DPA in the supernatant fluid was measured by monitoring the OD270 as described previously (6, 30). The initial DPA content in dormant spores was measured similarly. No significant DPA release was observed when spores were incubated in 25 mM Tris-HCl (pH 7.4) at 60°C for 1 h (data not shown).
Hexosamine release.
The release of hexosamine-containing fragments of cortex PG into the germination medium was measured by germinating heat-activated spores at an OD600 of 25 in 100 mM KCl and 10 mM Tris-HCl (pH 7.4). After incubation for 2 h at 40°C, samples (1 ml) were centrifuged, and analyses of hexosamine in the supernatant fluid were carried out as described previously (12, 31). Analyses of hexosamine-containing material in dormant spores were carried out similarly.
RESULTS
Evaluation of expression of sle genes in C. perfringens SM101.
When the C. perfringens SM101 (23) genome was subjected to BLASTP analyses to identify genes encoding CLEs, two open reading frames (CPR2566 and CPR1311) encoding proteins with high identity (90 to 94%) to CLEs from C. perfringens strain S40 were identified. CPR2566 is predicted to encode a 438-amino-acid protein with 90% identity to SleC from C. perfringens S40 (Fig. 1A). CPR1311 is predicted to encode a 200-amino-acid protein with 94% identity to SleM from C. perfringens S40 (Fig. 1A). Both the sleC and the sleM genes appear likely to be monocistronic (18).
To assess whether the C. perfringens SM101 sle genes are expressed during sporulation, DNA upstream of each sle gene's coding sequence, including the intergenic regions between the sle genes and the preceding gene (Fig. 1A), which most likely contain these genes' promoters as shown for C. perfringens strain S40 (18), was fused to Escherichia coli gusA, and the GUS activity was measured after introducing the various fusions into C. perfringens SM101. No significant GUS activity was observed in vegetative cultures of strain SM101 carrying sleC-gusA or sleM-gusA (Fig. 1B and C). However, sporulating cultures of strains carrying these gusA fusions exhibited significant GUS activity (Fig. 1B and C), indicating that a sporulation-specific promoter is located upstream of each sle gene. Expression of GUS from the sleC-gusA fusion began ∼2 h after induction of sporulation, and the GUS specific activity increased until 12 h (Fig. 1B). Expression of GUS from the sleM-gusA fusion also began ∼2 h after the induction of sporulation and reached a maximum specific activity ∼6 h after the induction of sporulation (Fig. 1C). Collectively, these results agree with previous work on the expression of the sleC and sleM genes in C. perfringens S40 (18) and indicate that SleC and SleM could be involved in cortex hydrolysis during C. perfringens spore germination.
Effect of sleC and sleM mutations on spore germination in BHI broth and colony formation of spores on BHI plates.
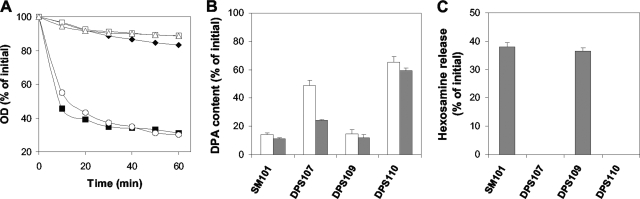
Several in vitro studies (7, 15, 45) indicate that C. perfringens SleC and SleM can specifically hydrolyze intact spore cortex or cortex fragments. To establish the roles of these CLEs during C. perfringens spore germination, we constructed strains carrying deletions of sleC (strain DPS107), sleM (strain DPS109), and both sleC and sleM (strain DPS110) (see Table S1 in the supplemental material and also Fig. 2A), and we examined the germination of spores of these strains in BHI broth (Fig. 2B). The decrease in OD600 of the sleM spores in BHI broth was significantly (P < 0.001) faster and reached a lower level than that of wild-type spores during the first 30 min of incubation for reasons that are not clear, but spores of both strains germinated to a similar extent after 60 min (Fig. 2B). Phase-contrast microscopy of these spores also showed similar levels (∼80%) of phase-dark spores (which is indicative of the completion of spore germination) after 60 min of incubation in BHI broth, with this number rising to ≥99% after 18 h (data not shown). However, only ∼10 and 20% of wild-type spores had become phase dark after incubation in phosphate buffer alone for 1 and 18 h, respectively (data not shown). In contrast to wild-type and sleM spores, sleC and sleC sleM spores exhibited only a small decrease in OD600 after 60 min of incubation in BHI broth (Fig. 2B), and this decrease was similar to that observed with sleC sleM spores incubated in phosphate buffer alone (data not shown). These results strongly suggest that SleC, but not SleM, is essential for hydrolysis of the PG cortex during C. perfringens spore germination and thus the large decrease in the OD600 during germination.
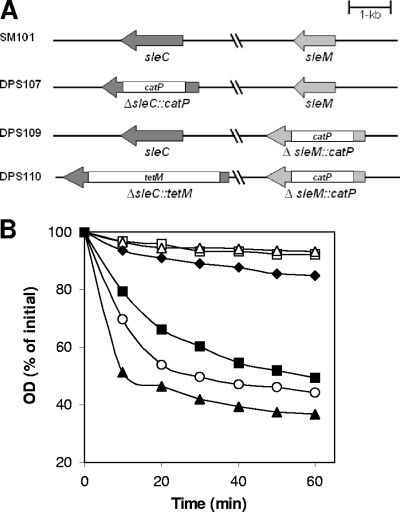
FIG. 2.
(A and B) C. perfringens sle deletion mutants and germination of their spores in BHI broth. (A) Arrangement of sleC and sleM in various C. perfringens deletion mutant strains. (B) Spores of C. perfringens strains SM101 (wild-type) (▪), DPS107 (sleC) (▵), DPS109 (sleM) (○), DPS110 (sleC sleM) (□), and DPS107(pDP138) (sleC strain complemented with wild-type sleC) (▴) were incubated at 40°C in BHI broth, and the OD600 was measured as described in Materials and Methods. Spores of C. perfringens strains SM101 (wild-type) (⧫) and DPS107 (sleC) were incubated at 40°C in 25 mM sodium phosphate buffer (pH 7.0), and the OD600 was measured as described in Materials and Methods. There was essentially no decrease in the OD600 for the sleC spores incubated in buffer alone (data not shown).
Phase-contrast microscopy further indicated that ∼50 and 30% of the sleC and sleC sleM spores, respectively, had become phase gray after 1 h of incubation in BHI broth, a finding indicative of some decrease in the spore core's refractive index, while after 18 h, ∼80 and 60% of the sleC and sleC sleM spores, respectively, had become phase gray (data not shown). However, ∼30 and 20% of the sleC and sleC sleM spores, respectively, had become phase gray after 1 h of incubation in phosphate buffer alone, and these values increased to ∼60% for both sleC and sleC sleM spores after 18 h. These data suggest that SleM may be important in conjunction with SleC in some way in effecting a fall in the spore core's refractive index during germination, perhaps by facilitating DPA release (see below).
The severe germination defects of sleC and sleC sleM spores suggested that the colony-forming efficiency of these spores might be lower than that of wild-type spores, since cortex hydrolysis is essential for the resumption of vegetative growth. No significant difference in colony-forming efficiency was observed between wild-type and sleM spores on BHI agar plates (Table 1) . In contrast, sleC spores exhibited ∼103-fold-lower colony formation efficiency than did wild-type spores, and the colony-forming efficiency of sleC sleM spores was ∼105- and 102-fold lower than that of the wild-type and sleC spores, respectively (Table 1). However, the colony-forming efficiencies of the sleC and sleC sleM spores were restored to wild-type levels when the spores were decoated and plated on BHI agar containing 1 μg of lysozyme/ml. Thus, the sleC and sleC sleM spores were completely viable but just unable to complete cortex hydrolysis and germination. These results (i) further support the essential role of SleC in cortex hydrolysis during the germination and outgrowth of C. perfringens spores and (ii) suggest that only in the absence of SleC does SleM have a role in the germination of C. perfringens spores, and even then it has only a secondary role.
TABLE 1.
Colony formation by spores of C. perfringens strainsa
| Strain/genotype | Spore titer (CFU/ml/OD600) onb:
|
|
|---|---|---|
| BHI | BHI + Lyzc | |
| SM101/wild-type | 4.0 × 107 | 8.9 × 107 |
| DPS107/sleC | 1.4 × 104 | 3.9 × 107 |
| DPS107(pDP138)/sleC carrying wild-type sleC | 2.7 × 107 | ND |
| DPS109/sleM | 3.4 × 107 | ND |
| DPS110/sleM sleC | 1.6 × 102 | 6.6 × 107 |
Heat-activated spores of various strains were plated on BHI agar, and colonies were counted after incubation at 37°C for 24 h.
Titers are the average of CFU/ml/OD600 determined in three experiments, and the variance was <25%.
Spores were decoated and plated on BHI plates containing 1 μg of lysozyme (Lyz)/ml. ND, not determined.
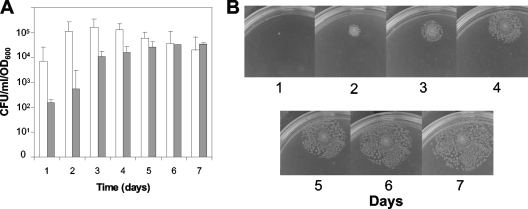
The appearance of at least the great majority of colonies from sleC and sleC sleM spores was not due to genetic reversion, since PCR analyses did not detect wild-type sleC and sleM genes in colonies obtained from sleC and sleC sleM spores after 24 h (data not shown). To test whether longer incubation of sleC and sleC sleM spores on BHI agar plates at 37°C would lead to increased colony formation efficiency, plates were incubated for up to 7 days, and colonies were counted every 24 h. As expected, >99% of the total colonies from wild-type spores appeared after only 24 h (data not shown). Surprisingly, when plates with sleC and sleC sleM spores were incubated for longer periods, 10- to 100-fold-higher colony counts appeared (Fig. 3A). However, these additional colonies only appeared surrounding colonies that arose during the first 24 h of incubation (Fig. 3B), and no new isolated colonies appeared (data not shown). While the cause of this behavior of the sleC and sleC sleM spores is not completely clear, it could be due to the diffusion of some cell wall hydrolase released by vegetative cells in the initial colony, with this hydrolase then degrading the cortex of a small population of sleC spores that have a defective coat. Indeed, the maximum number of colonies that appeared from sleC and sleC sleM spores during the 7 days of incubation was 10- to 100-fold lower than from wild-type spores (data not shown), suggesting that only a small percentage of C. perfringens spore populations gave rise to these late-appearing colonies. In addition, sleC spore viability went up only ∼10-fold in 48 h, whereas the sleC sleM spores required ∼5 days to reach their maximum level. Perhaps cortex hydrolysis by a cell wall hydrolase from vegetative cells on defective spores is accelerated by SleM.
FIG. 3.
(A and B) Germination of C. perfringens spores over long periods on BHI agar plates. (A) Spores of C. perfringens strains DPS107 (sleC) (□) and DPS110 (sleC sleM) (░⃞) were applied to BHI plates that were incubated at 37°C for 7 days, and the total number of colonies were counted every 24 h and expressed as CFU. (B) The plates described in panel A were photographed every 24 h. Similar results in the experiments in panels A and B were obtained with two different batches of spores, and error bars indicate the standard deviations.
DPA release by C. perfringens spores germinated with BHI broth.
The germination and viability defects noted above with sleC and sleC sleM spores suggest that these spores cannot complete the germination process because they cannot degrade cortex PG. However, since many of the sleC and sleC sleM spores incubated in BHI broth became phase gray in the phase-contrast microscope, this suggested that there was a decrease in these spores' core refractive index, perhaps because of DPA release and its replacement by water. As expected, wild-type and sleM spores released most of their DPA after 1 h of germination in BHI broth (Fig. 4), with slightly more released from the sleM spores, a finding consistent with the slightly more rapid germination of these spores, as measured by the decrease in OD600 (Fig. 2B), although after 18 h, both sleM and wild-type spores had released ≥90% of their DPA (Fig. 4). In contrast, sleC spores released only ∼50 and 85% of their DPA after 1 and 18 h of germination in BHI broth, respectively (Fig. 4). Although sleC sleM spores released DPA during incubation in BHI broth, the amount released was significantly less than that from wild-type, sleC, and sleM spores incubated similarly and was only slightly higher than the amount of DPA released from sleC sleM spores incubated in buffer alone (Fig. 4). Collectively, these results suggest that DPA release during C. perfringens spore germination does take place during germination of sleC spores in BHI broth but is significantly slowed in the absence of cortex hydrolysis, especially in the absence of both SleC and SleM. These results further suggest that SleM might have some role in DPA release but is clearly not sufficient for cortex hydrolysis.
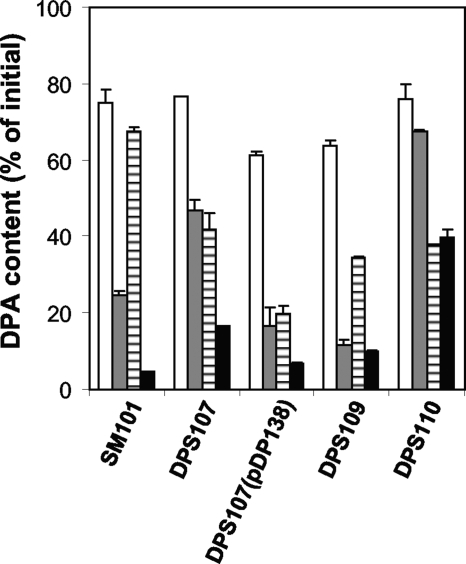
FIG. 4.
DPA release during the germination of C. perfringens spores with BHI broth. Heat-activated spores of strains SM101 (wild-type), DPS107 (sleC), DPS107(pDP138) (sleC strain complemented with wild-type sleC), DPS109 (sleM), and DPS110 (sleC sleM) were incubated at 40°C in BHI broth and, after 1 h (░⃞) and 18 h (▪), DPA release was measured as described in Materials and Methods. C. perfringens spores from various strains were also incubated in 25 mM sodium phosphate buffer (pH 7.0), and after 1 (□) or 18 h (▤) the DPA release was measured as described in Materials and Methods. The data represent the average of two independent experiments with two different spore preparations, and error bars indicate the standard deviations.
Complementation of sleC mutant with wild-type sleC.
To be certain that the phenotypes observed for sleC spores were due directly to deletion of sleC and not to any possible secondary effects due to strain construction, we constructed a suicide plasmid pDP138, carrying a mutated allele of the alpha-toxin gene (plc) in which a wild-type copy of sleC plus its promoter region (see Materials and Methods and Fig. 1) was inserted. The plc locus was chosen for insertion of the wild-type sleC into the chromosome of the sleC strain by a homologous single recombination event, assuming that plc disruption would not affect spore germination. Plasmid pDP138 was introduced into the sleC strain DPS107, and PCR with DNA from one DPS107(pDP138) transformant yielded products from wild-type plc, ΔsleC, and the Δplc::sleC allele, consistent with wild-type sleC in pDP138 being integrated into the plc locus by a single crossover event (data not shown). Spores of strain DPS107(pDP138) germinated like wild-type spores in BHI broth, as assessed by either a decrease in OD600 (Fig. 2B) or phase-contrast microscopy (data not shown). The low colony formation efficiency of sleC spores was also restored to that of wild-type spores when the sleC strain was complemented with wild-type sleC [strain DPS107(pDP138)] (Fig. 2B and Table 1). The slow DPA release from sleC spores during germination in BHI broth was also restored to the faster rate seen with wild-type spores in the complemented strain (Fig. 4). These results thus confirm that the phenotypes of sleC spores are due exclusively to deletion of sleC.
Effect of sleC and sleM mutations on C. perfringens spore germination with KCl.
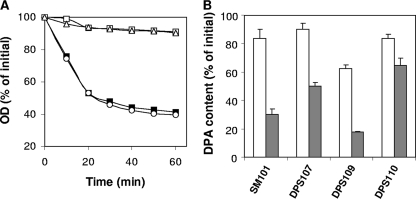
To gain further understanding of the roles of SleC and SleM in C. perfringens spore germination, wild-type, sleC, sleM, and sleC sleM spores were incubated with the germinant KCl (30). As expected, sleM spores germinated like wild-type spores with KCl, when germination was assessed by the decrease in OD600 (Fig. 5A). In contrast, sleC and sleC sleM spores showed only a minimal decrease in OD600 upon incubation with KCl (Fig. 5A). The small decrease in OD600 observed for the latter spores was even lower than that of wild-type spores incubated in sodium phosphate buffer (Fig. 5A). Phase-contrast microscopy indicated that ∼60 and ∼30% of the sleC and sleC sleM spores, respectively, incubated in 100 mM KCl had become phase gray after 1 h, whereas ∼10% of the wild-type spores incubated for 1 h in sodium phosphate buffer had become phase dark. This suggest that while the OD600 decrease of wild-type spores incubated in buffer alone was because a small percentage of the spores germinated spontaneously, thus becoming phase dark, the OD600 decrease of sleC and sleC sleM spores incubated in KCl was due to the release of DPA by a fraction of the spore population, thus slightly reducing these spores' refractive index.
FIG. 5.
(A to C) Germination of spores of C. perfringens strains with KCl. (A) Heat-activated C. perfringens spores of strains SM101(wild-type) (▪), DPS107 (sleC) (▵), DPS109 (sleM) (○), and DPS110 (sleC sleM) (□) were germinated with KCl, and the OD600 was measured as described in Materials and Methods. Heat-activated spores of strain SM101 (wild-type) (⧫) were also incubated in 25 mM sodium phosphate buffer (pH 7.0) alone, and the OD600 was measured. (B) DPA release during C. perfringens spore germination with KCl. Heat-activated spores of C. perfringens strains were germinated with KCl, and after 1 h (□) and 18 h (░⃞) the DPA content of the spores was measured as described in Materials and Methods. DPA release of C. perfringens spores from various strains in 25 mM sodium phosphate buffer (pH 7.0) was as shown in Fig. 4. (C) Release of hexosamine-containing material during C. perfringens spore germination with KCl. Heat-activated spores of C. perfringens strains were germinated with KCl and, after 2 h, the hexosamine-containing material released into the medium was measured as described in Materials and Methods. Values for hexosamine-containing material released are expressed relative to the amount of hexosamine in dormant spores that was defined as 100%. The data represent the average of two independent experiments with two different spore preparations, and error bars indicate the standard deviations.
As expected, there were no significant differences in DPA release during KCl germination between sleM and wild-type spores, since the majority of these spores' DPA was released in 1 h (Fig. 5B). The sleC spores also released DPA during incubation in KCl, and while this release was faster than that for spores incubated in buffer alone, it was significantly slower than for wild-type or sleM spores incubated in KCl (Fig. 4 and 5B). DPA release from sleC sleM spores incubated in KCl was even slower and no faster than from sleC sleM spores incubated in buffer alone (Fig. 4 and 5B).
In addition to DPA release, a second major event in spore germination is the hydrolysis of the spore's PG cortex and release of PG fragments into the medium. The hydrolysis of the spore's PG cortex during germination can thus be monitored by measuring the release of hexosamine-containing material into the medium (33, 44). As expected, wild-type and sleM spores released similar amounts of hexosamine into the medium after 2 h of germination with KCl: 35 to 40% of total spore hexosamine (Fig. 5C). However, no detectable release of hexosamine-containing material was observed from sleC and sleC sleM spores after a similar incubation (Fig. 5C). These results are similar to those found with germinating wild-type and cwlJ sleB spores of B. subtilis and B. megaterium (40, 41).
Ca-DPA germination of C. perfringens spores.
In addition to nutrients, spores of Bacillus and Clostridium species are germinated by exogenous Ca-DPA (30, 42). Ca-DPA triggers the germination of B. megaterium and B. subtilis spores by activation of the CLE CwlJ (28, 41). To determine whether a C. perfringens CLE is also activated by Ca-DPA, the Ca-DPA germination of wild-type, sleC, sleM, and sleC sleM spores was examined (Fig. 6). As expected, sleC and sleC sleM spores did not exhibit the large decrease in OD600 in Ca-DPA germination seen with wild-type and sleM spores (Fig. 6A), and no significant decrease in OD600 was observed when spores of these four strains were incubated in 25 mM Tris-HCl (pH 8.0) (data not shown). However, phase-contrast microscopy found that after 1 h of incubation with Ca-DPA, ∼50 to 30% of the sleC and sleC sleM spores, respectively, had become phase gray, whereas only ∼20% of sleC and sleC sleM spores had become phase gray after 1 h of incubation with 25 mM Tris-HCl (pH 8.0) (data not shown); only ∼10% of wild-type spores had become phase dark after 1 h with 25 mM Tris-HCl (pH 8.0) (data not shown).
FIG. 6.
(A and B) Germination of C. perfringens spores with Ca-DPA. (A) Heat-activated C. perfringens spores of strains SM101 (wild-type) (▪), DPS107 (sleC) (▵), DPS109 (sleM) (○), and DPS110 (sleC sleM) (□) were germinated with 50 mM Ca-DPA, and the OD600 was measured as described in Materials and Methods. (B) Heat-activated spores of C. perfringens strains were germinated with 50 mM Ca-DPA (░⃞) and 25 mM Tris-HCl buffer (pH 8.0) (□) for 1 h, and the DPA remaining in the spores was measured as described in Materials and Methods. The data represent the average of two independent experiments with two different spore preparations, and error bars indicate the standard deviations.
Measurements of DPA release (Fig. 6B) showed that, as found with BHI broth and KCl incubations, wild-type and sleM spores incubated for 1 h with Ca-DPA released significantly more DPA than did sleC spores, although DPA release from the sleC spores was much greater than during incubation with buffer alone (Fig. 6B). However, the amount of DPA released from sleC sleM spores during incubation with Ca-DPA was only slightly higher than the amount released during incubation in buffer (Fig. 6B). Unfortunately, the precipitation of the exogenous Ca-DPA after 1 h precluded assessment of DPA release over longer times.
Dodecylamine germination of C. perfringens spores.
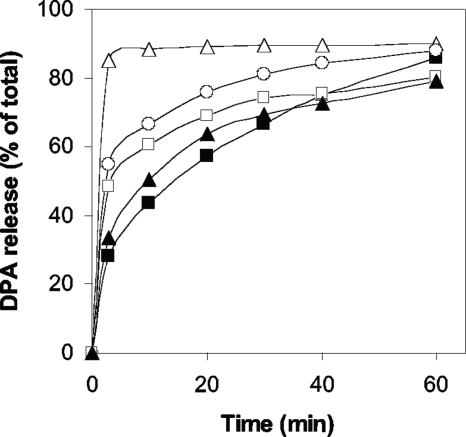
In addition to Ca-DPA, KCl, and nutrients, C. perfringens spores are also germinated by the cationic surfactant dodecylamine (30). While the precise mechanism of dodecylamine germination is not known, the absence of CLEs does not have any effect on DPA release during the dodecylamine germination of the spores of at least two Bacillus species (41, 42). Similarly, wild-type, sleM, and sleC sleM C. perfringens spores and spores of the sleC strain complemented with wild-type sleC exhibited relatively similar rates and extents of DPA release during dodecylamine germination, while sleC spores released their DPA even faster (Fig. 7). Thus, cortex hydrolysis is also not essential for normal DPA release during dodecylamine germination of C. perfringens spores.
FIG. 7.
Dodecylamine germination of C. perfringens spores. C. perfringens spores of strains SM101 (wild-type) (▪), DPS107 (sleC) (▵), DPS109 (sleM) (○), DPS110 (sleC sleM) (□), and DPS107(pDP138) (sleC strain complemented with wild-type sleC) (▴) were germinated with dodecylamine, and at various times the DPA release was measured as described in Materials and Methods.
DISCUSSION
Spores of Bacillus species contain three CLEs, with either of two of these enzymes being essential (7, 17, 41, 42). In contrast, SleC is the single essential CLE in C. perfringens spores, with SleM playing only an auxiliary role. However, SleM can contribute to cortex hydrolysis during spore germination, as shown by the lower viability of sleC sleM spores than sleC spores. SleC and SleM are active on cortex PG fragments in vitro, but SleM, unlike SleC, is inactive on the cortex in intact but decoated spores (7, 22). This suggests that SleM, unlike SleC, is normally inactive on cortex PG in intact spores that may be under strain because of the high osmotic pressure of the poorly hydrated spore core (42). By this reasoning, only SleC would be essential for cortex hydrolysis during spore germination. However, perhaps in a minority of sleC spores the cortex is under less stress, and SleM can hydrolyze this “low-stress” cortex. Alternatively, SleM may act in a small percentage of sleC spores that have a defective cortex, perhaps due to partial cortex hydrolysis during spore formation by an enzyme that acts on sporulating cell PG to allow release of the dormant spore. Indeed, if a few sleC spores have defective coats, a small amount of hydrolytic enzyme action during sporulation on such defective spores might generate spores whose cortex is susceptible to hydrolysis by SleM alone.
Although the loss of SleC eliminated cortex hydrolysis during C. perfringens spore germination, sleC spores still released their DPA in response to germinants that act through the spore's germinant receptors (30). Spores of Bacillus species lacking both essential CLEs also release their DPA during nutrient germination (41, 42). These observations suggest that full cortex hydrolysis is not essential for DPA release during spore germination and that at least the initiation of DPA release likely precedes cortex hydrolysis. However, DPA release from sleC spores germinating with BHI broth or KCl was slower than from germinating wild-type or sleM spores, with the rate of DPA release from germinating sleC sleM spores being even slower and at most only slightly greater than from spores incubated in buffer. This further indicates that while cortex hydrolysis is not essential for the initiation of DPA release during spore germination, cortex hydrolysis can accelerate DPA release. This effect of cortex hydrolysis of the rate of DPA release during germinant receptor-mediated spore germination has also been seen with B. megaterium and B. subtilis spores in which the loss of CwlJ but not of SleB slows DPA release significantly during nutrient germination (13, 41). Unfortunately, why cortex hydrolysis should accelerate DPA release from spores during germinant receptor-mediated germination and why SleM should contribute to this effect are not clear.
Another notable observation in the present study concerns the effects of the loss of CLEs on Ca-DPA germination of C. perfringens spores. With B. megaterium and B. subtilis spores, Ca-DPA triggers germination by activation of the CLE CwlJ and bypasses the spore's germinant receptors (41, 42). In contrast, Ca-DPA appears to trigger C. perfringens spore germination through the GerK germinant receptor (30). This latter finding leads to the prediction that Ca-DPA will trigger DPA release from C. perfringens spores lacking SleC, and this is what was observed, even though DPA release during Ca-DPA germination of sleC C. perfringens spores was slower than from wild-type or sleM spores. The similar response of rates of Ca-DPA release during Ca-DPA, BHI broth, and KCl germination to the absence of SleC and SleC plus SleM further suggests that DPA is released by the same mechanism during spore germination triggered by these three agents. Unfortunately, the mechanism for DPA release during spore germination is not known, although it may involve the SpoVA proteins (29, 46).
The final notable finding was that DPA release by C. perfringens spores during dodecylamine germination was not slowed by the absence of SleC or SleC and SleM. Indeed, sleC spores released their DPA faster during dodecylamine germination than did wild-type spores. DPA release during dodecylamine germination of B. megaterium and B. subtilis spores is also not slowed by the absence of these spores' redundant CLEs (41, 42). The mechanism of spore germination by dodecylamine is not known, but the germinant receptors are likely not involved (30, 42). There is also evidence with B. subtilis spores that DPA release during dodecylamine germination involves the SpoVA proteins thought to be involved in DPA release during nutrient germination (46). It has been suggested that dodecylamine may activate a DPA channel present in the spore's inner membrane (42, 46), but this has not been proven.
Hydrolysis of cortex PG is the culminating event in bacterial spore germination and is essential for resumption of enzymatic activity in the spore core and eventual vegetative growth (42). As a consequence, cortex hydrolysis is essential for spores of pathogenic organisms such as C. perfringens to cause disease. The identification of SleC as the major essential CLE in C. perfringens spores thus makes this enzyme of interest for the development of inhibitors, since such compounds would block spore germination and thus the ability of spores to cause disease. Cortex hydrolysis also makes the now fully germinated spore much less resistant to common decontamination procedures. Consequently, a drug that could rapidly activate SleC in spores would also be useful, since this would allow decontamination of germinated C. perfringens spores under less harsh conditions than needed for destruction of the more resistant dormant spores.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Army Research Office to M.R.S. and P.S., by a National Institutes of Health grant (GM19698) to P.S., and by a fellowship from MIDEPLAN (Chile) to D.P.-S.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 18828-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih, A., P. Zollner, G. Allmaier, and S. J. Foster. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 1786173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 1841219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland, F. M., A. Atrih, H. Chirakkal, S. J. Foster, and A. Moir. 2000. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 14657-64. [DOI] [PubMed] [Google Scholar]
- 5.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 1001316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera-Martinez, R. M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 1852457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., S. Miyata, S. Makino, and R. Moriyama. 1997. Molecular characterization of a germination-specific muramidase from Clostridium perfringens S40 spores and nucleotide sequence of the corresponding gene. J. Bacteriol. 1793181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirakkal, H., M. O'Rourke, A. Atrih, S. J. Foster, and A. Moir. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 1482383-2392. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, A. E., D. E. Koppel, B. Setlow, and P. Setlow. 2003. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc. Natl. Acad. Sci. USA 1004209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 643301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, C. L., and D. H. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 1682-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghuysen, J. M., D. J. Tipper, and J. L. Strominger. 1966. Enzymes that degrade bacterial cell walls. Methods Enzymol. 8685-699. [Google Scholar]
- 13.Ishikawa, S., K. Yamane, and J. Sekiguchi. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 1801375-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 322533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumazawa, T., A. Masayama, S. Fukuoka, S. Makino, T. Yoshimura, and R. Moriyama. 2007. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci. Biotechnol. Biochem. 71884-892. [DOI] [PubMed] [Google Scholar]
- 16.Lambert, E. A., and D. L. Popham. 2008. The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization. J. Bacteriol. 1907601-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino, S., and R. Moriyama. 2002. Hydrolysis of cortex peptidoglycan during bacterial spore germination. Med. Sci. Monit. 8RA119-RA127. [PubMed] [Google Scholar]
- 18.Masayama, A., K. Hamasaki, K. Urakami, S. Shimamoto, S. Kato, S. Makino, T. Yoshimura, M. Moriyama, and R. Moriyama. 2006. Expression of germination-related enzymes, CspA, CspB, CspC, SleC, and SleM, of Clostridium perfringens S40 in the mother cell compartment of sporulating cells. Genes Genet. Syst. 81227-234. [DOI] [PubMed] [Google Scholar]
- 19.McClane, B. A. 2007. Clostridium perfringens, p. 423-444. In M. P. Doyle and L. R. Beuchat (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC.
- 20.McDonnell, J. L. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p. 477-517. In F. Dorner and J. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, Oxford, England.
- 21.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 1824491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata, S., R. Moriyama, N. Miyahara, and S. Makino. 1995. A gene (sleC) encoding a spore-cortex-lytic enzyme from Clostridium perfringens S40 spores; cloning, sequence analysis and molecular characterization. Microbiology 1412643-2650. [DOI] [PubMed] [Google Scholar]
- 23.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 161031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 1834823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura, S., K. Urakami, M. Kimata, T. Aoshima, S. Shimamoto, R. Moriyama, and S. Makino. 2000. The N-terminal prepeptide is required for the production of spore cortex-lytic enzyme from its inactive precursor during germination of Clostridium perfringens S40 spores. Mol. Microbiol. 37821-827. [DOI] [PubMed] [Google Scholar]
- 26.Orsburn, B., S. B. Melville, and D. L. Popham. 2008. Factors contributing to heat resistance of Clostridium perfringens endospores. Appl. Environ. Microbiol. 743328-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paidhungat, M., and P. Setlow. 2000. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 1822513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 1834886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paredes-Sabja, D., B. Setlow, P. Setlow, and M. R. Sarker. 2008. Characterization of Clostridium perfringens spores that lack SpoVA proteins and dipicolinic acid. J. Bacteriol. 1904648-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes-Sabja, D., J. A. Torres, P. Setlow, and M. R. Sarker. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 1901190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 1774721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popham, D. L., S. Sengupta, and P. Setlow. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 613633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. USA 9315405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 1786451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham, D. L. 2002. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol. Life Sci. 59426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raju, D., M. Waters, P. Setlow, and M. R. Sarker. 2006. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotman, Y., and M. L. Fields. 1967. A modified reagent for dipicolinic acid analysis. Anal. Biochem. 22168. [DOI] [PubMed] [Google Scholar]
- 38.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33946-958. [DOI] [PubMed] [Google Scholar]
- 39.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 40.Setlow, B., E. Melly, and P. Setlow. 2001. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J. Bacteriol. 1834894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setlow, B., L. Peng, C. A. Loshon, Y.-Q. Li, G. Christie, and P. Setlow. Characterization of the germination of Bacillus megaterium spores lacking enzymes that degrade the spore cortex. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 42.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6550-556. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tennen, R., B. Setlow, K. L. Davis, C. A. Loshon, and P. Setlow. 2000. Mechanisms of killing of spores of Bacillus subtilis by iodine, glutaraldehyde and nitrous acid. J. Appl. Microbiol. 89330-338. [DOI] [PubMed] [Google Scholar]
- 45.Urakami, K., S. Miyata, R. Moriyama, K. Sugimoto, and S. Makino. 1999. Germination-specific cortex-lytic enzymes from Clostridium perfringens S40 spores: time of synthesis, precursor structure and regulation of enzymatic activity. FEMS Microbiol. Lett. 173467-473. [DOI] [PubMed] [Google Scholar]
- 46.Vepachedu, V. R., and P. Setlow. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J. Bacteriol. 1891565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warth, A. D., and J. L. Strominger. 1969. Structure of the peptidoglycan of bacterial spores: occurrence of the lactam of muramic acid. Proc. Natl. Acad. Sci. USA 64528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warth, A. D., and J. L. Strominger. 1972. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry 111389-1396. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.