Abstract
The similarity of BldG and the downstream coexpressed protein SCO3548 to anti-anti-sigma and anti-sigma factors, respectively, together with the phenotype of a bldG mutant, suggests that BldG and SCO3548 interact as part of a regulatory system to control both antibiotic production and morphological differentiation in Streptomyces coelicolor. A combination of bacterial two-hybrid, affinity purification, and far-Western analyses demonstrated that there was self-interaction of both BldG and SCO3548, as well as a direct interaction between the two proteins. Furthermore, a genetic complementation experiment demonstrated that SCO3548 antagonizes the function of BldG, similar to other anti-anti-sigma/anti-sigma factor pairs. It is therefore proposed that BldG and SCO3548 form a partner-switching pair that regulates the function of one or more sigma factors in S. coelicolor. The conservation of bldG and sco3548 in other streptomycetes demonstrates that this system is likely a key regulatory switch controlling developmental processes throughout the genus Streptomyces.
Streptomyces coelicolor A3(2), the well-studied model organism for processes of bacterial multicellular development and antibiotic production, possesses a large genome (8.67 Mbp) with a high degree of regulatory complexity (6). A large proportion of the coding sequence (12.3%) is predicted to encode the multitude of regulatory factors required to support a complex life cycle, involving the formation of sporulating aerial hyphae, that responds to the changing soil environment. Of particular note is the presence of 64 sigma factors, which are thought to play a critical role in the modulation of gene expression; this group is comprised of 4 housekeeping sigma factors, as well as 50 extracytoplasmic function sigma factors and 9 group 3 subfamily sigma factors (6, 23). The activity of alternative sigma factors is typically regulated by a number of mechanisms, including phosphorylation-dependent partner switching by antagonistic proteins. The best-studied examples of this regulatory mechanism, which is active against the group 3 sigma factors, are found in Bacillus subtilis, where partner switching controls the activity of both the sporulation-specific factor σF and the general stress response factor σB (1, 17, 18, 44, 50, 57, 59). In these systems, an anti-sigma factor protein (SpoIIAB and RsbW, respectively) sequesters the cognate sigma factor, preventing the expression of target genes. Upon sensing some activating signal, an anti-sigma factor antagonist (or anti-anti-sigma factor; SpoIIAA and RsbV, respectively) binds to the anti-sigma factor, mediating release of the active sigma factor to direct regulon transcription.
Partner-switching systems are also thought to play a critical role in sigma factor regulation in S. coelicolor. The first characterized example in this organism is the RsbV-RsbA partner-switching pair that controls the activity of the osmotic-stress-responsive factor σB (36). Many genes encoding additional putative paralogues of these regulatory factors are present in the S. coelicolor genome, including genes encoding 45 RsbW orthologues and 18 RsbV orthologues (45). One cluster of such genes is found at the bldG locus, which was originally identified as one of a group of key pleiotropic regulators, collectively termed the bld genes, that control both antibiotic production and aerial hypha formation in S. coelicolor (10, 11, 43). The bldG gene encodes an orthologue of the RsbV and SpoIIAA anti-anti-sigma factors (9). Immediately downstream of bldG is the open reading frame (ORF) sco3548 (http://strepdb.streptomyces.org.uk/) (previously referred to as orf3 [9]), which encodes an orthologue of the RsbW and SpoIIAB anti-sigma factors. Much like the genes in the B. subtilis systems, the bldG and sco3548 genes are cotranscribed, although, unlike the equimolar expression of the B. subtilis systems, bldG transcripts are always expressed in excess of sco3548 transcripts in S. coelicolor (9). Also unlike the B. subtilis operons, no cognate sigma factor is encoded at the bldG locus, and therefore the biochemical target of BldG regulation is unknown.
The high level of similarity between BldG and its B. subtilis orthologues suggests, however, that BldG functions in a similar partner-switching mechanism. This hypothesis is supported by the presence of a sulfate transporter and anti-sigma factor antagonist (STAS) domain in BldG, which is known to form a key surface for the interaction of anti-sigma factor antagonists with their cognate anti-sigma factors (3). Contiguous with this STAS domain in the B. subtilis SpoIIAA anti-anti-sigma factor is a phosphorylated serine residue known to be essential for the posttranslational control of the interaction with its cognate anti-sigma factor; the phosphorylation event drives the partner-switching mechanism (2, 41). This serine residue is conserved not only among related Bacillus anti-anti-sigma factors but also in BldG. Furthermore, BldG has been shown to be reversibly phosphorylated on its conserved serine, and this phosphorylation is essential for the regulation of morphological differentiation and antibiotic production (7). On the basis of these similarities, it is predicted that BldG is involved in a phosphorylation-dependent partner-switching interaction. Because of the proximity and coexpression of bldG and sco3548, it was predicted that SCO3548 is the antagonistic partner of BldG.
The purpose of this study was to test the hypothesis that BldG and SCO3548 are involved in an antagonistic protein interaction. To this end, a variety of biochemical and genetic experimental approaches were used to identify potential BldG-containing protein complexes, to characterize partners interacting with BldG, and to examine the antagonistic nature of the interactions in S. coelicolor development.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli and S. coelicolor strains used in this study are listed in Table 1. The growth conditions and media used for E. coli cultures have been described previously (52). Plasmid-containing cultures were supplemented as required with an antibiotic(s) as follows: 100 μg/ml ampicillin (Sigma), 50 μg/ml kanamycin (Sigma), and 50 μg/ml apramycin (Provel). S. coelicolor strains were grown in R2YE liquid medium or on R2YE agar as described previously (31). Plasmid-containing cultures were supplemented with antibiotic(s) as follows: 50 μg/ml apramycin, 200 μg/ml kanamycin, 25 μg/ml chloramphenicol (Sigma), and 25 μg/ml nalidixic acid (Sigma). For induction of gene expression from the ptipA promoter on recombinant plasmids, 30 μg/ml thiostrepton (Sigma) was used unless otherwise indicated.
TABLE 1.
Strains used in this study
| Strain | Description | Relevant genotype or description | Source and/or reference |
|---|---|---|---|
| E. coli strains | |||
| DH5α | Host for plasmid cloning and propagation | F− φ80ΔlacZΔM15 recA1 endA1 gyrA96 thi-1 λ−hsdR17(rR− mR+) supE44 relA1 Δ(lacZYA-argF)U169 phoA | Invitrogen |
| BL21(DE3) | Host for expression of recombinant proteins | F−ompT hsdS(rB− mB−) dcm+gal met λ(DE3) | Stratagene |
| BTH101 | Reporter strain for bacterial two-hybrid analysis | F−cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | 29 |
| ET12567 | Nonmethylating host strain | F−dam13::Tn9 dcm6 hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 lacY1 xyl5 leuB6 thi-1 tonA31 rpsL136 hisG4 tsx78 mtl-1 glnV44 | D. MacNeil, Merck Sharp and Dohme Research Laboratories (39) |
| S. coelicolor strains | |||
| M145 | Wild type | Prototrophic, SCP1− SCP2− Pgl+ | John Innes Centre (6) |
| ΔbldG 1DB | bldG null mutant | M145 derivative with an in-frame deletion in bldG | 7 |
DNA manipulations.
E. coli and S. coelicolor plasmids used in this study are listed in Table 2. The standard protocols used for in vitro DNA manipulation have been described previously (52). PCR was performed using the Expand high-fidelity PCR system (Roche) and DNA sequencing was performed using the DYEnamic ET system (Amersham), both using oligonucleotide primers listed in Table 3. Recombinant plasmids were routinely introduced into E. coli ET12567(pUZ8002) by electroporation and subsequently transferred to S. coelicolor ΔbldG 1DB by intergeneric conjugation (31).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics and/or use | Resistance | Source and/or reference(s) |
|---|---|---|---|
| pUZ8002 | Encodes transfer functions for mobilization of oriT-containing vectors from E. coli to Streptomyces by conjugative transfer | Kanamycin | M. Buttner, John Innes Centre (31) |
| pIJ6902 | Expression vector that contains thiostrepton-inducible ptipA promoter; integrates into the φC31 attB site in S. coelicolor | Apramycin | John Innes Centre (24) |
| pAU316 | pIJ6902 harboring bldG; for expression of BldG in S. coelicolor | Apramycin | This study |
| pAU317 | pIJ6902 harboring sco3548; for expression of SCO3548 in S. coelicolor | Apramycin | This study |
| pAU318 | pET30(a)+ derivative containing sco3548 cloned downstream of a 10-histidine tag; for overexpression of His10-SCO3548 in E. coli | Kanamycin | This study |
| pAU365 | pIJ6902 derivative containing the entire bldG and sco3548 coding regions and their native intergenic region; for expression of BldG and SCO3548 in S. coelicolor | Apramycin | This study |
| pGEX-2TK | Expression vector that contains IPTG-inducible tac promoter; for overexpression of protein with an N-terminal GST tag in E. coli | Ampicillin | Amersham Biosciences |
| pAU375 | pGEX-2TK harboring bldG; for overexpression of GST-BldG in E. coli | Ampicillin | This study |
| pAU376 | pGEX-2TK harboring sco3548; for overexpression of GST-SCO3548 in E. coli | Ampicillin | This study |
| pKT25 | pSU40 derivative that carries the T25 fragment of B. pertussis adenylate cyclase (residues 1 to 224) with a multicloning sequence at the 3′ end of T25 | Kanamycin | 28, 30 |
| pUT18 | pUC19 derivative that carries the T18 fragment of B. pertussis adenylate cyclase (residues 225 to 399) with a multicloning sequence at the 5′ end of T18 | Ampicillin | 28 |
| pUT18C | pUC19 derivative that carries the T18 fragment of B. pertussis adenylate cyclase with a multicloning sequence at the 3′ end of T18 | Ampicillin | 28 |
| pKT25Zip | pKT25 harboring the T25 fragment fused in frame to the 35-codon leucine zipper domain of the yeast GNC4 activator | Kanamycin | 30 |
| pUT18CZip | pUT18C harboring the 35-codon leucine zipper domain of the yeast GNC4 activator fused in frame to the T18 fragment | Ampicillin | 28 |
| pAU377 | pKT25 harboring bldG; for expression of T25-BldG in BTH101 | Kanamycin | This study |
| pAU378 | pUT18 harboring bldG; for expression of BldG-T18 in BTH101 | Ampicillin | This study |
| pAU379 | pUT18C harboring bldG; for expression of T18-BldG in BTH101 | Ampicillin | This study |
| pAU380 | pKT25 harboring sco3548; for expression of T25-SCO3548 in BTH101 | Kanamycin | This study |
| pAU381 | pUT18 harboring sco3548; for expression of SCO3548-T18 in BTH101 | Ampicillin | This study |
| pAU382 | pUT18C harboring sco3548; for expression of T18-SCO3548 in BTH101 | Ampicillin | This study |
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Restriction site | Sequence (5′-3′)a | Application |
|---|---|---|---|
| AP3 | BamHI | ATATAGGATCCCGTGGACCTGTCCCTG | Forward primer for creation of bldG-containing pKT25-, pUT18C-, and pUT18-based constructs |
| AP4 | KpnI | ATATAGGTACCCGTCAGTCGGTGGCCGC | Reverse primer for creation of bldG-containing pKT25- and pUT18C-based constructs |
| AP11 | KpnI | ATATAGGTACCCGGTCGGTGGCCGCCAC | Reverse primer for creation of bldG-containing pUT18-based constructs |
| AP12 | BamHI | ATATAGGATCCCATGGCCACCGTCGAACTCCGT | Forward primer for creation of sco3548-containing pKT25-, pUT18C-, and pUT18-based constructs |
| AP13 | KpnI | ATATAGGTACCCGTCAGATCGGTGGCAGCACCGC | Reverse primer for creation of sco3548-containing pKT25- and pUT18C-based constructs |
| AP14 | KpnI | ATATAGGTACCCGGATCGGTGGCAGCACCGCCGA | Reverse primer for creation of sco3548-containing pUT18-based constructs |
| AP7 | GGCGCGCAGTTCGGTGACCAGCGGC | Forward primer for sequencing pKT25 inserts | |
| AP8 | GGGATGTGCTGCAAGGCGATTAAG | Reverse primer for sequencing pKT25 inserts | |
| AP9 | TTTATGCTTCCGGCTCGTATGTT | Forward primer for sequencing pUT18 inserts | |
| AP10 | CAAGTCGATGCGTTCGCGAT | Reverse primer for sequencing pUT18 inserts | |
| AP30 | CTCGCCGGATGTACTGGAAAC | Forward primer for sequencing pUT18C inserts | |
| AP31 | CGGGGCTGGCTTAACTATGC | Reverse primer for sequencing pUT18C inserts | |
| BKL62 | EcoRI | CGGCGAATTCGTCTGCAACCAGGAGCGC | Forward primer for pAU365 construction |
| DBG3 | XbaI | GCGCTCTAGAGTTCGACGGTGGCCATG | Reverse primer for pAU316 construction |
| DBG33 | XbaI | GCGCTCTAGATCCCATGCCATTGATCGTGAA | Reverse primer for pAU317 construction |
| DBG34 | NdeI | GCGCCATATGGCCACCGTCGAACTCCGT | Forward primer for pAU317 construction |
| DBG35 | NdeI | GCGCCATATGGACCTGTCCCTGTCGAC | Forward primer for pAU316 construction |
| KC61 | XbaI | CGCGAGATCTGTCCGCACGGCACCAATG | Reverse primer for pAU365 construction |
Restriction enzyme sites are underlined.
Preparation of S. coelicolor cell lysates.
For preparation of S. coelicolor crude cell lysates, strains were grown in liquid cultures to mid-exponential phase and were harvested by centrifugation. Cell pellets were washed with double-distilled water and resuspended in Tris sonication buffer (50 mM Tris-Cl [pH 8.5], 300 mM NaCl, 1× Complete EDTA-free protease inhibitor cocktail [Roche]). Alternatively, strains were grown on R2YE agar overlaid with cellophane disks (75-mm 325P disks; Courtalds Films), harvested by scraping them directly from plates, and resuspended in HEPES sonication buffer (50 mM HEPES [pH 7.2], 1× Complete EDTA-free protease inhibitor cocktail). Mycelia were lysed by sonication, the resulting lysate was clarified by centrifugation at 4°C, and the protein content of the lysate was quantified using Bio-Rad protein assay dye reagent concentrate according to the manufacturer's instructions. Western analysis of both crude cell lysates and purified proteins was performed as described previously (7).
Construction of BldG and SCO3548 expression plasmids.
For construction of bldG and sco3548 expression plasmids, the corresponding ORFs were amplified by PCR using primers DBG35 and DBG3 (bldG) or DBG34 and DBG33 (sco3548). The resulting PCR products were digested with NdeI and XbaI and were cloned into similarly digested pIJ6902. The resulting plasmid constructs were verified by DNA sequencing and were designated pAU316 for bldG expression and pAU317 for sco3548 expression.
Construction of a BldG-SCO3548 coexpression plasmid.
In order to clone the entire bldG locus under control of the ptipA promoter, pAU316 was digested with BglII to remove and discard the distal end of the bldG ORF. Separately, the distal end of the bldG gene linked to the entire sco3548 ORF by the native intergenic region was PCR amplified from the H5 cosmid (51) using the KC61 and BKL62 primers and was similarly digested with BglII. The digested KC61-BKL62 fragment was then ligated into the prepared pAU316 vector containing the proximal end of the bldG ORF under control of the ptipA promoter. The sequence integrity and insert orientation of the resulting recombinant plasmid were verified by DNA sequencing, and the plasmid was designated pAU365.
Overexpression and purification of recombinant proteins from E. coli.
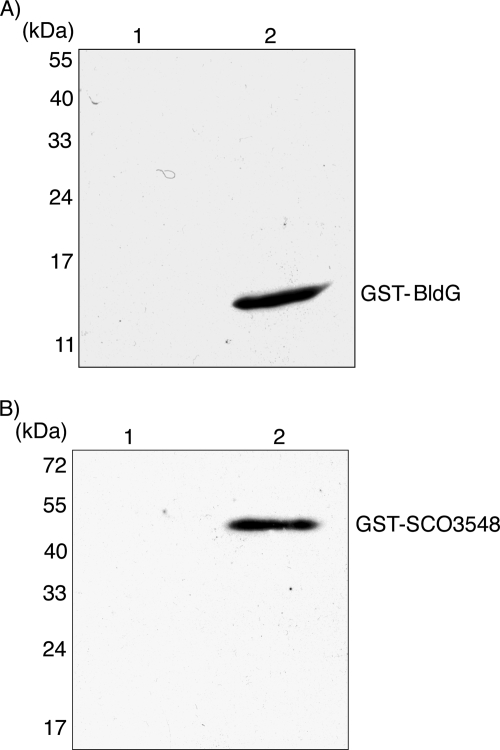
In order to express BldG and SCO3548 as glutathione S-transferase (GST) fusions, the bldG and sco3548 coding regions were isolated from plasmids pAU316 and pAU317, respectively, as NdeI-EcoRI fragments and were cloned into similarly digested vector pGEX-2TK (Amersham Biosciences). The resulting recombinant plasmids, designated pAU375 (GST-BldG) and pAU376 (GST-SCO3548), were transformed into E. coli BL21(DE3), and expression of the fusion proteins was induced in mid-logarithmic-phase cultures by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The resulting soluble protein was released by sonication in lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1× Complete EDTA-free protease inhibitor cocktail), and the GST fusion proteins were purified by glutathione-Sepharose affinity chromatography (Amersham Biosciences) according to the manufacturer's recommendations. Eluted fusion protein samples were desalted (PD-10 column; Amersham Biosciences), and the protein was quantified using the Bio-Rad protein assay dye reagent concentrate. As a control, GST was expressed from unmodified pGEX-2TK and was similarly purified from E. coli BL21(DE3).
In order to express SCO3548 as a His10-tagged fusion protein, the sco3548 coding region was isolated from the pAU317 plasmid as an NdeI-EcoRI fragment and was cloned into similarly digested vector pET30(a)+. The resulting recombinant plasmid, designated pAU318, and the pET30(a)+ control were transformed into E. coli BL21(DE3).
Bacterial two-hybrid analysis of protein interactions.
Bacterial two-hybrid analysis of protein interactions was performed as described by Karimova et al. (27, 29, 30). To construct recombinant plasmids for use in bacterial two-hybrid analysis, the bldG and sco3548 ORFs were amplified by PCR from the pAU316 and pAU317 plasmids, respectively, using the corresponding oligonucleotide primers (AP3 and AP4 or AP3 and AP11 for bldG and AP12 and AP13 or AP12 and AP14 for sco3548). To create in-frame fusions to the T25 and T18 fragments of the adenylate cyclase gene, all amplified fragments were digested with BamHI and KpnI; AP3-4 and AP12-13 fragments were cloned into similarly digested vectors pKT25 and pUT18C, and AP3-11 and AP12-14 fragments were cloned into similarly digested pUT18. The resultant recombinant plasmids (pAU377 to pAU382 [Table 2]) were verified by DNA sequencing using vector-based primers AP7 and AP8 for pKT25 constructs, AP9 and AP10 for pUT18 constructs, and AP30 and AP31 for pUT18C constructs.
For analysis of protein interactions, pKT25-derived plasmids and pUT18 or pUT18C-derived plasmids, as well as empty vector controls, were cotransformed into the adenylate cyclase-deficient strain E. coli BTH101 in all possible combinations. Leucine zipper fragments cloned into pKT25 and pUT18C were used as a positive control for protein interaction (28). The primary assay for protein interactions was conducted by plating cotransformants either on LB agar containing 0.5 mM IPTG and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) or on MacConkey-maltose agar; both media contained ampicillin and kanamycin to select for plasmid maintenance. The plates were then incubated at 30°C for a maximum of 36 h.
The secondary assay for protein interactions involved quantitative determination of β-galactosidase activity (55). Five individually isolated replicate cotransformants were grown overnight at 37°C in LB medium containing ampicillin and kanamycin. The cultures were then diluted 1:50 in fresh LB medium containing antibiotics and grown until an optical density at 600 nm of 0.5 was reached. Cells were harvested by centrifugation and permeabilized using chloroform and sodium dodecyl sulfate (SDS), and β-galactosidase activity was measured with a microtiter plate assay using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate.
Affinity purification of interacting protein complexes.
To examine the interaction between BldG and SCO3548, E. coli BL21(DE3) strains containing the recombinant plasmid pAU375 (GST-BldG) or pAU318 (His10-SCO3548) were grown to mid-logarithmic phase, and fusion protein expression was induced by addition of IPTG to a final concentration of 1 mM. After incubation for 3 h, cells were harvested by centrifugation, and the pellets were resuspended in GST extraction buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA; pH 8.5) and lysed by sonication. Equal volumes of the two cell lysates (one containing GST-BldG and one containing His10-SCO3548) were mixed together and were incubated at 4°C for 1 h. The mixture was subsequently combined with a 50% slurry of glutathione-Sepharose 4B in GST extraction buffer and was incubated at 4°C for 2 h before application to a gravity flow Poly-Prep chromatography column (Bio-Rad). Columns were washed sequentially with 10 bed volumes of GST extraction buffer containing 0.5% Triton X-100, 10 bed volumes of GST extraction buffer containing 0.1% Triton X-100, and 5 bed volumes of GST extraction buffer alone. Bound proteins were eluted with GST extraction buffer containing 20 mM reduced glutathione and were examined by Western analysis using anti-His antibody (Amersham Biosciences).
To examine self-interaction of SCO3548, E. coli BL21(DE3) strains containing recombinant plasmid pAU318 (His10-SCO3548) or pAU376 (GST-SCO3548) were grown, and fusion protein expression was induced as described above. After incubation for 3 h, cells were harvested by centrifugation, and the pellets were resuspended in Ni-nitrilotriacetic acid (NTA) buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0], 1× Complete EDTA-free protease inhibitor cocktail) and lysed by sonication. Equal volumes of the two cell lysates (one containing His10-SCO3548 and one containing GST-SCO3548) were mixed together and were incubated at 4°C for 1 h. The mixture was subsequently combined with a 50% slurry of Ni-NTA agarose (Qiagen) in Ni-NTA buffer and was incubated at 4°C for 2 h before application to a gravity flow Poly-Prep chromatography column. The columns were washed with 10 bed volumes of Ni-NTA buffer containing 50 mM imidazole, and bound proteins were eluted with Ni-NTA buffer containing 250 mM imidazole (pH 8.0) and were examined by Western analysis using anti-GST antibody (Amersham Biosciences).
Far-Western analysis.
To examine BldG protein interactions, cell lysates of S. coelicolor ΔbldG 1DB containing either pAU316 or pIJ6902 were separated by 15% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. After equilibration in wash buffer (50 mM Tris [pH 7.5], 100 mM sodium acetate, 350 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 0.2% Tween 20), membranes were blocked with 3% skim milk in wash buffer. To examine the BldG-SCO3548 interaction, membranes were subsequently incubated with purified GST-SCO3548 fusion protein (final concentration, 2.5 μg/ml) for 2 h at room temperature. To examine BldG self-interaction, membranes were similarly incubated with purified GST-BldG fusion protein. In both cases, unbound protein was removed by gentle agitation with wash buffer, the membranes were examined by Western analysis using anti-GST antibody (Amersham Biosciences) (7).
RESULTS
BldG interacts with SCO3548.
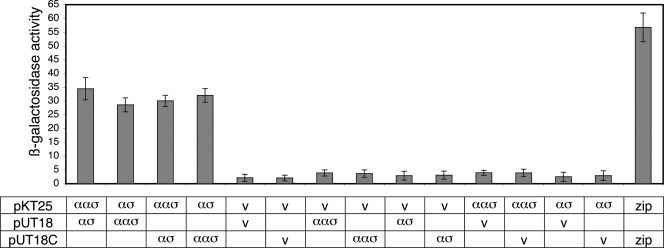
Orthologues of BldG typically interact with antagonistic protein partners; therefore, experiments to assess the ability of BldG to form protein complexes were performed. Preliminary chemical cross-linking experiments performed with BldG-containing S. coelicolor cell extracts demonstrated that BldG was present in several higher-molecular-weight complexes (major bands at ∼30 and 70 kDa), as well as in its monomeric form (predicted molecular mass, 12.3 kDa) (not shown), indicating that BldG indeed forms protein complexes. Given that in orthologous B. subtilis systems the cotranscribed gene encodes the antagonistic protein partner (5, 26, 53, 58), SCO3548 was therefore the most likely candidate to form a complex with BldG. To assess this potential interaction, we first used a bacterial two-hybrid system based on interaction-mediated reconstitution of the Bordetella pertussis adenylate cyclase enzyme in adenylate cyclase-deficient E. coli strain BTH101 (27, 28). Full-length bldG and sco3548 coding sequences were cloned into plasmids pKT25, pUT18, and pUT18C, and the resulting recombinant constructs were cotransformed into BTH101 in all possible combinations. Primary screening by color development of cotransformants on LB (with IPTG and X-Gal) and MacConkey-maltose media suggested that there was an interaction between BldG and SCO3548 (not shown). A secondary assay of the protein interaction was performed by quantitatively determining β-galactosidase activity. Coexpression of BldG and SCO3548 resulted in β-galactosidase activity comparable to that observed for the well-established leucine zipper interaction and at least sevenfold higher than that observed for cotransformants containing either two empty vectors or one empty vector and one recombinant construct, confirming that there is an interaction between BldG and SCO3548 (Fig. 1).
FIG. 1.
BldG-SCO3548 interaction was detected by bacterial two-hybrid analysis. The bldG (αασ) and sco3548 (ασ) ORFs were cloned into plasmids pKT25, pUT18, and pUT18C and were cotransformed into E. coli BTH101 in all possible combinations. Either two empty vectors (v) or one empty vector and one recombinant plasmid were used as negative controls. Plasmids containing leucine zipper fragments (zip) were used as the positive controls. The β-galactosidase activities of five individually isolated replicate liquid cultures of each cotransformant were determined by a microtiter plate assay using ONPG as a substrate. β-Galactosidase activity was expressed as (units per unit of optical density at 600 nm × ml of cell suspension × 103), where 1 U was defined as 1 μmol of o-nitrophenol formed per min. The error bars indicate the standard deviation from the mean.
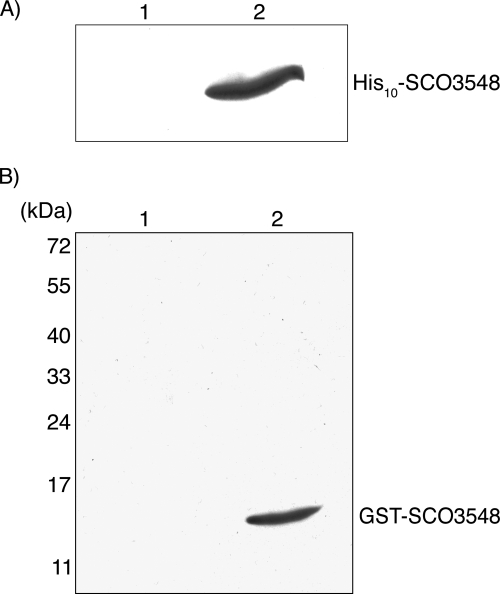
As a further test of the direct interaction between BldG and SCO3548, affinity purification of BldG protein complexes was performed. Recombinant plasmids pAU375 and pAU318, encoding GST-BldG and His10-SCO3548 fusion proteins, respectively, were introduced separately into E. coli BL21(DE3). Protein expression was induced in the resulting strains, and equal volumes of cell lysates of the strains were combined and incubated to allow protein interaction. Protein complexes were then captured by addition of glutathione-Sepharose 4B, were eluted with glutathione-containing buffer, and were examined by Western analysis with anti-His antibody. His10-SCO3548 was not detected in a control experiment in which native GST was expressed from the parent plasmid pGEX-2TK (Fig. 2A), indicating that His10-SCO3548 does not interact with GST alone and does not associate nonspecifically with glutathione-Sepharose 4B. Affinity purification of His10-SCO3548 in complex with GST-BldG, therefore, confirmed that there was an interaction between the two proteins (Fig. 2A).
FIG. 2.
BldG-SCO3548 interaction was detected in vitro by affinity purification and far-Western analysis. (A) Cell lysate from E. coli BL21(DE3) containing pGEX-2TK (GST control protein) (lane 1) or containing pAU375 (GST-BldG) (lane 2) was mixed with cell lysate from E. coli BL21(DE3) containing pAU318 (His10-SCO3548) at a ratio of 1:1 (vol/vol). Glutathione-Sepharose 4B beads were added to capture GST-containing protein complexes, which were then eluted with 20 mM reduced glutathione. Eluted proteins were analyzed by SDS-PAGE and Western analysis using anti-His antibody. (B) Cell lysates from S. coelicolor ΔbldG 1DB containing pIJ6902 (vector control) (lane 1) or pAU316 (bldG expression) (lane 2) were subjected to SDS-PAGE, and the separated proteins were transferred to a PVDF membrane. For far-Western analysis, the membrane was incubated with purified GST-SCO3548, washed thoroughly, and probed with anti-GST antibody. The positions of molecular mass markers are indicated on the left.
Lastly, direct interaction between BldG and SCO3548 was investigated by far-Western analysis. Cell lysates of S. coelicolor ΔbldG 1DB containing either pAU316 or pIJ6902 were separated by SDS-PAGE and immobilized on a PVDF membrane. The membrane was probed first with purified GST-SCO3548 and then with anti-GST antibody to detect protein interactions. In the lane containing cell lysate from the bldG-expressing strain, GST-SCO3548 was detected on the membrane at a position corresponding to the apparent molecular weight of BldG (Fig. 2B). This interaction was not observed in the lane containing cell lysate from the vector control strain (Fig. 2B) or in a control experiment in which the membrane was probed with native GST (not shown). These results further confirm the finding described above that BldG and SCO3548 form a complex both in vitro and in E. coli.
SCO3548 antagonizes BldG function in S. coelicolor.
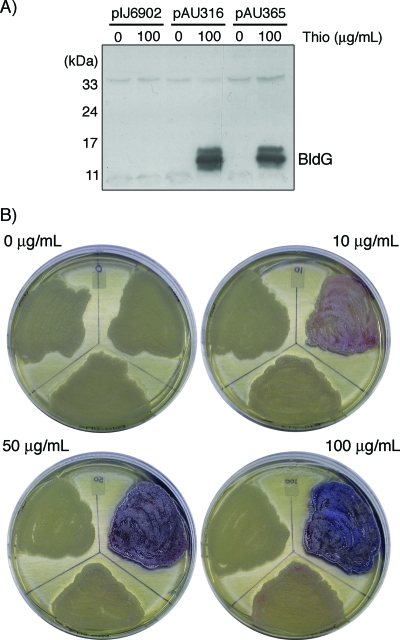
Based on analogy to the corresponding systems in B. subtilis, the BldG-SCO3548 interacting pair was predicted to be functionally antagonistic in vivo. In order to examine this prediction, the pAU365 coexpression construct was created such that the entire bldG locus (bldG ORF, native intergenic region, and sco3548 ORF) was expressed from the thiostrepton-inducible ptipA promoter. The pAU365 construct was then introduced into S. coelicolor ΔbldG 1DB, which lacks a functional bldG gene but expresses the wild-type sco3548 gene from the native bldG promoter, producing a recombinant strain that expresses an abnormally high proportion of SCO3548 relative to BldG. S. coelicolor ΔbldG 1DB strains containing the pAU365 coexpression construct, the pAU316 bldG expression construct, or the pIJ6902 vector control were grown in the presence or absence of thiostrepton, and crude cell lysates were examined by Western analysis with anti-BldG antibody. The bldG expression and bldG-sco3548 coexpression strains were found to accumulate equivalent amounts of BldG protein upon thiostrepton induction (Fig. 3A), whereas no BldG protein was detected in the vector control strain or in the expression strains in the absence of induction.
FIG. 3.
Inducible coexpression of BldG and SCO3548 does not complement the S. coelicolor bldG mutant phenotype. (A) S. coelicolor ΔbldG 1DB containing pIJ6902 (vector control), pAU316 (bldG expression), or pAU365 (bldG-sco3548 coexpression) was grown on R2YE agar containing 0 or 100 μg/ml thiostrepton (Thio) at 30°C for 40 h. Crude cell lysates were prepared and examined by SDS-PAGE, followed by Western analysis with anti-BldG antibody. (B) S. coelicolor ΔbldG 1DB containing pIJ6902 (top left region of each plate), pAU316 (top right region), or pAU365 (bottom center region) was grown at 30°C on R2YE agar containing 0, 10, 50, or 100 μg/ml thiostrepton and photographed after 80 h.
To determine the effect of increased levels of SCO3548 on the ability of plasmid-expressed BldG to complement the ΔbldG 1DB mutant phenotype, the strains described above were examined by growing them in the presence of increasing amounts of thiostrepton inducer (Fig. 3B). The pAU316 construct restored aerial hypha formation and pigmented antibiotic production to S. coelicolor ΔbldG 1DB as the amount of thiostrepton inducer was increased; at the highest thiostrepton concentrations, the bldG mutant phenotype was completely complemented. In contrast, the pAU365 coexpression construct was unable to complement the bldG mutant phenotype at even the highest thiostrepton concentration, despite accumulation of the BldG protein to levels equivalent to that in the strain containing pAU316. The increased expression of SCO3548 in the coexpression strain therefore abrogated the ability of the BldG protein to complement the bldG mutant phenotype, indicating that these two proteins have antagonistic functions in vivo in the regulation of aerial hypha formation and secondary metabolism.
BldG exhibits self-interaction.
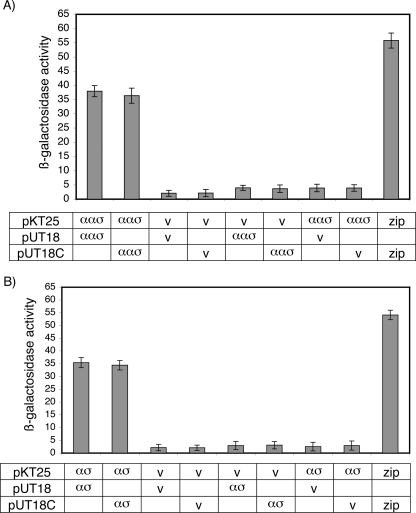
The similarity of BldG to proteins known to form homodimers (22, 38, 48), together with preliminary identification by chemical cross-linking of a BldG-containing complex whose size was consistent with dimerization (not shown), led to examination of BldG self-interaction. First, bacterial two-hybrid analysis was performed using bldG-containing recombinant plasmids (pAU377 to pAU379). Primary screening of cotransformants showed that there was an interaction between the T25- and T18-BldG fusion proteins. Secondary assays confirmed this interaction, demonstrating that there was at least ninefold-higher β-galactosidase activity in the bldG-expressing cotransformants than in controls containing either two empty vectors or one empty vector and one recombinant construct (Fig. 4A).
FIG. 4.
BldG and SCO3548 self-interactions were detected by bacterial two-hybrid analyses. The bldG ORF (αασ) (A) or the sco3548 ORF (ασ) (B) was cloned into plasmids pKT25, pUT18, and pUT18C, which were cotransformed into E. coli BTH101 in all possible combinations. Either two empty vectors (v) or one empty vector and one recombinant plasmid were used as negative controls. Plasmids containing leucine zipper fragments (zip) were used as the positive control. The β-galactosidase activities of five individually isolated replicate liquid cultures of each cotransformant were determined by a microtiter plate assay using ONPG as a substrate, as described in the legend to Fig. 1.
BldG self-interaction was further examined in vitro by far-Western analysis. Cell lysates from S. coelicolor ΔbldG 1DB containing either pAU316 or pIJ6902 were separated by SDS-PAGE and immobilized on a PVDF membrane. The membrane was probed first with purified GST-BldG and then with anti-GST antibody to detect protein interactions. In the lane containing cell lysate from the bldG-expressing strain, GST-BldG was detected on the membrane at a position corresponding to the apparent molecular weight of BldG (Fig. 5A). This interaction did not occur in the lane containing cell lysate from the vector control strain (Fig. 5A) or in a control experiment in which the membrane was probed with native GST (not shown). These results, combined with those described above, demonstrate that BldG exhibits self-interaction.
FIG. 5.
BldG and SCO3548 self-interactions were detected by far-Western analysis and affinity purification, respectively. (A) For far-Western analysis, cell lysates from S. coelicolor ΔbldG 1DB containing pIJ6902 (vector control) (lane 1) or pAU316 (bldG expression) (lane 2) were subjected to SDS-PAGE, and the separated proteins were transferred to a PVDF membrane. The membrane was incubated with purified GST-BldG, washed thoroughly, and probed with anti-GST antibody. (B) For affinity purification, cell lysate from E. coli BL21(DE3) containing pET30(a)+ (vector control) (lane 1) or containing pAU318 (His10-SCO3548) (lane 2) was mixed with cell lysate from E. coli BL21(DE3) containing pAU376 (GST-SCO3548) at a ratio of 1:1 (vol/vol). Ni-NTA agarose was added to capture His10-containing protein complexes, which were then eluted with 250 mM imidazole. Eluted proteins were analyzed by SDS-PAGE and Western analysis using anti-GST antibody. The positions of molecular mass markers are indicated on the left.
SCO3548 exhibits self-interaction.
Anti-sigma factors typically form homodimers (16, 21, 48, 57); therefore, based on similarity to this family of proteins, SCO3548 would be predicted to self-interact. Bacterial two-hybrid analysis, using the sco3548-containing recombinant plasmids pAU380 to pAU382, was employed to examine this interaction. Primary screening of cotransformants showed that there was interaction between the T25- and T18-SCO3548 fusion proteins. Secondary assays confirmed this interaction, demonstrating that there was at least 11-fold-higher β-galactosidase activity in the sco3548-expressing cotransformants than in controls containing either two empty vectors or one empty vector and one recombinant construct (Fig. 4B).
To further examine SCO3548 self-interaction, affinity purification of SCO3548 protein complexes was performed. Recombinant plasmids pAU376 and pAU318, encoding the GST-SCO3548 and His10-SCO3548 fusion proteins, respectively, were introduced separately into E. coli BL21(DE3). Protein expression was induced in the resulting strains, and equal volumes of their cell lysates were combined and incubated to allow protein interaction. Protein complexes were then captured by addition of Ni-NTA, were eluted with imidazole-containing buffer, and were examined by Western analysis with anti-GST antibody. The possibility of a nonspecific interaction of GST-SCO3548 with Ni-NTA was ruled out by the results of a control experiment using crude cell lysate from a BL21(DE3) derivative containing the pET30(a)+ vector control (Fig. 5B). Affinity purification of GST-SCO3548 in complex with His10-SCO3548 therefore confirmed SCO3548 self-interaction (Fig. 5B).
DISCUSSION
On the basis of similarity to the prototypical Bacillus systems, BldG was predicted to exert its regulatory function through interaction with the downstream-encoded protein SCO3548. This prediction was experimentally supported by the results of two-hybrid, affinity purification, and far-Western analyses, which conclusively demonstrated that there is a direct interaction between BldG and SCO3548. Preliminary cross-linking analysis also revealed the presence of a BldG-containing complex consistent with the predicted size of a BldG dimer (not shown). A combination of two-hybrid and far-Western analyses was therefore employed to demonstrate the self-interaction of BldG. Although RsbV and SpoIIAA do not form homodimers in solution (16, 34, 35, 57), other homologous anti-anti-sigma factors, including TM1442 of Thermotoga maritima (22, 38) and several examples in Mycobacterium tuberculosis (48), exist in solution in a dimeric form. Based on our results, we therefore propose that BldG also forms homodimers.
Similarly, many anti-sigma factors, including RsbW and SpoIIAB (16, 21, 57) and several from M. tuberculosis (48), form homodimers in solution. Results of two-hybrid analysis and affinity purification demonstrated that SCO3548 also forms homomultimers, which we propose to be homodimers based on similarity to the examples mentioned above. In all examined partner-switching systems, the two protein components associate at a 1:1 ratio to form a heterotetrameric complex (14, 16, 19, 20, 40, 42). The identification of a high-molecular-weight BldG-containing complex by preliminary chemical cross-linking analysis (∼70 kDa) (not shown) suggests that BldG and SCO3548 may form a similar heterotetrameric complex. These results are therefore consistent with the prediction that BldG and SCO3548 form a partner-switching regulatory complex with stoichiometry similar to that of previously characterized orthologous systems.
Without an identified sigma factor target for the BldG-SCO3548 pair, the existence of a partner-switching mechanism cannot be examined further biochemically. Furthermore, the apparently lethal nature of an sco3548 deletion (not shown) restricted our ability to address the in vivo role of SCO3548. Complementation analysis was therefore performed to examine the antagonistic relationship between BldG and SCO3548. The results of this analysis demonstrated that an increase in expression of SCO3548 abrogated the ability of BldG to complement the phenotype of a ΔbldG null mutant. This result confirms that the interaction between BldG and SCO3548 affects the regulation of morphological and physiological differentiation in vivo and supports our hypothesis that BldG and SCO3548 likely form an antagonistic partner-switching pair. Given these results, we propose that sco3548 should be given the more descriptive gene designation apgA (for antagonistic partner of BldG).
Furthermore, under wild-type conditions, the bldG monocistronic transcript is present at a two- to three-fold molar excess over the bldG-apgA polycistronic transcript (9). The fact that introduction of a single additional copy of apgA completely antagonizes BldG function therefore indicates that the regulatory function of this system requires an excess of BldG, suggesting that BldG may have additional anti-sigma factor interacting partners. Although a similar system, with an anti-anti-sigma factor regulating two or more anti-sigma factors, has not been identified so far, there are other complex systems which deviate from the B. subtilis paradigm. In M. tuberculosis, σF activity is regulated by a single coexpressed anti-sigma factor, which is in turn antagonized by two distantly encoded anti-anti-sigma factors (4). As well, the σF sporulation factor of S. coelicolor is regulated by an anti-sigma factor (RsfA) and at least two anti-anti-sigma factors, all encoded at separate loci (32). The presence of 45 additional RsbW orthologues in the S. coelicolor genome (45) provides a plethora of possible alternative binding partners for BldG. The possibility that the BldG system involves additional anti-sigma factors therefore must be examined further. As previously discussed (32), analysis of these interactions may prove to be difficult due to the apparent promiscuity of the interactions between the many anti-anti-sigma factors and anti-sigma factors in S. coelicolor.
The identity of the regulatory target(s) of the BldG-ApgA pair also remains to be determined. Similarity to the Rsb and SpoIIA systems suggests that the likely target is one of the nine group 3 subfamily sigma factors of S. coelicolor; however, current work (13, 15, 33, 37, 49, 54, 56) has not identified a single sigma factor whose mutant phenotype can solely account for the regulatory effect of BldG. It is therefore likely that more than one sigma factor is affected by BldG and/or ApgA. A similar situation was observed by Kim et al. (32), who found that the phenotypes of an rsfA anti-sigma factor mutant and the corresponding sigF sigma factor mutant were not completely complementary, suggesting that RsfA regulates additional sigma factor targets. Further characterization of the BldG-ApgA system is therefore necessary to identify its regulatory targets and to determine the roles that its interactions play in regulating morphological and physiological differentiation in S. coelicolor.
Finally, reversible phosphorylation of the conserved serine residue of BldG has been shown to be critical for regulating the onset of aerial hypha formation and antibiotic production (7) and therefore is predicted to play a critical role in the proposed partner-switching mechanism between BldG and ApgA. In B. subtilis, phosphorylation of the anti-anti-sigma factor is carried out by the cognate anti-sigma factor (RsbW and SpoIIAB), which possesses kinase activity (1, 17, 18, 46). A similar relationship does not exist between BldG and ApgA; sequence alignment of ApgA with the B. subtilis anti-sigma factors showed that ApgA lacks the conserved residues required for kinase function (9), and protein domain analysis did not reveal the presence of any kinase domain in ApgA (45). Moreover, a bldG in-frame deletion mutant does not possess BldG kinase activity (7), despite the fact that apgA is known to be expressed in bldG mutants. A similar situation has been observed in M. tuberculosis, in which the anti-sigma factor UsfX lacks both the conserved kinase domain and the ability to phosphorylate its cognate anti-anti-sigma factor RsfB (4). These observations clearly demonstrate that an additional unidentified kinase must be involved in the phosphorylation of BldG. Candidates for this unknown kinase are plentiful; a plethora of proteins possessing the necessary HATPase_c kinase domain are present in the S. coelicolor genome (45). Identification of this BldG kinase is a critical avenue for future investigation, as the regulation of the phosphorylation state of BldG likely plays a critical role in controlling the antagonistic partner-switching mechanism of BldG and ApgA and thereby in controlling the activity of their unidentified direct regulatory target(s).
Close orthologues of BldG and ApgA have been found in syntenic regions of the genomes in all sequenced Streptomyces spp. examined to date (Streptomyces avermitilis [25], Streptomyces griseus [47], and Streptomyces scabies [http://www.sanger.ac.uk/Projects/S_scabies/]), as well as in the partially sequenced species Streptomyces clavuligerus (8). In addition, transcriptional regulation of the bldG locus is conserved in S. clavuligerus and is known to play a similar key role in regulating both morphological and physiological differentiation (8). This system therefore appears to be a key developmental regulatory switch common to members of the genus Streptomyces, and understanding it may increase our ability to understand and manipulate a variety of industrially important Streptomyces species. Furthermore, the conservation of bldG and apgA in Thermobifida fusca (12) suggests that the bldG locus may be important not only in Streptomyces but also in a wide range of other actinobacteria.
Acknowledgments
This work was supported by the National Sciences and Engineering Research Council of Canada and the Alberta Ingenuity Fund.
We thank Troy Locke for technical assistance.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260165-177. [DOI] [PubMed] [Google Scholar]
- 2.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77195-205. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and anti-sigma-factor antagonists. Curr. Biol. 10R53-R55. [DOI] [PubMed] [Google Scholar]
- 4.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-σ factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 451527-1540. [DOI] [PubMed] [Google Scholar]
- 5.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 1752347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 7.Bignell, D. R., L. H. Lau, K. R. Colvin, and B. K. Leskiw. 2003. The putative anti-anti-sigma factor BldG is post-translationally modified by phosphorylation in Streptomyces coelicolor. FEMS Microbiol. Lett. 22593-99. [DOI] [PubMed] [Google Scholar]
- 8.Bignell, D. R., K. Tahlan, K. R. Colvin, S. E. Jensen, and B. K. Leskiw. 2005. Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob. Agents Chemother. 491529-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bignell, D. R., J. L. Warawa, J. L. Strap, K. F. Chater, and B. K. Leskiw. 2000. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 1462161-2173. [DOI] [PubMed] [Google Scholar]
- 10.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 1701168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4667-673. [DOI] [PubMed] [Google Scholar]
- 12.Chater, K. F., and G. Chandra. 2006. The evolution of development in Streptomyces analysed by genome comparisions. FEMS Microbiol. Rev. 30651-672. [DOI] [PubMed] [Google Scholar]
- 13.Cho, Y. H., E. J. Lee, B. E. Ahn, and J. H. Roe. 2001. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42205-214. [DOI] [PubMed] [Google Scholar]
- 14.Clarkson, J., I. D. Campbell, and M. D. Yudkin. 2001. NMR studies of the interactions of SpoIIAA with its partner proteins that regulate sporulation in Bacillus subtilis. J. Mol. Biol. 314359-364. [DOI] [PubMed] [Google Scholar]
- 15.Dalton, K. A., A. Thibessard, J. I. Hunter, and G. H. Kelemen. 2007. A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64719-737. [DOI] [PubMed] [Google Scholar]
- 16.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J. Bacteriol. 1845583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Erringston, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 82653-2663. [DOI] [PubMed] [Google Scholar]
- 18.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 1761813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufour, A., U. Voelker, A. Voelker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 1783701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan, L., S. Alper, and R. Losick. 1996. SpoIIAA governs the release of the cell-type specific transcription factor σF from its anti-sigma factor SpoIIAB. J. Mol. Biol. 260147-164. [DOI] [PubMed] [Google Scholar]
- 21.Duncan, L., and R. Losick. 1993. SpoIIAB is an anti-σ factor that binds to and inhibits transcription by regulatory protein σF from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 902325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha, K. S., J. E. Kwak, B. W. Han, J. Y. Lee, J. Moon, B. I. Lee, and S. W. Suh. 2001. Crystallization and preliminary X-ray crystallographic analysis of the TM1442 gene product from Thermotoga maritima, a homologue of Bacillus subtilis anti-anti-sigma factors. Acta Crystallogr. D Biol. Crystallogr. 57276-278. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, M. Y., J. B. Bae, J. H. Park, and J. H. Roe. 2003. Isolation and characterization of Streptomyces coelicolor RNA polymerase, its sigma, and anti-sigma factors. Methods Enzymol. 37073-82. [DOI] [PubMed] [Google Scholar]
- 24.Huang, J., J. Shi, V. Molle, B. Sohlberg, D. Weaver, M. J. Bibb, N. Karoonuthaisiri, C. J. Lih, C. M. Kao, M. J. Buttner, and S. N. Cohen. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 581276-1287. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21526-531. [DOI] [PubMed] [Google Scholar]
- 26.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 1725575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 1872233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 955752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karimova, G., A. Ullmann, and D. Ladant. 2000. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 32859-73. [DOI] [PubMed] [Google Scholar]
- 30.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 373-82. [PubMed] [Google Scholar]
- 31.Kieser, T., M. J. Bibb, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Colney, Norwich, United Kingdom.
- 32.Kim, E. S., J. Y. Song, D. W. Kim, K. F. Chater, and K. J. Lee. 2008. A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. J. Bacteriol. 1907559-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kormanec, J., D. Homerova, I. Barak, and B. Sevcikova. 1999. A new gene, sigG, encoding a putative alternative sigma factor of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 172153-158. [DOI] [PubMed] [Google Scholar]
- 34.Kovacs, H., D. Comfort, M. Lord, I. D. Campbell, and M. D. Yudkin. 1998. Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor σF of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 955067-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovacs, H., D. Comfort, M. Lord, M. Yudkin, I. D. Campbell, and M. Nilges. 2001. NMR studies of the sporulation protein SpoIIAA: implications for the regulation of the transcription factor σF in Bacillus subtilis. J. Biomol. NMR 19293-304. [DOI] [PubMed] [Google Scholar]
- 36.Lee, E. J., Y. H. Cho, H. S. Kim, B. E. Ahn, and J. H. Roe. 2004. Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 1868490-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, E. J., N. Karoonuthaisiri, H. S. Kim, J. H. Park, C. J. Cha, C. M. Kao, and J. H. Roe. 2005. A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 571252-1264. [DOI] [PubMed] [Google Scholar]
- 38.Lee, J. Y., H. J. Ahn, K. S. Ha, and S. W. Suh. 2004. Crystal structure of the TM1442 protein from Thermotoga maritima, a homolog of the Bacillus subtilis general stress response anti-anti-sigma factor RsbV. Proteins 56176-179. [DOI] [PubMed] [Google Scholar]
- 39.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 11161-68. [DOI] [PubMed] [Google Scholar]
- 40.Magnin, T., M. Lord, J. Errington, and M. D. Yudkin. 1996. Establishing differential gene expression in sporulating Bacillus subtilis: phosphorylation of SpoIIAA (anti-anti-σF) alters its conformation and prevents formation of a SpoIIAA/SpoIIAB/ADP complex. Mol. Microbiol. 19901-907. [DOI] [PubMed] [Google Scholar]
- 41.Magnin, T., M. Lord, and M. D. Yudkin. 1997. Contribution of partner switching and SpoIIAA cycling to regulation of σF activity in sporulating Bacillus subtilis. J. Bacteriol. 1793922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda, S., K. S. Murakami, S. Wang, C. A. Olson, J. Donigian, F. Leon, S. A. Darst, and E. A. Campbell. 2004. Crystal structures of the ADP and ATP bound forms of the Bacillus anti-σ factor SpoIIAB in complex with the anti-anti-σ SpoIIAA. J. Mol. Biol. 340941-956. [DOI] [PubMed] [Google Scholar]
- 43.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96299-315. [DOI] [PubMed] [Google Scholar]
- 44.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74735-742. [DOI] [PubMed] [Google Scholar]
- 45.Mittenhuber, G. 2002. A phylogenomic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. Biotechnol. 4427-452. [PubMed] [Google Scholar]
- 46.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 1772912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 1904050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parida, B. K., T. Douglas, C. Nino, and S. Dhandayuthapani. 2005. Interactions of anti-sigma factor antagonists of Mycobacterium tuberculosis in the yeast two-hybrid system. Tuberculosis (Edinburgh) 85347-355. [DOI] [PubMed] [Google Scholar]
- 49.Potuckova, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 1737-48. [DOI] [PubMed] [Google Scholar]
- 50.Price, C. W. 2002. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 51.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 2177-96. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 53.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, Jr., and R. Losick. 1990. Control of developmental transcription factor σF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 879221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sevcikova, B., O. Benada, O. Kofronova, and J. Kormanec. 2001. Stress-response sigma factor σH is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 17798-106. [DOI] [PubMed] [Google Scholar]
- 55.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 1734039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viollier, P. H., G. H. Kelemen, G. E. Dale, K. T. Nguyen, M. J. Buttner, and C. J. Thompson. 2003. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47699-714. [DOI] [PubMed] [Google Scholar]
- 57.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. The yeast two-hybrid system detects interactions between Bacillus subtilis σB regulators. J. Bacteriol. 1787020-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, J. J., M. G. Howard, and P. J. Piggot. 1989. Regulation of transcription of the Bacillus subtilis spoIIA locus. J. Bacteriol. 171692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yudkin, M. D., and J. Clarkson. 2005. Differential gene expression in genetically identical sister cells: the initiation of sporulation in Bacillus subtilis. Mol. Microbiol. 56578-589. [DOI] [PubMed] [Google Scholar]