Abstract
The family Rhizobiaceae contains plant-associated bacteria with critical roles in ecology and agriculture. Within this family, many Rhizobium and Sinorhizobium strains are nitrogen-fixing plant mutualists, while many strains designated as Agrobacterium are plant pathogens. These contrasting lifestyles are primarily dependent on the transmissible plasmids each strain harbors. Members of the Rhizobiaceae also have diverse genome architectures that include single chromosomes, multiple chromosomes, and plasmids of various sizes. Agrobacterium strains have been divided into three biovars, based on physiological and biochemical properties. The genome of a biovar I strain, A. tumefaciens C58, has been previously sequenced. In this study, the genomes of the biovar II strain A. radiobacter K84, a commercially available biological control strain that inhibits certain pathogenic agrobacteria, and the biovar III strain A. vitis S4, a narrow-host-range strain that infects grapes and invokes a hypersensitive response on nonhost plants, were fully sequenced and annotated. Comparison with other sequenced members of the Alphaproteobacteria provides new data on the evolution of multipartite bacterial genomes. Primary chromosomes show extensive conservation of both gene content and order. In contrast, secondary chromosomes share smaller percentages of genes, and conserved gene order is restricted to short blocks. We propose that secondary chromosomes originated from an ancestral plasmid to which genes have been transferred from a progenitor primary chromosome. Similar patterns are observed in select Beta- and Gammaproteobacteria species. Together, these results define the evolution of chromosome architecture and gene content among the Rhizobiaceae and support a generalized mechanism for second-chromosome formation among bacteria.
The family Rhizobiaceae (order Rhizobiales) of the Alphaproteobacteria includes the plant pathogens of the genus Agrobacterium and the nitrogen-fixing plant mutualists of the genera Rhizobium and Sinorhizobium. Members house single and multiple chromosome arrangements, linear replicons, and plasmids of various sizes. Genes of pathogenicity, mutualism, and other symbiotic properties are primarily encoded on large transmissible plasmids. Given the promiscuous nature of these elements, different genomic lineages within the Rhizobiaceae exhibit a variety of symbiotic phenotypes that range from pathogenesis to nitrogen-fixing mutualism.
Agrobacterium taxonomy and phylogeny display a marked disparity. Empirically, organisms of the genus Agrobacterium are grouped into five species based on the disease phenotype associated with the resident disease-inducing plasmid: A. tumefaciens causes crown gall on dicotyledonous plants, including stone fruit and nut trees; A. rubi causes crown gall on raspberries; A. vitis causes gall formation that is limited to grapes; A. rhizogenes causes hairy root disease; and A. radiobacter is avirulent. An alternative classification scheme groups Agrobacterium organisms into three biovars based on physiological and biochemical properties without consideration of disease phenotype. Whole-genome and molecular marker comparisons indicate that Agrobacterium strains are derived from multiple chromosomal lineages (see below) (19, 26, 47, 48). The species and biovar classification schemes do not coincide well, in large part because the disease-inducing plasmids are readily transmissible. The history of Agrobacterium classification was recently reviewed by Young (48).
Representative genomes from all three Agrobacterium biovars are now available. The genome of the biovar I strain A. tumefaciens C58 (C58) was previously sequenced (19, 47). The genomes for representatives of the two remaining biovars have now been sequenced and are available as indicated in Materials and Methods. Agrobacterium radiobacter K84 (K84), an avirulent biovar II strain, is a widely used biological control agent for preventing crown gall disease in the field (25, 35). A. vitis S4 (S4), a virulent biovar III strain, is phenotypically distinct from strains of A. tumefaciens in two significant ways. First, whereas A. tumefaciens infects many host species, A. vitis causes crown gall only on grapevines (2, 4). Second, A. vitis induces necrosis on grapevine roots and a hypersensitive response on nonhost plants (3, 22).
This study examines the evolution of genome architecture among agrobacteria, selected sequenced members of the Rhizobiales, and additional bacteria that harbor multiple chromosomes. The biovar I genome of C58 harbors a linear chromosome II derived from a plasmid to which large blocks of DNA, including rRNA operons and other essential genes, have transferred from chromosome I (19, 47). While the sequencing of S4 and K84 was motivated by the need to have full genomic sequences for at least one biovar II representative and at least one biovar III representative, we have found that their genomes, as well as those of C58 and other Rhizobiales species, enabled us to infer a general model for bacterial genome evolution. Crucial for this inference is the complex (for bacteria) replicon architecture of all three Agrobacterium genomes. The data provided here and additional evidence (40, 46) support our model as a generalized mechanism of genome evolution among bacteria that harbor multiple chromosomes.
MATERIALS AND METHODS
DNA sequencing and assembly.
Two DNA libraries (insert sizes, 2 to 4 kbp and 4 to 8 kbp) were generated for each Agrobacterium genome by mechanical shearing of DNA and cloning into pUC18, followed by a shotgun sequencing approach. The reads (∼87,000 for K84 and ∼82,000 for S4) were assembled and edited by using Phred, Phrap, and Consed (13, 14, 20). Gaps were closed by sequencing specific products. All rRNA operons were amplified with specific flanking primers, sequenced, and assembled individually. All nucleotides with Phred scores of less than 40 were resequenced using an independent PCR fragment as template. The error rate is estimated to be less than 1:10,000.
Comparative genomics analyses.
Ortholog families were obtained with orthoMCL (32). Ortholog alignments were obtained with custom Perl scripts. Circular representations of these alignments were obtained with the tool genomeViz (17). Analysis of potential intragenome transfers (http://www.agrobacterium.org; see Tables S6 to S22 in the supplemental material) involved the Multi-Genome Homology Comparison (38) and Phylogenetic Profiler (33) Web-based tools. Completed bacterial genomes listed in NCBI as having more than one chromosome were initially examined, and only those cases where the additional chromosome(s) carried a substantial number of essential genes were maintained. Within this subset, three cases in which two or more closely related genera appear to share a common origin of additional chromosomes were analyzed in greater detail. If intragenome transfer is a robust explanation for the origin of additional chromosomes, then the “transferred” genes should occur in clusters within which the synteny from the initial ancestral chromosome I was maintained. The additional chromosomes of two related genera, A and B, were searched for such shared gene clusters that are present on chromosome I of a unichromosomal relative C but are no longer found on chromosome I in genera A and B. An initial lower limit of similarity of 60% identity was used, and once clusters were identified, the lower limit was adjusted to 40% identity to determine the fullest extent of each shared gene cluster. Preliminary versions of Tables S6 to S22 (see the supplemental material) were checked against the ortholog alignments, and minor corrections and additions were done to obtain the final set of tables.
Analysis of the repABC systems of Agrobacterium organisms.
The RepA, RepB, and RepC protein sequences from Agrobacterium tumefaciens were used as a query against the NCBI database as of May, 2007, using the NCBI BlastP program (1). The top 100 matches were used for analysis. The sequences of each protein were aligned by using the MUSCLE program (11). Phylogenetic and molecular evolutionary analyses were conducted by using MEGA, version 4 (42).
Phylogenetic comparisons among the Rhizobiaceae.
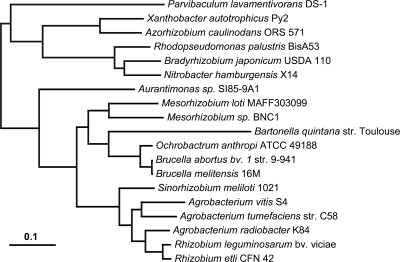
Phylogenetic analysis was performed on a data set of 507 homologous protein groups selected from 19 species of Rhizobiales organisms (Fig. 1; taxa are listed in Table S4 in the supplemental material). The genes were selected strictly from the primary chromosome of each genome. It was allowed that one or two genomes could be missing the gene. Three hundred seventeen homologous groups contained all genomes, 146 were missing one genome, and 45 missed two genomes (see Table S5 in the supplemental material). Homolog groups with more than one entry for a genome were not used. Sequences in each homolog group were trimmed by fit to a hidden Markov model using the HMMer package (10) and then aligned using MUSCLE (11) with the default parameters, as described previously (43). The concatenation of 119,758 aligned positions was analyzed by using the program RaxML (41) using the GAMMA-distributed WAG substitution model. Bootstrapping was performed using the nonparametric (slow) method for 100 replicates.
FIG. 1.
Phylogenetic tree relating 19 genomes in the Rhizobiales. The tree was inferred from 119,758 aligned protein positions from 507 genes located strictly on the primary chromosome in each genome. Bootstrap support was 100% for all nodes except that linking Bradyrhizobium and Nitrobacter genomes, which was 98 out of 100. Bar, number of amino acid substitutions per site.
Comparative single protein analysis.
Genomes were clustered by gene content by constructing a matrix of pairwise distances between bacterial proteomes (results are in Fig. S1 in the supplemental material). Pairwise distances were estimated by using the following procedure. Using NCBI BlastP, each protein in genome A was compared to the proteome of genome B. The similarity of the top hit in genome B was noted for each protein in genome A. All such A-to-B comparisons were summarized by calculating the percentage of the proteins in A which had a match in B of at least a certain similarity. That is, a table was created showing what fraction of proteome A had matches of at least 100% identity in B, what fraction had matches of at least 99% identity, what fraction had 98%, and so on. If A and B were the same proteome, this table would contain values of 1.0 for all percentages of identity from 100% to 1%. A histogram was generated for each genomic comparison, and the area above the histogram was measured. This represents the sum of the differences between the actual fractions observed and those which would arise from having identical proteomes; this is a distance measure. The proteomic comparison was repeated for all possible pairwise comparisons. To generate the actual distances used in the phylogeny reconstruction, we compared the pairs of organisms in both directions (A→B and B→A) and averaged the histogram areas.
A dendrogram illustrating how the genomes cluster (and their relative distances) with this scheme can be easily derived from the matrix of pairwise distances by using the neighbor-joining method implemented in PHYLIP (15). This proteomic comparison method also appears robust with respect to highly divergent and even largely disjoint protein sets: the archaeon Pyrococcus furiosus branches deeply from the genus Gammaproteobacteria, while the small genome of Buchnera aphidicola, for example, clusters with the genus Wigglesworthia (data not shown). In spirit, this clustering method is similar to the more rigorous average amino acid identity measure proposed by Konstantinidis and Tiedje (31); like theirs, our method shows that entire-proteome comparisons largely recapitulate standard 16S rRNA phylogeny and yet provide insights into the correlation of genome and ecological role, as well as highlighting possible horizontal gene transfer.
Nucleotide sequence accession numbers.
The annotated genome sequences of both A. vitis S4 (GenBank CP000633 through CP000639) and A. radiobacter K84 (GenBank CP000628 through CP000632) are available from GenBank and from the Agrobacterium genome database (http://agro.vbi.vt.edu/public).
RESULTS
Sequencing and annotation of representative genomes from Agrobacterium biovars II and III.
Representative genomes from all three Agrobacterium biovars are now available. The genome sequence of the biovar I strain A. tumefaciens C58 (C58) was sequenced by our group and has been recently revised and updated (19, 47; S. Slater, J. C. Setubal, B. Goodner, Y. Zhou, K. Houmiel, J. Sun, B. S. Goldman, S. K. Farrand, W. M. Huang, S. Casjens, R. Kaul, Q. Chen, T. Burr, E. Nester, R. Kadoi, T. Ostheimer, N. Nicole Pride, A. Allison Sabo, E. Erin Henry, E. Erin Telepak, L. Lindsey Wilson, A. Alana Harkleroad, and D. Wood, submitted for publication). The genome sequences for representatives of the two remaining biovars are available as indicated in Materials and Methods.
Table 1 compares the general features of C58, K84, and S4, and Tables S1 to S3 in the supplemental material provide a more detailed picture of each genome. The three sequenced Agrobacterium biovars have distinct genome architectures. The genomes of C58 and S4 contain two true chromosomes, which we define as replicons containing both rRNA operons and genes essential for prototrophic growth. C58, however, has one circular and one linear chromosome (19, 47), while S4 has two circular chromosomes. In both strains, the larger chromosome (chromosome I) contains an origin of replication that is similar to other chromosomal origins within the Alphaproteobacteria (24), while chromosome II has a repABC origin of replication typical of the large plasmids within the Rhizobiaceae. C58 contains two plasmids, pTiC58 and pAtC58 (Table 1; see Table S1 in the supplemental material) (19, 47), whereas S4 has five plasmids (Table 1; see Table S2 in the supplemental material). K84, in contrast, has a single circular chromosome, a second 2.65-Mbp replicon, and three plasmids (Table 1; see Table S3 in the supplemental material): pAgK84 (44 kbp) (30), pAtK84b (185 kbp, pNOC) (7), and pAtK84c (388 kbp, pAgK434) (9). Like the second chromosomes of C58 and S4, the 2.65-Mbp replicon contains a plasmid-type repABC origin. However, it lacks the rRNA operons and does not contain the extensive sets of essential metabolic genes found on the second chromosomes of C58 and S4. It does contain at least one gene that is likely to be essential, an l-seryl-tRNA selenium transferase gene (Arad7947).
TABLE 1.
Summary of genome features from sequenced Agrobacterium strains
| Characteristic | A. tumefaciens C58 | A. radiobacter K84 | A. vitis S4 |
|---|---|---|---|
| Biovariant | Biovar I | Biovar II | Biovar III |
| Genome size (bp) | 5,674,260 | 7,273,300 | 6,320,946 |
| % G+C content | 59.0 | 59.9 | 57.5 |
| No. of chromosomesa | 2 | 1 | 2 |
| No. of plasmids | 2 | 4b | 5 |
| No. of indicated protein-coding genes | |||
| Total | 5,385 | 6,752 | 5,479 |
| With functionality assigned | 3,516 | 5,099 | 3,897 |
| Conserved hypothetical | 1,287 | 1,201 | 1,282 |
| Hypothetical | 582 | 452 | 300 |
| Pseudogenes | 28 | 68 | 90 |
| No. of indicated RNA genes | |||
| rRNA operons | 4 | 3 | 4 |
| tRNAs | 56 | 51 | 54 |
| Other RNAsc | 26 | 23 | 30 |
| Genomic islands | |||
| Total no. | 38 | 59 | 20 |
| Avg size (kb) | 23.3 | 28.2 | 33.0 |
A chromosome is defined here as a replicon harboring rRNA operons and essential genes.
The 2.65-Mbp replicon, which does not meet our definition of a chromosome, is included here (see Results).
Includes transfer-messenger RNAs, signal recognition particle RNAs, suhB, riboswitches, and miscellaneous features.
Multiprotein phylogeny of new genomes shows Agrobacterium organisms to be paraphyletic.
The relationships among C58, S4, and K84 and 16 previously sequenced genomes in the order Rhizobiales were investigated by maximum likelihood phylogenetic analysis. Protein alignments were performed for 507 single-copy orthologous gene families located on primary chromosomes that are likely to have tracked the vertical component of ancestry (Fig. 1; see Tables S4 and S5 in the supplemental material). Analysis of the concatenated data set produces a single topology with 100% a posteriori support for all branches within the Rhizobiaceae, which is consistent with the results of Williams et al. (45). This phylogenetic reconstruction finds S4 to group with C58 and K84 to group with two Rhizobium genomes (R. leguminosarum and R. etli). The lineage uniting K84 with the genus Rhizobium has a substantial branch length, while S4 and C58 appear to have separated soon after the divergence of the genus Sinorhizobium.
Whole-genome similarity plots support these findings (see Fig. S1 in the supplemental material). The neighbor-joining tree of the distances measured from these plots gives the same topology and similar relative branch lengths within the Rhizobiaceae as the maximum likelihood tree analysis (see Fig. S2 in the supplemental material). These large-scale investigations provide a well-defined phylogenetic basis for uniting biovar II (represented by K84) with the genus Rhizobium.
RepABC replication origins are not linearly descendant among secondary chromosomes and large plasmids.
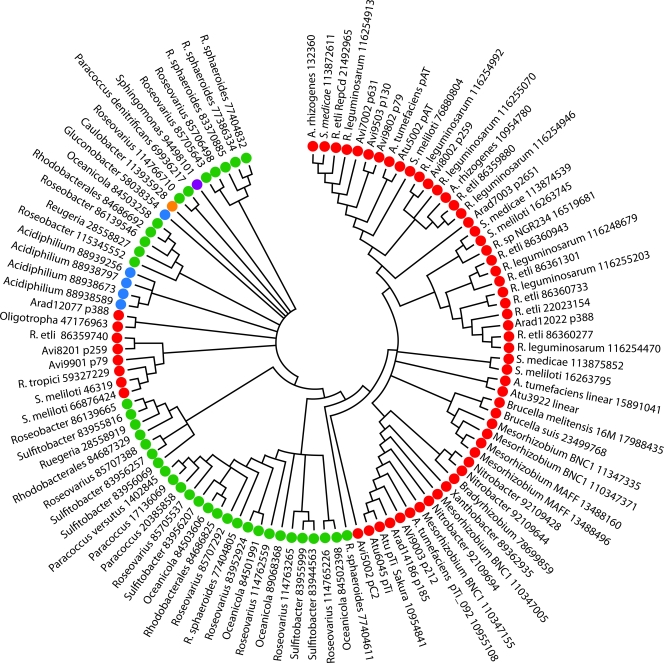
Plasmid replication among the Rhizobiaceae is generally under the control of the RepABC system (5, 44). Plasmid origins of replication are typically considered characteristic of a plasmid, since replication is required for transmission. Thus, we would predict that the repABC genes, which are generally found in an operon, evolved as a single unit on the plasmids and second chromosomes for which they mediate replication. Phylogenetic analyses of these gene lineages, however, indicate a lack of evolutionary congruence with the species tree (Fig. 1) among the repABC systems of plasmids and of the second-largest replicons of the three biovars (see Fig. S5 to S6 in the supplemental material). Therefore, one cannot infer an ancestry for repABC genes that does not invoke continuous horizontal gene transfer of these genes. Individual repABC genes show a similar lack of evolutionary congruence within replicons (the RepA and RepB trees, while congruent to each other, are not congruent to the RepC tree [see Fig. S7 and S8 in the supplemental material]), suggesting that plasmid evolution is mediated both by the frequent movement of plasmids among strains and by exchange of the individual repABC genes within replicons. In a wider evolutionary perspective, congruence among repABC genes generally does hold. For example, even though the repC genes appear to move easily within families, they move less easily within orders and rarely outside of an order (Fig. 2). These findings are consistent with recent work by Cevallos et al. (5) and confirm that the intragenomic movement of genes across replicons includes the replication systems.
FIG. 2.
Phylogenetic analysis of RepC proteins among the Rhizobiaceae. The organism name is followed by the NCBI accession number. Red indicates membership in the Rhizobiales, purple in the Sphingomonadales, blue in the Rhodospirillales, green in the Rhodobacterales, and orange in the Caulobacterales. Arad, A. radiobacter; Avi, A. vitis; Atu, A. tumefaciens; A., Agrobacterium; S., Sinorhizobium; R. etli, Rhizobium etli; R. leguminosarum, Rhizobium leguminosarum; R. sphaeroides, Rhodabacter sphaeroides.
Conservation of gene content and order is much greater on primary chromosomes than on secondary chromosomes.
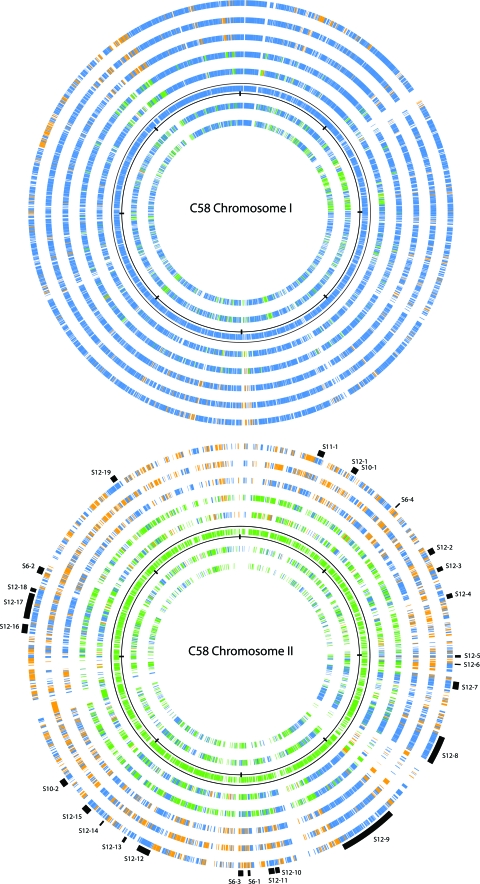
The C58 chromosome I shares large-scale synteny with the chromosome of Sinorhizobium meliloti 1021 and with the chromosome of the more distantly related Mesorhizobium loti MAFF303099 (19, 47). Subsequent analyses show conservation of gene order and content among primary ancestral chromosomes of other Rhizobiales (Brucella, Bradyrhizobium, Mesorhizobium, and Rhizobium strains, Ochrobactrum anthropi, and Azorhizobium caulinodans) (37, 40). Given these relationships, we might expect the secondary chromosomes and large replicons within the genus Agrobacterium and across the order Rhizobiales to display similar syntenic relationships. Although some conservation of gene content is apparent, these replicons lack the large-scale conservation of gene order seen among the primary chromosomes (Fig. 3). Where gene order has been retained, it is limited to small blocks of genes. These contrasting findings led us to examine the origins of the large secondary replicons.
FIG. 3.
Gene conservation among replicons of the Rhizobiales. Graphic depicts ortholog gene alignments shown from the outer circle and moving inward as follows (GenBank accession numbers are in parentheses): Sinorhizobium meliloti 1021 (NC_003047.1), Rhizobium leguminosarum biovar viciae 3841 (NC_008380.1), Rhizobium etli CFN42 (NC_007761.1), K84, S4, C58, Ochrobactrum anthropi ATCC 49188 (NC_009668.1), and Brucella suis 1330 (NC_004310.3). Top, the alignment is anchored by C58 chromosome I; bottom, the alignment is anchored by C58 chromosome II. The anchor replicons themselves are represented by the circles bordered by scales with marks every one-eighth of their total size. Each gene is colored according to its replicon of origin: blue for the primary chromosome, green for secondary chromosomes (including the K84 2.65-Mbp replicon), and orange for plasmids. Note that in all circles except the anchor, the location of a gene in the figure is not tied to physical position in that genome. At higher resolution (http://agro.vbi.vt.edu/public), it is possible to see that many genes in the nonanchor circles occur consecutively in their respective replicons, thus representing syntenic blocks or clusters. The positions of clusters that occur in C58 are listed in Tables S9 and S13 to S15 in the supplemental material and are indicated in the figure by the outermost-arc sections colored black. Each such arc is labeled as Sx-y, where x is the number of the table in the supplemental material and y is the order of the cluster in the table. The alignment in the top panel is predominantly blue, suggesting the high degree of conservation among Rhizobiales primary chromosomes. The alignment in the bottom panel is a mixture of blue, green, and orange, suggesting the mosaic nature of chromosome II and hinting at the various genomic transfers hypothesized to have taken place, as explained in the text.
Secondary chromosomes originated from intragenomic transfers from primary chromosomes to ancestral plasmids.
In spite of the lack of large-scale synteny across the secondary chromosomes and large replicons of the Rhizobiales, evidence supports a common origin for chromosome II of C58 and S4 and the 2.65-Mbp replicon of K84. Of the 3,382 genes shared by all three genomes, 291 are located on chromosome II of C58 and S4 and on the 2.65-Mbp replicon of K84. This represents 16%, 27%, and 12% of the genes on each of the respective DNA molecules (http://agro.vbi.vt.edu/public). In addition, six gene clusters are shared by chromosome II of C58 and S4, by the 2.65-Mbp replicon of K84, and by plasmids p42e of R. etli and pRL11 of R. leguminosarum (Fig. 3; see Tables S6 and S10 in the supplemental material).
Comparisons among the Rhizobiales suggest that gene transfer from primary chromosomes to ancestral plasmids resulted in secondary chromosomes. Because these transfers occur within the same genome (and can potentially occur between any pair of replicons), we term them “intragenomic gene transfers.” Under this model, translocated genes would be expected to occur in clusters that retain synteny with the ancestral chromosome, and this is clearly observed (Fig. 3). All fully sequenced genomes of the Brucella/Ochrobactrum clade (five sequenced strains), two members of the genus Sinorhizobium, and the mixed Agrobacterium/Rhizobium clade (five sequenced strains) possess multiple chromosomes or a large replicon with some chromosomal characteristics. Moreover, except for the genus Brucella, all these members carry one or more plasmids.
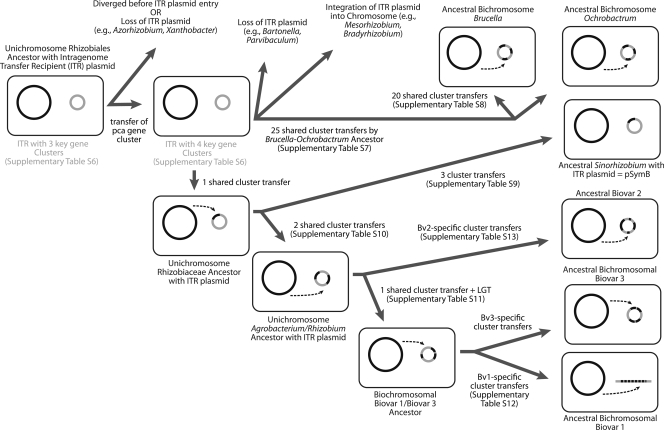
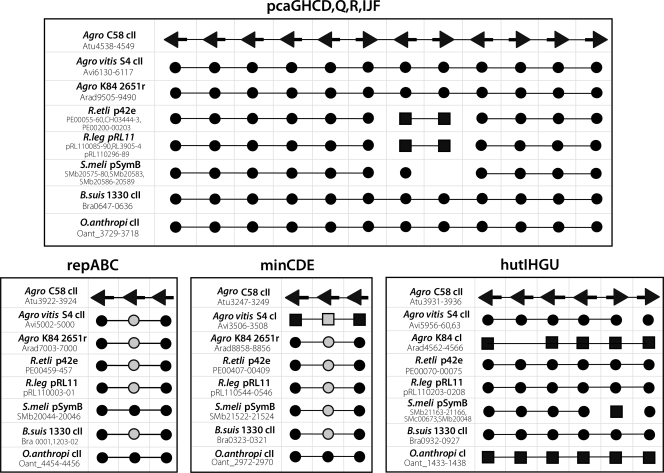
All fully sequenced Rhizobiales species that harbor multiple replicons have at least one RepABC replicon. We suggest that the common ancestor of this order was a unichromosomal strain that acquired a single ancestral plasmid of this class, here referred to as the Intragenomic Translocation Recipient (ITR) (Fig. 4). The best evidence for the existence of this ancestral plasmid is three gene clusters shared by almost all fully sequenced Rhizobiales (in addition to repABC). As shown in Fig. 5 and Table S6 in the supplemental material, in 29 out of 32 cases these four clusters are found in secondary large replicons. The three exceptions (A. vitis [minCDE], O. anthropi [hutIHGU], and A. radiobacter [hutIHGU]) can be explained by subsequent retrotransfers to the primary chromosome from the ITR, based on analysis of adjacent syntenic regions shared with chromosome II of their nearest sequenced relatives. Moreover, three of these clusters (minCDE, hutIHGU, and repABC) are not seen in the unichromosomal genome of Azorhizobium caulinodans, a Rhizobiales member, suggesting that the ITR plasmid brought those genes to the ancestral strain and that the fourth gene cluster (pca) later moved from the ancestral chromosome to the ITR plasmid.
FIG. 4.
Reconstruction of the origin of secondary chromosomes and related large replicons within the Rhizobiales through transfers of gene clusters from the primordial chromosome to what originally was a repABC-type plasmid (called here the ITR plasmid). LGT, lateral gene transfer.
FIG. 5.
Key gene clusters present on ITR plasmid progenitor of chromosome II and related large replicons during evolution of Rhizobiales. C58 is the reference, and its genes are represented as arrows consistent with the strand they are found on in the deposited genome sequence. Genes for the other genomes were aligned with the C58 genes and are represented with circles or squares. Circles/squares are connected with lines when corresponding genes are consecutive. A black or gray circle means that the gene represented is in a secondary chromosome or plasmid; a black or gray square means that the gene represented is in the primary chromosome. A black circle or square means that the alignment to the C58 ortholog covered 80% or more of both genes; a gray circle or square means the alignment covered less than 80%. Gene numbering is shown for C58 (Agro C58), S4 (Agro vitis S4), K84 (Agro K84), R. etli CFN42, R. leguminosarum biovar viciae 3841 (R.leg), S. meliloti 1021 (S.meli), B. suis 1330, and O. anthropi ATCC49188.
At some point the Brucella/Ochrobactrum clade diverged from the lineage that gave rise to the family Rhizobiaceae (Fig. 1). The transfer of chromosomal genes to the ITR plasmid took place independently in the Brucella/Ochrobactrum clade (also hypothesized in reference 36) and in the Rhizobiaceae family. In the Brucella/Ochrobactrum clade, there have been 25 intragenomic transfers from the primary chromosome to the ITR plasmid, as shown by the facts that these 25 clusters are shared by all of the sequenced members of the Brucella/Ochrobactrum clade (see Table S7 in the supplemental material) and that these clusters are still found in the primary chromosome of S. meliloti. Twenty more transfers occurred since Brucella diverged away from Ochrobactrum (see Table S8 in the supplemental material). In fact, the recent sequencing of the genome of Brucella suis ATCC 23445 (NC_010169.1) shows that another 220-kbp section, found in chromosome I for all other fully sequenced members of the genus Brucella, is now part of its chromosome II (A. R. Wattam, K. P. Williams, E. E. Snyder, N. F. Almeida, Jr., M. Shukla, A. W. Dickerman, O. R. Crasta, R. Kenyon, J. Lu, J. M. Shallom, H. Yoo, T. A. Ficht, R. M. Tsolis, C. Munk, R. Tapia, C. S. Han, J. C. Detter, D. Bruce, T. S. Brettin, B. W. Sobral, S. M. Boyle, and J. C. Setubal, submitted for publication). In Sinorhizobium meliloti, the ancestral ITR plasmid evolved into the pSymB plasmid, with one intragenomic transfer event from the chromosome to the ITR plasmid occurring prior to its divergence from the Agrobacterium/Rhizobium clade and three events after (see Table S9 in the supplemental material).
Among the Rhizobiaceae, at least two gene clusters transferred to the ancestral ITR plasmid prior to the divergence of the clade that includes the biovar I/III strains from the biovar II clade that includes K84, Rhizobium etli CFN42, and R. leguminosarum biovar viciae 3841. These transfers include a cluster containing genes encoding a glutamate synthase and glutamine synthetase III (Fig. 3, bottom panel; see Table S10 in the supplemental material). After this divergence, there was at least one intragenomic transfer to the ITR plasmid before it became chromosome II for Agrobacterium biovar I/III strains (Fig. 3, bottom panel; see Table S11 in the supplemental material). Subsequently, transfers to chromosome II have occurred that are unique to biovars I or III (19). For example, there have been at least seven large-scale gene transfer events, ranging from 10 kbp to 220 kbp, and a few smaller transfer events between the ancestral chromosome and chromosome II of C58 that did not occur in S4 (Fig. 3, bottom panel; see Table S12 in the supplemental material). In a separate but parallel track, there was at least one intragenomic transfer to the ITR plasmid ancestral to K84 (2.65-Mbp replicon), R. etli (plasmid p42e), and R. leguminosarum (plasmid pRL11) (see Table S13 in the supplemental material). None of the secondary replicons in this branch has reached chromosome status yet.
We observe that among Rhizobiales, another evolutionary path seems to be that of integration of the ancestral ITR plasmid into the main chromosome. The best example of this path is Bradyrhizobium strains. All fully sequenced Bradyrhizobium strains have very large chromosomes (B. japonicum USDA 110 has a single chromosome larger than 9 Mbp [29]), and only one strain (Bradyrhizobium sp. strain BTAi1) has a plasmid that might serve to nucleate a second chromosome. However, the presence of ITR plasmid gene clusters and other plasmid genes in the chromosomes of these species (also seen in Mesorhizobium main chromosomes) suggests the integration of one or more plasmids into the ancestral chromosome (Fig. 4).
Intragenomic flow from chromosomes to large plasmids mediates second-chromosome formation in other bacteria.
A plasmid-based mechanism of secondary chromosome formation was first proposed with the genome sequence of the two chromosomes of Vibrio cholerae, based solely on the presence of plasmid replication functions (12). The extensive data for the Rhizobiales just described goes well beyond just replication functions, and we now provide evidence for two more examples of extensive intragenomic gene transfer to a new chromosome based on published genome sequences.
First, among the Gammaproteobacteria, the example of the genus Vibrio is much older and more complex than first proposed. Strains of Photobacterium were once considered to be within the genus Vibrio, and multiple lines of evidence support Vibrio and Photobacterium as sister genera. Both genera have two chromosomes, and sequences are available for P. profundum and four Vibrio species. Phylogenetic analysis of several conserved proteins showed that among the available sequenced genomes, Aeromonas hydrophila is the closest relative with a single chromosome. Comparative analyses support 6 gene cluster transfers from the ancestral chromosome I to the plasmid progenitor of chromosome II (itself defined by 7 unique gene clusters) prior to the divergence of the sister genera Photobacterium and Vibrio, 7 additional gene cluster transfers to chromosome II of the common ancestor of all the sequenced Vibrio strains, and 29 transfers unique to the Photobacterium side (see Fig. S3 and Tables S14 to S17 in the supplemental material).
Second, in the Betaproteobacteria, the genus Burkholderia was subdivided several years ago, with some members of Burkholderia along with some stragglers from other genera reclassified into the genus Ralstonia. Several lines of evidence support a very close relationship between Burkholderia and Ralstonia, and they each consist of species with two or three chromosomes. The most closely related sequenced genomes with a single chromosome are those from the genus Bordetella; B. bronchiseptica was used as the comparison genome for this analysis. Using chromosome II sequences from five different Burkholderia species and three different Ralstonia species for analysis, the second chromosomes of Burkholderia and Ralstonia are seen to share a common origin, with 11 gene cluster transfers from the ancestral chromosome to a plasmid progenitor (defined by two unique gene clusters) (see Fig. S4 and Tables S18 and S19 in the supplemental material). After the divergence of these two clades, 12 additional transfers to chromosome II are unique to the Burkholderia bichromosome ancestor and 24 to the Ralstonia bichromosome ancestor (see Fig. S4 and Tables S20 and S21 in the supplemental material). Within a subset of Burkholderia strains, there is a third plasmid-based chromosome to which four gene clusters were transferred from either chromosome I or chromosome II (see Fig. S4 and Table S22 in the supplemental material). Taken together, these data support a generalized mechanism of secondary chromosome formation among bacteria.
DISCUSSION
Within the Rhizobiaceae, the available evidence strongly supports a mixed Agrobacterium/Rhizobium clade containing two subclades. One subclade includes the biovar II agrobacteria (e.g., K84) and certain of the fast-growing rhizobia, including R. etli and R. leguminosarum. The second subclade includes the biovar I (e.g., C58) and III (e.g., S4) lineages that separated after diverging from the biovar II lineage. Linearization of the biovar I chromosome appears to have been a seminal event in this radiation (S. Slater et al., submitted).
Analysis of complete genome sequences within the Rhizobiales allows a more precise definition of phylogenetic relationships. While it has long been known that gene transfer can occur between organisms, the picture that results from our study shows a group characterized by composite genomes in which genes of all classes are not only migrating between organisms (19, 47) but also intracellularly among chromosomal and plasmid replicons. In the Rhizobiaceae, such movements, as well as chromosomal rearrangements, have not completely disrupted the backbone of the ancestral chromosome. In contrast, while second chromosomes and evolving plasmid-based large replicons have some overlapping gene content, they display significant loss of gene order. In biovar I and III agrobacteria, these movements produced second chromosomes derived from plasmids, while in the biovar II strain K84, the plasmid-based replicon has yet to reach second-chromosome status.
Although it is clear that the 2.65-Mbp replicon of K84, second chromosomes of C58 and S4, and large plasmids in other members of the Rhizobiales have evolved from a common plasmid ancestor, the repABC genes involved in replication initiation, copy number control, and partition on these molecules are phylogenetically distinct even within a single organism. These findings show that repABC genes, like other genes, are being exchanged among replicons. This may reflect selective pressure to move from incompatibility to coexistence in genomes with multiple repABC-based replicons. It also means there is no internal standard by which to directly compare replicon lineages among these plasmids.
Our data show a common mechanism of secondary chromosome formation in Rhizobiaceae and other bacteria. A prerequisite for this evolution is the intracellular presence of a second replicon capable of stably and efficiently replicating large DNA molecules. The repABC-type replicons that are widely distributed among the Rhizobiales fall into this class and have produced second chromosomes in addition to large replicons, such as the 2.65-Mbp K84 replicon and the Sym plasmids of nitrogen-fixing members of the Rhizobiaceae (6, 8, 16, 18, 19, 21, 37, 47, 49). In A. tumefaciens, it has been shown that chromosome II is replicated concurrently with chromosome I; such overall genome synchrony probably allowed intragenomic transfers to be maintained (27, 28). Most of the large gene movements have been from the ancestral chromosome to plasmid replicons, with only rare retrotransfers. While plasmids can undergo large gene rearrangements and losses/insertions, the available evidence suggests that there are some constraints to large-scale rearrangements of the bacterial chromosome (23, 34, 39).
The advantage of multiple chromosomes is unclear, but we speculate that they may permit further accumulation of genes when the primary replicon cannot support further chromosome enlargement. Within the Rhizobiaceae, different species appear to handle gene accumulation in different ways. Bradyrhizobium and Mesorhizobium species have very large chromosomes with few, if any, relatively small plasmids. In contrast, Agrobacterium and Rhizobium strains have multiple chromosomes or large replicons that show gene accumulation, as well as anywhere from one to six plasmids. These differences may suggest that chromosomal origins have differing abilities to replicate molecules larger than about 5 or 6 Mbp, with multiple chromosomes providing an alternative reservoir for newly acquired DNA. Alternatively, the initial movement of a few essential gene clusters to a plasmid replicon may be simply a historical contingency with no attached selective advantage. Additional essential gene transfers would simply solidify the essential nature of the new replicon. An evaluation of the selective-advantage hypothesis is needed, but regardless of the reason, it is clear that the genetic organization of even essential genes in bacteria is much more complex and fluid than has been imagined.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grants 0333297 and 0603491 to E.W.N. and 0736671 to S.C.S., grants from the M. J. Murdock Charitable Trust Life Sciences program (2004262:JVZ and 2006245:JVZ) to D.W.W., by a science education grant from the Howard Hughes Medical Institute to B.G. (52005125), by a Conselho Nacional de Desenvolvimento Científico e Tecnológico fellowship to N.F.A. (no. 200447/2007-6), and by the Monsanto Company.
Special thanks to the more than 450 undergraduate students at Hiram College, Oregon State University, Seattle Pacific University, Arizona State University, the University of North Carolina, Washington University in St. Louis, and Williams College who contributed to the deep annotation of all three Agrobacterium genomes between 2004 and 2008.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burr, T. J., C. Bazzi, S. Sule, and L. Otten. 1998. Crown gall of grape: biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis. 821288-1297. [DOI] [PubMed] [Google Scholar]
- 3.Burr, T. J., A. L. Bishop, B. H. Katz, L. M. Blanchard, and C. Bazzi. 1987. A root-specific decay of grapevine caused by Agrobacterium tumefaciens and A. radiobacter biovar 3. Phytopathology 771424-1427. [Google Scholar]
- 4.Burr, T. J., and L. Otten. 1999. Crown gall of grape: biology and disease management. Annu. Rev. Phytopathol. 3753-80. [DOI] [PubMed] [Google Scholar]
- 5.Cevallos, M. A., R. Cervantes-Rivera, and R. M. Gutierrez-Rios. 2008. The repABC plasmid family. Plasmid 6019-37. [DOI] [PubMed] [Google Scholar]
- 6.Chain, P. S., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect. Immun. 738353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clare, B. G., A. Kerr, and D. A. Jones. 1990. Characteristics of the nopaline catabolic plasmid in Agrobacterium strains K84 and K1026 used for biological control of crown gall disease. Plasmid 23126-137. [DOI] [PubMed] [Google Scholar]
- 8.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donner, S. C., D. A. Jones, N. C. McClure, G. M. Rosewarne, M. E. Tate, A. Kerr, N. N. Fajardo, and B. G. Clare. 1993. Agrocin 434, a new plasmid encoded agrocin from the biocontrol Agrobacterium strains K84 and K1026, which inhibits biovar 2 agrobacteria. Physiol. Mol. Plant Pathol. 42185-194. [Google Scholar]
- 10.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14755-763. [DOI] [PubMed] [Google Scholar]
- 11.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan, E. S., M. A. Fogel, and M. K. Waldor. 2005. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol. Microbiol. 561129-1138. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1989. PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 16.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F.-J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 17.Ghai, R., and T. Chakraborty. 2007. Comparative microbial genome visualization using GenomeViz. Methods Mol. Biol. 39597-108. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 1033834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. W. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 2942323-2328. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 21.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herlache, T. C., H. S. Zhang, C. L. Ried, S. A. Carle, P. Basaran, M. Thaker, A. T. Burr, and T. J. Burr. 2001. Mutations that affect Agrobacterium vitis-induced grape necrosis also alter its ability to cause a hypersensitive response on tobacco. Phytopathology 91966-972. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, D. 2000. Evaluating genome dynamics: the constraints on rearrangements within bacterial genomes. Genome Biol. 1REVIEWS0006.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis, P., J. C. Hotopp, P. Sapountzis, S. Siozios, G. Tsiamis, S. R. Bordenstein, L. Baldo, J. H. Werren, and K. Bourtzis. 2007. New criteria for selecting the origin of DNA replication in Wolbachia and closely related bacteria. BMC Genomics 8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. A., M. H. Ryder, B. G. Clare, S. K. Farrand, and A. Kerr. 1991. Biological control of crown gall using Agrobacterium strains K84 and K1026, p. 161-170. In H. Komada, K. Kiritani, and J. Bay-Peterson (ed.), The biological control of plant diseases. Food and Fertilizer Technology Center for the Asian and Pacific Region, Taipei, Taiwan.
- 26.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 1802749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 1833065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahng, L. S., and L. Shapiro. 2003. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J. Bacteriol. 1853384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. G., B. K. Park, S. U. Kim, D. Choi, B. H. Nahm, J. S. Moon, J. S. Reader, S. K. Farrand, and I. Hwang. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. USA 1038846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis, K. T., and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 1876258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 132178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz, V. M., E. Szeto, K. Palaniappan, Y. Grechkin, K. Chu, I. M. Chen, I. Dubchak, I. Anderson, A. Lykidis, K. Mavromatis, N. N. Ivanova, and N. C. Kyrpides. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36D528-D533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miesel, L., A. Segall, and J. R. Roth. 1994. Construction of chromosomal rearrangements in Salmonella by transduction: inversions of non-permissive segments are not lethal. Genetics 137919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, L. W., and G. Warren. 1979. Agrobacterium radiobacter strain K84 and biological control of crown gall. Annu. Rev. Phytopathol. 17163-179. [Google Scholar]
- 36.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90209-227. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha, E. P. 2006. Inference and analysis of the relative stability of bacterial chromosomes. Mol. Biol. Evol. 23513-522. [DOI] [PubMed] [Google Scholar]
- 40.Setubal, J. C., D. Wood, T. Burr, S. Farrand, B. Goldman, B. Goodner, L. Otten, and S. Slater. 2009. The genomics of Agrobacterium: insights into pathogenicity, biocontrol, and evolution., p. 91-112. In R. Jackson (ed.), Plant pathogenic bacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 41.Stamatakis, A., T. Ludwig, and H. Meier. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21456-463. [DOI] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Tian, Y., and A. W. Dickerman. 2007. GeneTrees: a phylogenomics resource for prokaryotes. Nucleic Acids Res. 35D328-D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weaver, K. E. 2007. Emerging plasmid-encoded antisense RNA regulated systems. Curr. Opin. Microbiol. 10110-116. [DOI] [PubMed] [Google Scholar]
- 45.Williams, K. P., B. W. Sobral, and A. W. Dickerman. 2007. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 1894578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong, K., and G. B. Golding. 2003. A phylogenetic analysis of the pSymB replicon from the Sinorhizobium meliloti genome reveals a complex evolutionary history. Can. J. Microbiol. 49269-280. [DOI] [PubMed] [Google Scholar]
- 47.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, L. Woo, Y. C. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. N. Wu, P. Romero, D. Gordon, S. P. Zhang, H. Y. Yoo, Y. M. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
- 48.Young, J. M. 2008. Agrobacterium: taxonomy of plant-pathogenic Rhizobium species, p. 184-220. In T. Tzfira and V. Citovsky (ed.), Agrobacterium: from biology to biotechnology. Springer, New York, NY.
- 49.Young, J. P., L. C. Crossman, A. W. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.