Abstract
Aeromonas caviae Sch3N possesses a small genomic island that is involved in both flagellin glycosylation and lipopolysaccharide (LPS) O-antigen biosynthesis. This island appears to have been laterally acquired as it is flanked by insertion element-like sequences and has a much lower G+C content than the average aeromonad G+C content. Most of the gene products encoded by the island are orthologues of proteins that have been shown to be involved in pseudaminic acid biosynthesis and flagellin glycosylation in both Campylobacter jejuni and Helicobacter pylori. Two of the genes, lst and lsg, are LPS specific as mutation of them results in the loss of only a band for the LPS O-antigen. Lsg encodes a putative Wzx flippase, and mutation of Lsg affects only LPS; this finding supports the notion that flagellin glycosylation occurs within the cell before the flagellins are exported and assembled and not at the surface once the sugar has been exported. The proteins encoded by flmA, flmB, neuA, flmD, and neuB are thought to make up a pseudaminic acid biosynthetic pathway, and mutation of any of these genes resulted in the loss of motility, flagellar expression, and a band for the LPS O-antigen. Furthermore, pseudaminic acid was shown to be present on both flagellin subunits that make up the polar flagellum filament, to be present in the LPS O-antigen of the A. caviae wild-type strain, and to be absent from the A. caviae flmD mutant strain.
Mesophilic Aeromonas strains are being increasingly recognized as important bacterial pathogens. They are widely distributed in the environment and cause gastrointestinal and wound infections in healthy humans and, less commonly, septicemia in immunocompromised patients (15). In particular, Aeromonas caviae is reported to be the most prevalent pediatric enteropathogenic species of the genus (30, 46). A range of putative virulence factors have been described for the aeromonads, from the hemolytic toxin aerolysin and cytotonic toxins to capsules and extracellular enzymes (44). The process of adherence of aeromonads is still poorly understood, although a number of factors have been implicated, such as long wavy pili, outer membrane proteins, lipopolysaccharide (LPS) O-antigen, and the polar flagellum (1, 44). The mesophilic aeromonads are interesting as most strains express two distinct flagellum systems (10, 34). They have a polar flagellum for swimming in liquid and express separate lateral flagella for swarming over surfaces. Investigations have revealed that both the polar and lateral flagellum systems of the mesophilic aeromonads are involved in adherence to both biotic and abiotic surfaces (20).
Previously, we showed that transposon mutations in the flm locus of A. caviae greatly reduced adherence of this organism to the human epithelial cell line HEp-2. In addition, mutation of this locus caused losses of motility, flagella, and the LPS O-antigen (12). In A. caviae Sch3N the flmA and flmB genes were clustered together in a locus with neuA, flmD, and neuB. All of these genes appeared to encode proteins associated with polysaccharide biosynthesis. Only flmA and flmB were found in the other Aeromonas strains investigated, such as Aeromonas hydrophila AH-3 (12). However, recently, the flmD, neuB, and neuA genes have been reported to be present in A. hydrophila AH-3 (4). Mutation of the A. hydrophila AH-3 flmA, flmB, flmD, neuA, and neuB genes affected only motility and flagellar expression and did not cause any alteration in the LPS (4, 12). This suggested that the flm gene products in A. caviae have two roles, a role in the biosynthesis of the LPS O-antigen and a role in flagellar assembly.
The orthologues of the flm genes were originally reported to be involved in flagellar assembly in Caulobacter crescentus, possibly through glycosylation of the flagellin or other flagellar proteins (23). Moreover, as observed for A. hydrophila AH-3, mutation of the C. crescentus flm genes did not result in any LPS defects. More recently, there have been a number of reports describing related loci in several bacterial pathogens that have associated the orthologous gene products with glycosylation of proteins important in pathogenesis, such as flagella, pili, and adhesins (33). There are similar genetic localizations and arrangements of these glycosylation genes in several bacterial species, such as Campylobacter jejuni, Helicobacter pylori, and C. crescentus (16, 33). A number of the related glycosylation gene products have been shown to play a role in the biosynthesis of a nine-carbon sugar related to sialic acid (Neu5Ac) called pseudaminic acid (Pse5Ac7Ac) that is present on the flagellins of C. jejuni and H. pylori (11, 37, 43). Our laboratory and other laboratories have also shown that both the polar flagellins of A. caviae are glycosylated with Pse5Ac7Ac. It has been suggested that the flm gene cluster plays a role in this process (10, 33, 34).
In this study, we describe a genetic island in A. caviae that contains the genes required for biosynthesis of the sugar Pse5Ac7Ac, which has been shown to be present both in the LPS O-antigen and on the polar flagellins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) Miller broth and on LB Miller agar, while Aeromonas strains were grown in tryptic soy broth, on tryptic soy agar, or in brain heart infusion broth (BHIB) (Oxoid). Ampicillin (50 μg/ml), nalidixic acid (50 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (25 μg/ml) were added to the different media when necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Reference or source |
|---|---|---|
| Aeromonas caviae strains | ||
| Sch3N | Sch3, spontaneous Nalr | 12 |
| IAG75 | Sch3N flmD::mini-Tn5Cm, Nalr Cmr | 12 |
| AAR269 | flaA::Kmr | 34 |
| AAR27 | flaB::Kmr | 34 |
| SMT50 | rmlB::Kmr | This study |
| SMT137 | flmA::Kmr | This study |
| SMT40 | flmB::Kmr | This study |
| SMT62 | neuA::Kmr | This study |
| SMT15 | flmD::Kmr | This study |
| SMT166 | neuB::Kmr | This study |
| SMT18 | lsg::Kmr | This study |
| SMT145 | lst::Kmr | This study |
| Escherichia coli strains | ||
| CC118λpir | Δ(ara leu)7697 araD139 ΔlacX74 galE galK phoA20 thi-1 rspE rpoB(Rfr) argE(Am) recA1 λpir+ | 14 |
| S17-1λpir | hsdR pro recA, RP4-2 in chromosome, Km::Tn7 (Tc::Mu) λpir, Tpr Smr | 28 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac [F′ proAB, lacIqZΔM15 Tn10(Tcr)] | Stratagene |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector, MCS, Ampr | Gibco BRL |
| pUC4-KIXX | Source of Tn5-derived nptII gene (Kmr) | Pharmacia |
| pBBR1MCS | Broad-host-range vector, IncP, -W, -Q, ColE1 and p15A compatible, contains pBluescript IIKS-lacZα-polylinker, Cmr | 22 |
| pBBR1MCS-2 | Broad-host-range vector, IncP, -W, -Q, ColE1 and p15A compatible, carries Kmr | 21 |
| pKNG101 | oriR6K mobRK2 strAB sacBR, 6.8 kb, Smr | 17 |
| pTZ110 | Broad-host-range vector, ori1600 carrying promoterless lacZ gene, Ampr | 41 |
| pKAGb-2(−) | Cmr derivative of pTZ110 | M. S. Thomas, unpublished |
| pMJW400 | pKAGb-2(−) with A. caviae flaA promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pMJW400 | pKAGb-2(−) with A. caviae flaB promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pBRSMT300 | neuB from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT500 | flmB from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT900 | neuA from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1000 | flmD from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1200 | rmlB from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1300 | flmA from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1400 | lsg from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1500 | lst from A. caviae Sch3N inserted into pBBR1MCS, Cmr | This study |
| pBRSMT600 | neuB3 (Cj1317) from C. jejuni inserted into pBBR1MCS, Cmr | This study |
| pBRSMT700 | neuB2 (Cj1327) from C. jejuni inserted into pBBR1MCS, Cmr | This study |
| pBRSMT600 | neuB1 (Cj1141) from C. jejuni inserted into pBBR1MCS, Cmr | This study |
| pBRSMT1100 | flaA1 (HP0840) from H. pylori inserted into pBBR1MCS, Cmr | This study |
| pMJW103 | pBBR1MCS with A. caviae flmA, flmB, neuA, flmD, and neuB genes, Cmr | This study |
| pDI54 | pDSK519 with A. caviae flmB, neuA, flmD, and neuB genes, Kmr | 12 |
| pACYC-FLA2 | pACYC184 with A. hydrophila AH-3 neuA, flmD, and neuB genes, Tcr | 4 |
| pLA-FLA2 | pLA2917 with A. hydrophila AH-3 neuA, flmD, neuB, and flmH genes, Kmr | 4 |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance; MCS, multiple cloning site.
Whole-cell protein preparation, SDS-PAGE, and immunoblotting.
Aeromonas strains were grown statically overnight in BHIB at 37°C. Equivalent numbers of cells were harvested by centrifugation, and each cell pellet either was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer to obtain whole-cell proteins or was resuspended in 1 ml of phosphate-buffered saline (PBS) and then flagella were sheared off by passing the bacterial suspension through an 18-gauge needle 20 times. The bacteria were pelleted by centrifugation and discarded; 100 μl of the supernatant was added to 100 μl of SDS-PAGE loading buffer and boiled for 5 min. Protein samples were separated on SDS-polyacrylamide gels (12% acrylamide). For immunoblotting, proteins were transferred onto a Hybond-C (GE Healthcare) nitrocellulose membrane. Following transfer, membranes were blocked with 5% (wt/vol) powdered skim milk and probed with a polyclonal rabbit anti-polar flagellin antibody (1:500). The unbound antibody was removed by five washes in PBS, and a goat anti-rabbit peroxidase-conjugated secondary antibody (1:1,000) was added. The unbound secondary antibody was washed away with PBS as described above for the primary antibody. The bound conjugate was then detected using the ECL detection system (GE Healthcare).
Motility assay.
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of plates containing motility agar (1% tryptone, 0.5% NaCl, 0.3% agar). The plates were incubated face up at 37°C for 14 to 24 h, and motility was assessed by examining the migration of bacteria through the agar from the center toward the periphery of the plate.
LPS extraction and PAGE analysis.
LPS was purified and detected as described previously (12).
General DNA methods.
DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
Nucleotide sequencing and sequence analysis.
Double-stranded DNA sequencing was performed by using the Sanger dideoxy chain termination method with an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). DNA fragments were ligated into pUC19 and sequenced using an ABI Prism 377 DNA sequencer (Perkin-Elmer Corporation). The M13 universal primers were employed to sequence the ends of the DNA inserts. Following the first sequencing reaction and whenever required, primers were designed until the inserts' sequences were complete. Primers used for DNA sequencing were purchased from Eurofins. For chromosomal walking to extend the sequence into flanking regions, direct genomic sequencing was used. Custom 24-mer primers were designed for a known nucleotide sequence and were used with sheared A. caviae genomic DNA in a 99-cycle polymerase reaction using the BigDye Terminator mixture according to the manufacturer's instructions (PE-Applied Biosystems). The DNA sequence was translated in all six frames and analyzed as previously described (34).
Construction of defined insertion mutants.
Mutants were created by insertion of the Tn5-derived kanamycin resistance cartridge (nptII) from pUC4-KIXX (Pharmacia). This cartridge contains an outward-reading promoter that drives the transcription of downstream genes when it is inserted in the correct orientation. For each mutant the 1.4-kb SmaI-digested kanamycin resistance cartridge was inserted into a convenient restriction site in the middle of the gene. If a convenient site was not present, a site was created by spliced overlap extension PCR. Constructs containing the mutated genes were ligated into the suicide vector pKNG101 (17) and transferred into Aeromonas by conjugation. Conjugal transfer of the recombinant plasmids from E. coli S17-1λpir to A. caviae Sch3N was performed using a filter mating technique. Bacterial conjugation was allowed to proceed for 6 to 8 h at 37°C on sterile nitrocellulose filters (pore size, 0.45 μm) placed on an LB agar plate. Serial dilutions of the mating mixture were then plated on LB agar supplemented with nalidixic acid and kanamycin; the latter antibiotic was added in order to select for recombination. Colonies that were kanamycin resistant (Kmr) and streptomycin sensitive for pKNG101 derivatives (derivatives not likely to have retained the vector) were purified and probed for the kanamycin cartridge and the absence of any plasmid sequences by Southern hybridization. This demonstrated that a double recombination event and allelic exchange occurred.
Construction of lacZ transcriptional fusions.
The mobilizable broad-host-range lacZ promoter probe plasmid pKAGb-2(−) was used in this study (K. Agnoli and M. Thomas, unpublished); this vector is a derivative of pTZ110 (41) that encodes chloramphenicol resistance. The promoter regions of both the A. caviae flaA and flaB genes were amplified by PCR, and the resulting fragments were directionally ligated separately into pKAGb-2(−), yielding plasmids pMJW400 and pMJW500, respectively. These plasmids were introduced separately by conjugation into wild-type strain Sch3N, the neuB mutant SMT166, and the lsg mutant SMT18. The activities of the flaA and flaB promoters were determined as a function of β-galactosidase activity. A. caviae cultures were grown in triplicate to an optical density at 600 nm of 0.5 to 0.8 and were then chilled on ice for 15 min. Duplicate assays were performed at 30°C using 200 μl of cells for each culture in a 1-ml (total volume) mixture following permeabilization of the cells with chloroform-SDS (28). The values are expressed in Miller units (MU) below.
Use of RT-PCR to study gene expression in the flm locus.
To determine which of the flm genes were cotranscribed, reverse transcriptase PCR (RT-PCR) was used. Total RNA was isolated with TRIzol (Invitrogen) used according to the manufacturer's instructions from strain Sch3N grown overnight in BHIB. RNA was dissolved in pyrocarbonic acid diethyl ester-treated water and then treated with 20 U of RNase-free DNase for 30 min at 37°C. The DNase reaction was stopped by extracting the RNA with an equal volume of acid phenol (pH 4.3), followed by ethanol precipitation. The RNA concentration was determined spectrophotometrically. cDNA synthesis was carried out using an Access RT-PCR kit (Promega) with 1 to 3 μg of total RNA according to the manufacturer's instructions. The RNA was heat denatured for 10 min at 75°C and then at 48°C for 45 min to allow first-strand cDNA synthesis by avian myeloblastosis virus RT. The RNA-cDNA duplex and avian myeloblastosis virus RT were denatured at 96°C for 2 min. Second-strand synthesis and subsequent DNA amplification were carried out for 45 cycles with denaturation at 96°C for 45 s, primer annealing at 50°C for 45 s and primer extension at 72°C for 75 s. A final extension step consisting of one cycle at 72°C for 10 min was carried out. Table 2 shows the primers used, the regions amplified, and the expected DNA fragment sizes.
TABLE 2.
Primers used for RT-PCR
| Intergenic region amplified | Primer | Expected product size (bp) |
|---|---|---|
| rmlB-flmA | 5′-ACCGACGTTACGCCATTGATGCCG-3′ | 372 |
| 5′-ATCCCGTACCACCTGTGATTAATACTG-3′ | ||
| flmA-flmB | 5′-TCTGGAGACATCATCTCGGTGGTGA-3′ | 519 |
| 5′-CGCATACGCTGCGCCAGTATGGGT-3′ | ||
| flmB-neuA | 5′-TGCCGATGTTCCACGGCATGACAG-3′ | 292 |
| 5′-GCACCATATTCGAGTGCAACAGCAGCA-3′ | ||
| neuA-flmD | 5′-CCCAAGATCTGGAGGAAGCCTACC-3′ | 458 |
| 5′-CTGCAGCGTCTTGTTGCCATGGCA-3′ | ||
| flmD-neuB | 5′-CTGACCGAAAATATAGCATCGCAGCGC-3′ | 270 |
| 5′-CCCATAGCCCACCGTGAATTTGAA-3′ | ||
| neuB-lsg | 5′-GCGTAGCATTAGGCCCGGATTTGG-3′ | 587 |
| 5′-TTAATGGCCTCCCCTCGAATCTGC-3′ | ||
| lsg-lst | 5′-AGAGATAATTGGCTGGCTT-3′ | 657 |
| 5′-CTCTGCTCAACCCGGGATGTAGAGTTTCTATTT-3′ |
Virulence studies.
The virulence of A. caviae strains grown at 37°C was measured by determining the 50% lethal dose (LD50) by the method of Reed and Muench, as previously described (35). Separate lots of female albino Swiss mice (5 to 7 weeks old) were inoculated intraperitoneally with 0.25-ml portions of the test samples (range, approximately 5 × 109 to 5 × 105 viable cells). Mortality was recorded up to 10 days; all deaths occurred within 3 to 5 days.
Flagellin purification for MS.
Polar flagellins were purified from strains grown in 1 liter of BHIB that were harvested by centrifugation, washed in 10 mM potassium phosphate buffer (pH 7.2), and harvested again. Each bacterial pellet was resuspended in 100 ml of 10 mM potassium phosphate buffer (pH 7.2), and the suspension was passed through an 18-gauge needle 20 times to remove the flagella. Bacteria were removed by two further rounds of centrifugation (10,000 × g), and the supernatant was decanted and subjected to ultracentrifugation for 1 h at 100,000 × g. The resulting flagellum pellet was resuspended in 1% SDS. Purified flagellins were run on 10% precast gels (Invitrogen) and stained using Novex colloidal blue reagent (Invitrogen), and the desired proteins were excised, lyophilized, and digested with trypsin (EC 3.4.21.4; Promega) overnight. Peptides were extracted from gel pieces and purified using a C-8 microtrap peptide cartridge (Presearch) in preparation for analysis by mass spectrometry (MS).
LPS purification and derivatization for MS.
LPS were purified from strains grown in 1 liter of BHIB that were harvested by centrifugation, washed in 10 mM potassium phosphate buffer (pH 7.2), and harvested again. LPS was obtained after treatment with hot phenol, followed by incubation with DNase and then with proteinase K as described previously (12). The LPS was extracted with hot phenol once more before it was freeze-dried. For polysaccharide analysis, the core plus O-antigen was released from lipid A by mild hydrolysis of the LPS (1% acetic acid, 100°C, 2 h) and reduced (10 mg/ml NaBH4 in 2 M NH3, room temperature, 2 h). Reduced samples were loaded directly onto a Dowex H+ column [50W-X8 (H); 50 to 100 mesh; pretreated sequentially with 4 M HCl, water, and 5% acetic acid], and excess borates were removed by repeated addition (four times) of 10% acetic acid in methanol. Methylation using the sodium hydroxide procedure was performed, and the reaction products were purified using Sep-Pak C18 cartridges (Waters Corporation, Massachusetts) (8).
MALDI MS analysis.
Matrix-assisted laser desorption ionization (MALDI) MS was performed using a PerSeptive Biosystems Voyager DE STR mass spectrometer (Foster City, CA) in the reflectron mode with delayed extraction. Methylated samples were dissolved in methanol, and 1-μl aliquots were loaded onto a metal plate with 1 μl of the matrix 2,5-dihydrobenzoic acid. Sequazyme peptide mass standards were used as external calibrants (Applied Biosystems, California).
Electrospray ionization MS analysis.
Samples were dissolved in 30% acetonitrile-0.1% trifluoroacetic acid and sequenced by tandem MS (MS-MS) using a hybrid quadrupole orthogonal acceleration time of flight mass spectrometer (Micromass, United Kingdom). MS and MS-MS spectra were collected in the positive ion mode. The collision energies typically were 50 to 90 eV. Data were acquired and processed using Masslynx software (Micromass, United Kingdom). The instrument was precalibrated using a 1-pmol/μl solution of [Glu1]-fibrinopeptide B in acetonitrile-5% aqueous acetic acid (1:3, vol/vol).
Nucleotide sequence accession number.
The nucleotide sequence of the genes described here has been deposited in the GenBank database under accession number AF126256.
RESULTS
Isolation of new flm genes in A. caviae.
Previously, we described isolation of the flm locus of A. caviae Sch3N; this locus contained one partial (flmA) and four complete (flmB, neuA, flmD, and neuB) open reading frames (ORFs) and was organized in the order flmA-flmB-neuA-flmD-neuB (12). The DNA sequence both upstream and downstream of this locus was determined by direct genomic sequencing of the bacterial chromosomal DNA using custom-designed primers. This strategy extended the flanking sequence by 2.9 kb upstream and by 3.3 kb downstream, resulting in a locus consisting of more than 11 kb.
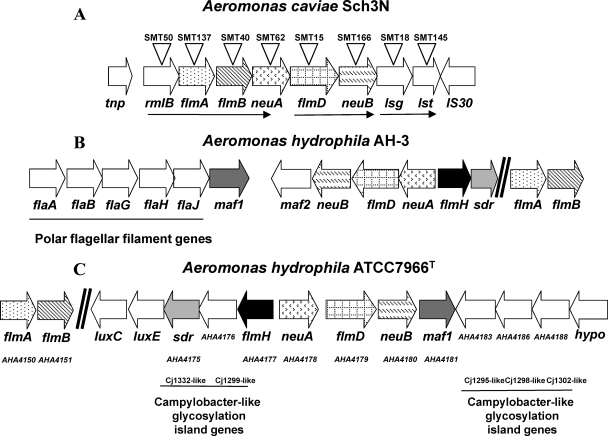
Sequence analysis extending upstream from the partial A. caviae flmA gene identified the rest of this gene in addition to two other ORFs, whereas downstream three further ORFs were revealed (Fig. 1A). Proteins homologous to the putative products of the ORFs were identified using the blastx program of the NCBI (Table 3). All of the ORFs with the exception of the last ORF were transcribed in the same direction. The predicted amino acid sequence encoded by the first ORF (tnp) showed homology to the sequences of enzymes belonging to the transposase 11 family. Downstream of tnp was a second ORF, which encoded a deduced amino acid sequence that had very high levels of identity to a series of RmlB proteins (Table 3). These proteins are dTDP-d-glucose-4,6-dehydratase enzymes that convert dTDP-d-glucose to dTDP-4-keto-6-deoxy-d-glucose, one of the steps in rhamnose biosynthesis (31). Following rmlB was the complete ORF representing flmA; this ORF encoded a protein which belongs to Pseudomonas aeruginosa WbpM subfamily 2 (3) and whose homologies we have described previously (12). The FlmA orthologue in H. pylori, FlaA1, was shown to have C6 dehydratase/C4 reductase activity specific for UDP-GlcNAc (7). In C. jejuni FlmA is called PseB and is involved in the Pse5Ac7Ac biosynthetic pathway converting UDP-GlcNAc to UDP-4-keto-4,6-dideoxy-β-l-AltNAc (25, 27, 40). Downstream of flmA were four ORFs that we have previously described, flmB, neuA, flmD, and neuB, and the deduced amino acid sequences encoded by these ORFs contained conserved domains of a pyridoxal-dependent aminotransferase for FlmB, a CMP-sugar synthetase for NeuA, and a sugar synthetase for NeuB. Closer analysis of the A. caviae FlmD protein showed that it belongs to a subfamily of these proteins that were larger than other proteins in the family, containing 505 amino acids rather than the 350 amino acids more typically seen. This indicated that the A. caviae protein has two domains, the characteristic glycosyltransferase domain that is present in the smaller proteins in the family and an RimL-like acetyltransferase domain that is found in another Flm protein, FlmH. It therefore appears that the A. caviae FlmD protein is a chimera of FlmD and FlmH, equivalent to the C. jejuni PseG and PseH proteins (Table 3 and see below).
FIG. 1.
Genetic organization of the A. caviae Sch3N (A), A. hydrophila AH-3 (B), and A. hydrophila ATCC 7966T (C) flm loci. Predicted ORFs were named after their homologues in other bacterial species and are indicated by open arrows, which indicate the direction of transcription. The same pattern in arrows indicates orthologous genes in the clusters. The ORF labeled “hypo” is a gene encoding a hypothetical protein. ORFs of interest in A. hydrophila ATCC 7966T are indicated by their gene locus numbers. Open triangles indicate the sites of insertion of the antibiotic resistance cassette, and the corresponding mutant designations are indicated above the triangles. The thin arrows indicate transcriptional units as determined by RT-PCR.
TABLE 3.
Properties of the putative ORFs of A. caviae Sch3N
| A. caviae ORFa | Length (nucleotides) | No. of amino acids | Molecular mass (kDa)a | Homologous protein | Homologue function | % Identity/% similarityb | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| rmlB | 1,064 | 321 | 36 | RmlB of A. hydrophila | dTDP-d-glucose-4,6-dehydratase | 77/85 | AF148126 |
| RmlB of Vibrio cholerae | dTDP-d-glucose-4,6-dehydratase | 73/82 | AY239001 | ||||
| RmlB of Salmonella enterica | dTDP-d-glucose-4,6-dehydratase | 71/83 | AF279619 | ||||
| flmA | 1,013 | 338 | 38 | PseB (Cj1293) of C. jejuni | Sugar nucleotide epimerase/dehydratase | 53/68 | NP282439 |
| FlaA1 of H. pylori | UDP-GlcNAc C6 dehydratase/ C4 reductase | 52/71 | NP207633 | ||||
| FlmA of C. crescentus | Flagellin modification | 51/68 | NP419052 | ||||
| flmBc | 1,164 | 387 | 43 | PseC (Cj1294) of C. jejuni | Pyridoxal-dependent aminotransferase | AF126256 | |
| neuAc | 687 | 228 | 25.8 | PseF (Cj1311) of C. jejuni | CMP-sugar synthetase | AF126256 | |
| flmD/Hc | 1,518 | 505 | 56.8 | PseG (Cj1312) of C. jejuni | Glycosyltransferase | AF126256 | |
| PseH (Cj1313) of C. jejuni | Acetyltransferase | ||||||
| neuBc | 1,059 | 352 | 38.6 | PseI (Cj1317) of C. jejuni | Sugar synthetase | AF126256 | |
| lsg | 1,262 | 420 | 46.5 | Lsg1 of H. influenzae | LPS biosynthesis protein | 28/46 | NP439842 |
| CpsL of Streptococcus agalactiae | Polysaccharide repeating unit transporter, “flippase” | 21/44 | AF349539 | ||||
| Wzx of P. aeruginosa | Polysaccharide repeating unit transporter, “flippase” | 24/42 | AAF24000 | ||||
| RfbX of Shigella flexneri | Polysccharide repeating unit transporter, “flippase” | 23/42 | JC4069 | ||||
| lst | 947 | 315 | 37.2 | WaaH of Salmonella enterica | UDP-glucose:glucosyl LPS, a 1,2 glucosyltranferase | 29/47 | AF519775 |
| Lst of Pasteurella multocida | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3 sialyltransferase | 27/46 | Q9CNC4 | ||||
| Lst of H. influenzae | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3 sialyltransferase | 28/43 | Q48211 | ||||
| Lst of Neisseria meningitidis | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3 sialyltransferase | 23/42 | P72097 |
ORFs and molecular masses were determined by ORF finder of NCBI.
Homologous proteins and levels of identity and similarity for the homologous regions were determined by using the Blastx program of NCBI.
Homology was determined previously, and putative functions are indicated.
Homologues of all these genes encode proteins that have been shown to be essential for Pse5Ac7Ac biosynthesis in C. jejuni and in this bacterium have been designated PseB (FlmA), PseC (FlmB), PseF (NeuA), PseG/H (FlmD/H), and PseI (NeuB) (Table 3) (25). Following neuB was an ORF encoding a deduced amino acid sequence that shared homology with a series of proteins belonging to the Wzx family of LPS O-antigen flippase transport proteins, the closest of which was encoded by the lsg locus of Haemophilus influenzae (9). The Lsg protein is predicted to have 12 transmembrane regions, which is characteristic for this family of proteins that typically contain 12 or 13 membrane-spanning regions and are involved in the transport of di- or trisaccharides across the cytoplasmic membrane (32). Downstream of lsg was an ORF designated lst, and the protein encoded by this ORF shared homology with a series of glucosyl- and sialyltransferases from a number of bacteria (Table 3). These proteins transfer the activated forms of glucose or sialic acid to other sugars. The final ORF encoded a protein that was similar to another series of transposase 8 enzymes (COG2826) belonging to the IS30-like element family. Interestingly, the average G+C content of the locus, not including the transposase genes (i.e., rmlB to lst), was 42%; this is significantly less than the average G+C content, 60%, which is typically seen in A. hydrophila ATCC 7966T (42). Although there is heterogeneity in the locations, organizations, and sequences of the flmA, flmB, neuA, flmD, and neuB genes in the aeromonads, all strains tested so far (n = 50) possess these genes (4, 12).
In a separate study we isolated the flm locus from A. hydrophila AH-3 through complementation of A. caviae mutants (4). The genetic organization of this locus is shown in Fig. 1B. Moreover, the genome sequence of A. hydrophila ATCC 7966T became available during this study (42), and the genetic organization of the flm locus of this strain is shown in Fig. 1C. The striking feature of comparisons of the flm loci of the three strains is the difference in the location within the chromosomes. In AH-3 the flm locus is downstream of the polar flagellin gene cluster (polar flagella region 2), whereas in ATCC 7966T it is downstream of the acyl coenzyme A synthetase genes luxC and luxE (Fig. 1C). In both A. hydrophila strains the flmA and flmB genes are located away from the other genes isolated in A. caviae (4, 42). Furthermore, the average G+C contents of both of the A. hydrophila loci are around 60%, which is typical for Aeromonas species.
Transcript mapping by RT-PCR.
Complementation analysis of the polar transposon insertions in our previous study, along with the transcriptional direction of the genes and the small gaps between the end of one ORF and the start of the next ORF, suggested that the genes formed part of a polycistronic operon. Therefore, RT-PCR was employed to test whether each gene was cotranscribed with its immediate partner downstream. Primer pairs that overlapped different genes in the locus were designed for the 3′ end of the upstream gene and near the 5′ end of the downstream gene (Table 2). This was done in order to amplify the intergenic region between the two genes that would be expressed if the two genes were cotranscribed. RT-PCR products of the expected sizes were detected (data not shown) for every gene pair with the exception of neuA-flmD and neuB-lsg. This suggests that there are three transcripts in the locus, the first containing rmlB, flmA, flmB, and neuA, the second containing flmD and neuB, and the third containing lsg and lst (Fig. 1A).
Creation of insertional mutations in the A. caviae flm locus genes.
To determine the roles of the identified genes in either motility or LPS biosynthesis or both, mutants were constructed for each gene, including the genes previously mutated by insertion of the polar transposon mini-Tn5Cm (12). A kanamycin resistance cassette was inserted in the same transcriptional orientation with respect to the target gene; the presence of an outward-reading promoter on the cassette ensures expression of downstream genes, thereby reducing any polar effects. However, such insertions may alter the regulation of the downstream genes. The construction of all mutants was verified by Southern hybridization of chromosomal DNA with the kanamycin cassette and vector probes (data not shown). The kanamycin cassette position and strain designations are shown in Fig. 1A.
The motility of the A. caviae mutant strains was assessed by examining static growth in BHIB and in semisolid motility plates. In liquid all mutant strains with the exception of the lsg mutant and the lst mutant grew at the bottom of the culture tube as a “loose” pellet and not as a turbid suspension like that seen for wild-type strain Sch3N (data not shown). This finding was supported by the inability of mutant cells (except for the lsg mutant and lst mutant cells) to swim in semisolid motility agar (Fig. 2A) and by the results of immunoblotting of the mutant whole-cell protein preparations for the polar flagellin proteins. In contrast to the parental strain, the lsg mutant, and the lst mutant preparations, the polar flagellins were not detected in the other mutant preparations, an observation suggesting that there was a loss of polar flagellin protein expression and thus that flagellum filaments were not present (Fig. 2B).
FIG. 2.
Analysis of motility, polar flagellin, and LPS for A. caviae Sch3N and isogenic mutants. (A) Motility as assessed by determining a strain's ability to swim in 0.3% semisolid motility agar. WT, wild type. (B) (Upper panel) Polar flagellin immunoblot of whole-cell proteins of A. caviae Sch3N (WT) and flm locus isogenic mutants. (Lower panel) Polar flagellin immunoblot of whole-cell proteins of flm locus isogenic mutants complemented with individual copies of the wild-type genes in pBBR1MCS (Table 1). The genes in which the knockout occurs are indicated above the lanes. Proteins were obtained from bacteria grown at 37°C in BHIB and were analyzed by SDS-PAGE (12%). They were transferred onto nitrocellulose membranes and immunoblotted with anti-polar flagellin antibodies (1:500). (C) Analysis of LPS isolated from A. caviae Sch3N (WT) and the flm locus isogenic mutants. (Upper panel) The genes in which the knockout occurs are indicated above the lanes. (Lower panel) flm locus isogenic mutants complemented with individual copies of the wild-type genes in pBBR1MCS (Table 1). LPS was extracted from bacteria grown at 37°C in BHIB, analyzed by SDS-PAGE (12%), and silver stained. The positions of LPS bands A and B are indicated on the left.
The possibility that the mutants had defects in LPS biosynthesis was also investigated. The wild-type strain and the mutants were grown overnight in BHIB at 37°C, and LPS was extracted and analyzed by PAGE. The wild-type strain produced a smooth LPS (O-antigen+) exhibiting two major bands (band A and band B) and a faint ladder of higher-molecular-weight bands, whereas all the mutants produced only major band A. On Tricine-PAGE gels either the LPS patterns for the mutants lacked one of the bands (band B) always present in the LPS pattern of the wild-type strain or the LPS produced a band at a lower molecular weight whose abundance was lower (Fig. 2C). The parental phenotypes of all of the mutants except the rmlB mutant (see below) were rescued when the mutants were complemented in trans using wild-type copies of each individual gene cloned into the broad-host-range vector pBBR1MCS (22) in an orientation that allowed expression from the lac promoter (Fig. 2B and 2C). Complementation analysis demonstrated that insertion of the kanamycin cassette into the rmlB mutant had a polar effect on downstream genes, as providing the rmlB gene alone in trans did not rescue the phenotype observed. However, introduction of the broad-host-range plasmid pMJW103 containing complete copies of flmA, flmB, neuA, flmD, and neuB (but not rmlB) into the rmlB mutant rescued motility and flagellum expression and restored band B to the LPS pattern (data not shown). It was concluded from the complementation data that mutation of the rmlB gene did not result in the phenotypes observed for the other mutants and that this gene was not involved in flagellum or LPS biogenesis in this strain. Therefore, no further characterization was carried out for this strain.
Sequence analysis suggested that the A. caviae FlmD protein was in fact a chimeric protein equivalent to both FlmD and FlmH from A. hydrophila AH-3. To test this hypothesis, plasmid pACYC-FLA2, which contains the A. hydrophila AH-3 neuA-like, flmD, neuB-like genes (4), was introduced into the A. caviae flmD mutant. However, motility or the O-antigen LPS band was not restored by this complementing plasmid. But when the pLA-FLA2 cosmid (40 kb) carrying the A. hydrophila AH-3 neuA-like, flmD, and neuB-like genes in addition to flmH (4) was introduced into the flmD mutant, motility and the O-antigen band of the A. caviae LPS were fully restored (data not shown). The results of this complementation analysis support the bioinformatic data concerning the dual role of the A. caviae FlmD protein (i.e., that it has both FlmD activity and FlmH activity).
As the mutations in the A. caviae flm locus affect two structures associated with aeromonad pathogenicity, we determined the LD50s of two of the mutants in a mouse model. The LD50 of wild-type strain Sch3N in mice is 106.8 cells, whereas the LD50s of the flmD and neuA mutants were >108.0 cells, which is an increase of at least 1 log unit.
Complementation and characterization of the A. caviae neuA, neuB, and flmA genes.
The A. caviae Sch3N neuA and neuB genes in pDI54 (12) were unable to complement their E. coli counterparts in E. coli mutant strains EV5 (neuA) and EV24 (neuB) (45). However, three neuB genes, termed neuB1 (Cj1141), neuB2 (Cj1327), and neuB3 (Cj1317), were shown to be present in the genome of C. jejuni NCTC 11168 (24). All three genes have previously been shown to encode Neu5Ac synthetase activity (24). In C. jejuni mutation of neuB1 resulted in loss of Neu5Ac from the lipooliogosaccharide of C. jejuni, whereas mutation of neuB2 was cryptic in strain NCTC 11168 but resulted in a reduction in the flagellin mass in strain G1 and mutation of neuB3 caused the cells to become aflagellate and nonmotile. NeuB3 has been shown to be involved in the biosynthesis of Pse5Ac7Ac, a nine-carbon sugar related to Neu5Ac (43), and has been designated PseI (25). Each of the C. jejuni neuB genes was cloned separately into the broad-host-range mobilizable vector pBBR1MCS (22) in an orientation that allowed expression from the lac promoter, and the constructs were then introduced into the A. caviae neuB mutant by conjugation. The vectors expressing neuB1 (pBRSMT800) and neuB2 (pBRSMT700) in trans were unable to complement the A. caviae neuB mutant, whereas when the plasmid expressing neuB3 (pBRSMT600) was introduced in trans into the A. caviae neuB mutant, the O-antigen band of the A. caviae LPS was produced (Fig. 3). On motility plates the wild-type strain had an average motility zone with a diameter of 16 ± 0.7 mm, which was significantly different (P = <0.01) from the diameters of the average motility zones of the neuB mutant SMT166 (2.1 ± 0.5 mm), SMT166 expressing C. jejuni neuB1 (1.9 ± 0.2 mm), and SMT166 expressing C. jejuni neuB2 (2.0 ± 0.3 mm). The SMT166 neuB mutant expressing C. jejuni neuB3 had a motility zone with a diameter of 12.8 ± 0.8 mm, which was significantly different (P = <0.01) from the results for the wild-type strain, the original mutant, and the complemented mutants, suggesting that there was only partial complementation for motility.
FIG. 3.
Analysis of motility and LPS isolated from the A. caviae neuB mutant SMT166 and derivative strains complemented with C. jejuni paralogues. (A) Motility as assessed by swimming in 0.3% semisolid motility agar for A. caviae Sch3N (WT), SMT166 (neuB mutant), and SMT166 containing pBBR1MCS expressing the C. jejuni paralogue neuB1, neuB2, or neuB3. (B) Analysis of LPS isolated from A. caviae Sch3N (WT), SMT166 (neuB mutant), and SMT166 containing pBBR1MCS expressing the C. jejuni paralogue neuB1, neuB2, or neuB3. LPS was extracted from bacteria grown at 37°C in BHIB, analyzed by SDS-PAGE (12%), and silver stained. The positions of LPS bands A and B are indicated on the left.
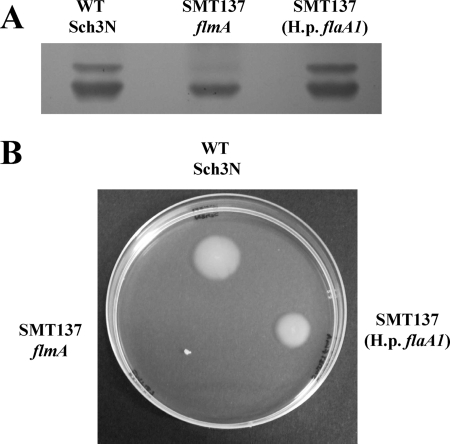
Previously, the FlaA1 protein of H. pylori has been shown to have bifunctional C6 dehydratase/C4 reductase activity for the conversion of UDP-GlcNAc to UDP-QuiNAc (7). However, recently, both the H. pylori FlaA1 (HP0840, FlmA orthologue) and C. jejuni PseB proteins have been shown to have C6 dehydratase and C5 epimerase activities, resulting in the production of UDP-2-acetamido-2,6-dideoxy-β-l-arabino-4-hexulose and UDP-2-acetamido2,6-dideoxy-α-d-xylo-4-hexulose; the former compound is the substrate for the C. jejuni FlmB homologue PseC (39). To test whether the H. pylori FlaA1 protein could functionally complement the A. caviae FlmA protein, the flaA1 gene was amplified by PCR, sequenced, and cloned into the broad-host-range mobilizable vector pBBR1MCS in an orientation that allowed expression from the lac promoter. The resulting construct, pBRSMT1200, was then introduced into the A. caviae flmA mutant by conjugation; expression of flaA1 in trans rescued both flagellin expression and LPS O-antigen (Fig. 4). However, on motility plates the wild-type strain had a motility zone with an average diameter of 15.8 ± 0.8 mm, which was significantly different (P = <0.01) from the results for both the flmA mutant strain SMT137 (2.0 ± 0.3 mm) and the complemented mutant strain (13.6 ± 0.9 mm), again suggesting that in the complemented strain motility was partially rescued.
FIG. 4.
Analysis of motility and LPS isolated from the A. caviae flmA mutant SMT137 and derivative strains complemented with the H. pylori orthologue flaA1. (A) Analysis of LPS isolated from A. caviae Sch3N (WT), SMT137 (flmA mutant), and SMT137 containing pBBR1MCS expressing the H. pylori orthologue flaA1 (H.p. flaA1). LPS was extracted from bacteria grown at 37°C in BHIB, analyzed by SDS-PAGE (12%), and silver stained. The lower and upper LPS bands are bands A and B, respectively. (B) Motility as assessed by swimming in 0.3% semisolid motility agar for A. caviae Sch3N (WT), SMT137 (flmA mutant), and SMT137 containing pBBR1MCS expressing the H. pylori orthologue flaA1.
Analysis of flagellin transcription in the flm locus mutants.
Mutations in most genes in the flm locus (all of the genes except lsg and lst) and equivalent loci in other bacteria result in a reduction in the ability or an inability to detect flagella by flagellin Western blotting of whole-cell proteins. To determine whether flagellin gene transcription is repressed in an A. caviae neuB mutant or lsg mutant background, the putative promoter regions of the polar flagellin genes flaA and flaB were transcriptionally fused to the promoterless reporter gene lacZ in pKAGb-2(−). DNA fragments that were 550 bp long for flaA and 716 bp long for flaB were amplified by PCR from A. caviae chromosomal DNA; these fragments represented the 5′ ends of the genes and their putative promoter regions (34). The PCR fragments were ligated into the broad-host-range lacZ expression vector pKAGb-2(−) in an orientation that allowed expression of the lacZ gene. The constructs were introduced into the A. caviae wild-type, neuB mutant, and lsg mutant strains separately. Quantitative assays for β-galactosidase activity with cells grown in BHIB showed that the parental A. caviae wild-type strain, neuB mutant, and lsg mutant and the strains containing only the vector had very low intrinsic β-galactosidase activities. Approximately equal levels of β-galactosidase activity (220 to 255 MU) were detected in all strains that contained the flaA promoter plasmid pMJW400, whereas the activities in strains with the flaB promoter plasmid pMJW500 were between 5- and 10-fold higher (1,287 to 2,447 MU) depending on the strain background (Fig. 5). The lowest level of activity for flaB promoter plasmid pMJW500 was detected with the neuB mutant background; this activity was around 62% of the activity of the wild-type strain containing the same plasmid.
FIG. 5.
β-Galactosidase activities of the flaA-lacZ and flaB-lacZ fusion plasmids in Sch3N, the nonmotile neuB mutant SMT166, and the motile lsg mutant SMT18. β-Galactosidase activity was assayed for A. caviae Sch3N (WT), SMT166 (neuB mutant), and SMT18 (lsg mutant) and the derivative strains containing only the vector or the flaA-lacZ and flaB-lacZ fusion plasmids, as indicated below the bars. The activity was assayed using bacteria grown at 37°C in BHIB. Assays were carried out in triplicate, and the values are the means ± standard deviations.
Structural analysis of the A. caviae polar flagellins.
Polar flagellin proteins were purified from the A. caviae wild-type strain, in which two flagellin subunits (FlaA and FlaB) are present, and flagellin was also purified from A. caviae strains with single mutations in either the flaA or flaB gene (34). The flagellins were analyzed by SDS-PAGE, which indicated that the molecular mass is approximately 35 kDa. This molecular mass is larger than that predicted from the coding sequence (34) and is thought to result from glycosylation since the flagellins gave a positive result in a glycan detection assay. Glycoprotein analysis was employed to ascertain the nature of the flagellin sugar moiety present in the A. caviae Sch3N strain used in this study. Thus, bands corresponding to flagellin were excised from the gel and digested with trypsin. Electrospray MS of the tryptic digests confirmed the presence of the FlaA and FlaB proteins and also identified a number of multiply charged signals not corresponding to the masses of predicted flagellin tryptic peptides. The m/z values of these signals are shown in Tables 4 and 5 for FlaA and FlaB, respectively. Each of these signals was selected for MS-MS (electrospray CAD MS-MS), and all of them had characteristic fragment ions at m/z 299 and m/z 317, consistent with Pse5Ac7Ac (43). Moreover, second-generation fragment ions at m/z 317 (data not shown) were fully consistent with the reported fragmentation pattern for Pse5Ac7Ac found for C. jejuni flagellin (43). For both FlaA and FlaB, between six and eight Pse5Ac7Ac residues were found in the serine- and threonine-rich domain spanning residues 159 to 232 (FlaA) and 146 to 232 (FlaB).
TABLE 4.
Assignment of glycopeptides obtained from electrospray MS-MS analysis of the A. caviae FlaA flagellin subunit
| Observed signal (m/z)a | Charge state of glycopeptide | Sequence of glycopeptide | Sequence position | No. of Pse5Ac7Ac residues |
|---|---|---|---|---|
| 1859 | [M+4H]4+ | AAGTTIDIVSGPAGSVTTATGISLIFVSGSAGGISISTQSKAQAV | 174-232 | 6 |
| 1487 | [M+5H]5+ | LAAADAMLEVVDGK | ||
| 1240 | [M+6H]6+ | |||
| 1063 | [M+7H]7+ | |||
| 1780 | [M+4H]4+ | AAGTTIDIVSGPAGSVTTATGISLIFVSGSAGGISISTQSKAQAV | 174-232 | 5 |
| 1424 | [M+5H]5+ | LAAADAMLEVVDGK | ||
| 1187 | [M+6H]6+ | |||
| 1136 | [M+3H]3+ | FQVGADANQTIGFSLSQAGGFSISGIAK | 146-173 | 2 |
| 1545 | [M+2H]2+ | FQVGADANQTIGFSLSQAGGFSISGIAK | 146-173 | 1 |
The observed signal corresponds to the average molecular mass.
TABLE 5.
Assignment of glycopeptides obtained from electrospray-MS-MS analysis of A. caviae FlaB flagellin subunit
| Observed signal (m/z)a | Charge state of glycopeptide | Sequence of glycopeptide | Sequence position | No. of Pse5Ac7Ac residues |
|---|---|---|---|---|
| 1859 | [M+4H]4+ | AAGTTIDIVSGPAGSVTTATGISLIFTGGSAGGISISTQSKAQAV | 174-232 | 6 |
| 1487 | [M+5H]5+ | LAAADAMLEVVDSK | ||
| 1240 | [M+6H]6+ | |||
| 1780 | [M+4H]4+ | AAGTTIDIVSGPAGSVTTATGISLIFTGGSAGGISISTQSKAQAV | 174-232 | 5 |
| 1424 | [M+5H]5+ | LAAADAMLEVVDSK | ||
| 1187 | [M+6H]6+ | |||
| 1334 | [M+4H]4+ | AAGTTIDIVSGPAGSVTTAT GISLIFTGGSAGGISISTQSK | 174-214 | 5 |
| 1035 | [M+2H]2+ | SLAQTGGFSISGIAK | 159-173 | 2 |
The observed signal corresponds to the average molecular mass.
Analysis of the A. caviae wild-type and flmD mutant LPS.
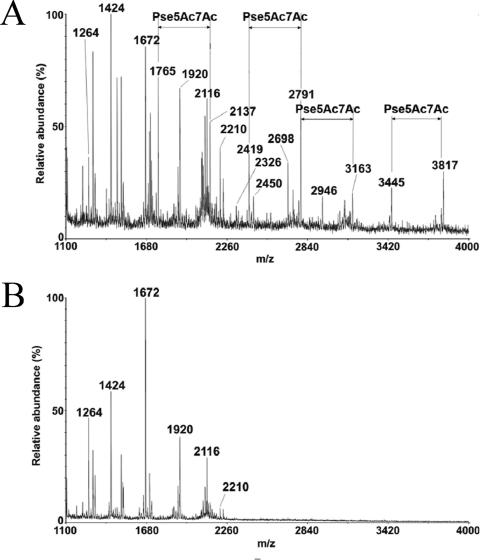
In order to gain further insight into the role of FlmD in LPS biosynthesis, LPS from both Sch3N and the flmD mutant were isolated and used for structural comparison by MS. The lipid A was removed from the samples by mild hydrolysis, and the core and O-antigen polysaccharides were reduced and analyzed by MALDI MS as permethylated derivatives. The signals up to m/z 2260 that are common to both the wild type and the flmD mutant (Fig. 6) are derived from the core oligosaccharide. The MS spectrum for the wild-type LPS contains an additional series of signals indicative of O-antigen repeat units containing Pse5Ac7Ac (mass increments of 372 U) (Fig. 6A). These signals are not present for the flmD mutant (Fig. 6B). Analysis of perdeuteromethylated LPS provided further evidence that Pse5Ac7Ac is a component of the Sch3N LPS but not the mutant LPS. Thus, MALDI MS screening of perdeuteromethylated wild-type polysaccharide (data not shown) revealed that the mass increments attributed to Pse5Ac7Ac increased from m/z 372 to m/z 384, in agreement with the expected mass of perdeutermethylated Pse5Ac7Ac. These signals were not present in the mutant.
FIG. 6.
MALDI MS analysis of permethylated LPS derived from A. caviae. Lipid A was released prior to derivatization and analysis of the core and O-antigen polysaccharide component. (A) Wild-type strain-derived core and O-antigen polysaccharide. Examples of signals corresponding to O-antigen repeat units differing by a Pse5Ac7Ac residue are labeled “Pse5Ac7Ac.” (B) flmD mutant-derived core oligosaccharide. Signals corresponding to the O-antigen repeat and Pse5Ac7Ac were not observed.
DISCUSSION
Orthologues of the A. caviae neuA, flmD, and neuB genes were previously thought not to be present in A. hydrophila (12) but have recently been isolated (4). In an earlier study, we were unable to detect them by either PCR or Southern blotting, probably due to the large degree of heterogeneity between A. caviae Sch3N and A. hydrophila AH-3 at the nucleotide level. In the present work we demonstrated that the flm locus of A. caviae Sch3N appears to have been acquired laterally. Several lines of evidence support this conclusion. First, the G+C content of the locus was much lower than the average G+C content for Aeromonas (42% versus 60%), unlike the G+C contents of the loci in A. hydrophila AH-3 and A. hydrophila ATCC 7966T, which are similar to the average Aeromonas G+C content. In addition, the Sch3N locus is flanked by transposon sequences; all of these sequences are markers of genes acquired from another source, and as both flagella and LPS have been linked to virulence in Aeromonas (1, 26, 34), this suggests that the A. caviae flm locus could be a horizontally acquired glycosylation and pathogenicity island. Furthermore, we demonstrated here that mutations in the flm locus do indeed increase the LD50 in the mouse model, thereby supporting its role in pathogenicity. However, the genes required for Pse5Ac7Ac biosynthesis are present in all mesophilic aeromonad strains tested so far. Mutation of each individual A. caviae gene resulted in loss of the LPS O-antigen bands for all mutants and the flagella for all of the mutants with the exception of the rmlB, lsg, and lst mutants. This finding supports our previous conclusion that most of the proteins encoded by the flm locus in A. caviae have dual roles in LPS biosynthesis and flagellum assembly, whereas A. hydrophila AH-3 appears to have the indigenous copies of some of the genes that are required only for flagellum glycosylation, as another locus contains genes for the LPS O-antigen (4). The rmlB gene that is present in the flm locus is thought to encode an enzyme required for rhamnose biosynthesis; this gene is probably a remnant from another LPS biosynthesis gene cluster that was acquired by A. caviae Sch3N along with the Pse5Ac7Ac biosynthetic genes by horizontal transfer from another bacterium. The A. caviae rmlB gene is not needed for Pse5Ac7Ac biosynthesis, nor is it required for flagellin glycosylation or LPS biosynthesis as rhamnose is not present in the O-antigen (J. G. Shaw, unpublished).
Two possible routes for bacterial protein glycosylation have been suggested. The first route utilizes and shares part of the LPS biosynthesis pathway, an example of which is pilus glycosylation in P. aeruginosa (5, 38). The second possible glycosylation route is distinct from the LPS pathway and is dedicated to only glycosylation, as seen for flagellin glycosylation in C. jejuni. In this and our previous studies we have shown that different strains in the same genus use two disparate pathways for flagellin glycosylation. For example, A. caviae Sch3N utilizes a shared LPS-glycosylation route, and A. hydrophila AH-3 uses a dedicated glycosylation system in which mutation affects flagellin expression but does not alter the strain's LPS (4, 12).
The dual functions of the A. caviae flm locus proteins encoded by flmA, flmB, neuA, flmD, and neuB suggested that they are involved in sugar biogenesis and that the sugars that they make are placed both on the flagella and in the LPS (Fig. 7); this was subsequently proved by MS, which demonstrated that mass increments both in flagellins and in the LPS O-antigen were consistent with Pse5Ac7Ac. Sugars for the LPS are linked to each other by glycosyltransferases, suggesting a role for Lst; these sugars are then transported across the cytoplasmic membrane by the Wzx-like flippase Lsg. However, similar Wzx-like proteins have also been shown to be involved in pilin glycosylation in Neisseria and P. aeruginosa (5) but do not appear to be involved in flagellin glycosylation. Although Aeromonas strains express type IV pili, it is not known whether they are glycosylated (19). Bacterial surface sugars need to be transported across the cytoplasmic membrane; this usually occurs by two main systems, based on the proteins Wzx-Wzy and Wzm-Wzt. Mutation of lsg affects only LPS biosynthesis, suggesting that flagellin glycosylation is Wzx independent. Therefore, flagellin glycosylation may be Wzm-Wzt dependent. However, our data support the more favored idea that glycosylation of the flagellar proteins occurs in the cytoplasm and the glycosylated flagellins are then exported to the cell surface for filament assembly and are not glycosylated once they are on the surface.
FIG. 7.
Hypothetical pathway for flagellin glycosylation and LPS modification in A. caviae Sch3N. The biosynthetic pathway to Pse5Ac7Ac is based on the predicted functions of the A. caviae proteins compared with those elucidated for C. jejuni and H. pylori proteins (25, 39). The activated form of Pse5Ac7Ac, CMP-Pse5Ac7Ac, is then either transferred onto the flagellin by an unknown mechanism or is predicted to be transferred onto a sugar-antigen carrier lipid (ACL) by Lst to create an LPS O-antigen unit, and this O-antigen unit is subsequently transported across the cytoplasmic membrane by Lsg.
Attempts to functionally characterize the A. caviae neuA and neuB genes through complementation of their E. coli mutant counterparts using pDI54 were unsuccessful, even though pDI54 is able to fully complement the neuA and neuB mutants of A. caviae (12). This suggests that the A. caviae neuA and neuB genes encode proteins not involved in Neu5Ac biosynthesis. C. jejuni strain NCTC 11168 contains three paralogues of neuB, and the proteins encoded by them have been demonstrated to have various levels of Neu5Ac synthetase activity (24). The presence of three genes that all encode the same enzyme does suggest that there is some functional redundancy if all three enzymes make the same product (i.e., Neu5Ac). Only the C. jejuni neuB3 gene was able to complement and completely rescue all of the phenotypes of the A. caviae neuB mutant. Mutation of the neuB3 gene in a number of C. jejuni strains does result in a phenotype similar to that of the A. caviae neuB mutant with regard to the loss of flagella and motility (24, 43). Additionally, the glycosyl modifications of the Campylobacter flagellin have been demonstrated to be variations of the nine-carbon sugar Pse5Ac7Ac, which is structurally similar to Neu5Ac, and NeuB3 has been shown to be involved in the biosynthesis of Pse5Ac7Ac (43) and is now designated PseI (25). The C. jejuni neuB3 and H. pylori flaA1 genes are able to complement their A. caviae counterparts. Furthermore, both H. pylori flaA1 and its orthologues in C. jejuni, pseB (Cj1293) and pseI (neuB3), have also been associated with Pse5Ac7Ac synthesis (6, 11, 37). This suggested that the A. caviae flm locus is involved in Pse5Ac7Ac biosynthesis; we subsequently confirmed this by our detection of Pse5Ac7Ac both on the flagellins and in the LPS O-antigen of A. caviae Sch3N. MS showed that Pse5Ac7Ac is present on both the flagellin subunits and in LPS of A. caviae. Glycosylation occurred only in the central domain of the A. caviae flagellin, as has been observed previously for both C. jejuni and H. pylori. It was recently demonstrated that A. caviae flagellin from another strain is glycosylated with Pse5Ac7Ac modified with an extra glycine residue (36). A number of related derivatives of Pse5Ac7Ac have also been reported to be present in C. jejuni (36, 37, 43). In this study of A. caviae Sch3N flagellin glycosylation, only Pse5Ac7Ac was observed. The additional modification of Pse5Ac7Ac by glycine reported by Schirm et al. (36) was not detected. However, additional campylobacter-like glycosylation island genes are present adjacent to the flm locus in the genome sequences of A. hydrophila ATCC 7966T and AH-3 (4, 42); these genes could be involved in generating derivatives of Pse5Ac7Ac similar to those seen on the flagellins of C. jejuni.
Based on enzymology, mutational analysis, and metabolomic studies, a pathway for the production of Pse5Ac7Ac has been proposed for C. jejuni (13, 25). Investigations of C. jejuni 81-176 have shown that the proteins essential for the production of Pse5Ac7Ac are PseB, PseC, PseF, PseG, PseH, and PseI, homologues of which are encoded by genes present in the A. caviae Sch3N flm locus. This suggests that this strain of A. caviae has acquired the minimum gene cluster required for the biosynthesis of Pse5Ac7Ac.
Activation of Pse5Ac7Ac through addition of CMP by the PseF homologue NeuA would result in CMP-Pse5Ac7Ac; this compound is probably the substrate of the A. caviae sialyltransferase-like protein Lst for addition of Pse5Ac7Ac to the LPS O-antigen (Fig. 7). As mutation of lst affects only LPS production and not flagella or motility, addition of Pse5Ac7Ac to the flagellin most likely involves another protein. One possible candidate is the Maf family of proteins (motility-associated factors), as the genes for these proteins are linked to, or found within, the flagellin and glycosylation loci of a number of bacteria, including H. pylori (HP0114), C. jejuni (Cj1318), A. hydrophila AH-3, and A. hydrophila ATCC 7966T (4, 18, 37, 42). Mutation of these genes results in a phenotype like that observed for the flm locus mutants, with the loss of motility and flagella (18, 36). Furthermore, a maf (Cj1318) orthologue has been found in A. caviae Sch3N directly downstream of the polar flagellin locus (J. G. Shaw, unpublished).
Mutation of genes in the flm locus in this study and our previous study has resulted in the absence of or a reduction in the amount of the flagellin subunit on whole-cell Western blots (4, 12). Here we showed that the expression from the putative flaA and flaB gene promoters transcriptionally fused to lacZ was reduced only for the flaB promoter in a neuB mutant background compared to the wild-type strain or the motile lsg mutant strain. The expression from the flaA promoter, although 10-fold lower, appeared to be the same in all the strain backgrounds tested. The small reduction in transcription of the flaB gene does not explain the large reduction in or absence of flagella on Western blots. However, this is similar to the situation in C. crescentus, where transcriptional and translational fusions to the fljK flagellin subunit gene were expressed at levels close to wild-type levels in flmA, flmD, and flmH mutant backgrounds (2). In H. pylori similar mutations greatly reduced the amount of detectable flagellin protein on Western blots (16, 37), but the flagellin genes were transcribed, as judged by RT-PCR (37). In A. hydrophila AH-3 mutations of flm locus genes stop the development of a functional flagellum filament and reduce the amount of flagellin present by around 50% (4). It therefore appears at this stage that in A. caviae Sch3N the flm genes are not part of the flagellar regulatory hierarchy, but at present the full regulatory hierarchy for the Aeromonas polar flagellum is not known. Furthermore, the pseB gene in Campylobacter has been shown to be coordinately regulated with flagellum genes (11).
Acknowledgments
We thank Brendan Wren and Dennis Linton for plasmids containing the three C. jejuni neuB genes and Eric Vimr for E. coli strains EV24, EV36, and EV5 and phage K1.
Part of this work was supported by grants from the Wellcome Trust, the Bardhan Research and Education Trust, and Plan Nacional de I+D and by FIS grants from REIPI (Ministerio de Educación, Cienca y Deporte and Ministerio de Sanidad, Spain) and from the Generalitat de Catalunya. M.W. is a predoctoral fellow from the Ministerio de Educación, Cienca y Deporte. The Imperial College laboratory is supported by funding from the Biotechnology and Biological Sciences Research Council (grants B19088 and SF19107).
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Aguilar, A., S. Merino, X. Rubirés, and J. M. Tomás. 1997. The influence of osmolarity on lipopolysaccharide and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37°C. Infect. Immun. 651245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. K., and A. Newton. 1997. Posttranscriptional regulation of Caulobacter flagellin genes by late flagellum assembly checkpoint. J. Bacteriol. 1792281-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows, L. L., R. V. Urbanic, and J. S. Lam. 2000. Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect. Immun. 68931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canals, R., S. Vilches, M. Wilhelm, J. G. Shaw, S Merino, and J. M. Tomás. 2007. Non-structural flagella genes affecting both polar and lateral flagella mediated motility in Aeromonas hydrophila. Microbiology 1531165-1175. [DOI] [PubMed] [Google Scholar]
- 5.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 27626479-26485. [DOI] [PubMed] [Google Scholar]
- 6.Creuzenet, C. 2004. Characterization of Cj1293, a new UDP-GlcNAc C dehydratase from Campylobacter jejuni. FEBS Lett. 559136-140. [DOI] [PubMed] [Google Scholar]
- 7.Creuzenet, C., M. J. Schur, J. Liu, W. W. Wakarchuk, and J. S. Lam. 2000. FlaA1, a new bifunctional UDP-GlcNAc C6 dehydratase/C4 reductase from Helicobacter pylori. J. Biol. Chem. 27534873-34880. [DOI] [PubMed] [Google Scholar]
- 8.Dell, A., A. J. Reason, K.-H. Khoo, M. Panico, R. A. McDowell, and H. R. Morris. 1994. Mass spectrometry of carbohydrate-containing biopolymers. Methods Enzymol. 230108-132. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, J. Bult, J.-F. Tomb, B. Dougherty, J. M. Merrick, G. Sutton, W. FitzHugh, C. Fields, J. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, L. C. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 10.Gavín, R., A. A. Rabaan, S. Merino, J. M. Tomás, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43383-397. [DOI] [PubMed] [Google Scholar]
- 11.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50659-671. [DOI] [PubMed] [Google Scholar]
- 12.Gryllos, I., J. G. Shaw, R. Gavín, S. Merino, and J. M. Tomás. 2001. Role of flm locus in mesophilic Aeromonas adherence. Infect. Immun. 6965-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerry, P., C. P. Ewing, M. Schirm, M. Lorenzo, J. Kelly, D. Pattarini, G. Majam, P. Thibault, and S. Logan. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing nonantibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27332-344. [DOI] [PubMed] [Google Scholar]
- 16.Josenhans, C., L. Vossebein, S. Friedrich, and S. Suberbaum. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210165-172. [DOI] [PubMed] [Google Scholar]
- 17.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109137-141. [DOI] [PubMed] [Google Scholar]
- 18.Karlyshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148473-480. [DOI] [PubMed] [Google Scholar]
- 19.Kirov, S. M., T. C. Barnett, C. Pepe, M. S. Strom, and M. J. Albert. 2000. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 684040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirov, S. M., M. Castrisios, and J. G. Shaw. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 721939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 22.Kovach, M. E., R. W. Phillips, P. H. Elzer., R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16800-802. [PubMed] [Google Scholar]
- 23.Leclerc, G., S. Wang, and B. Ely. 1998. A new class of Caulobacter crescentus flagellar genes. J. Bacteriol. 1805010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linton, D., A. V. Karlyshev, P. G. Hitchen, H. R. Morris, A. Dell, N. A. Gregson, and B. W. Wren. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 351120-1134. [DOI] [PubMed] [Google Scholar]
- 25.McNally, D. J., J. P. M. Hui, A. J. Aubry, K. K. K. Mui, P. Guerry, J.-R. Brisson, S. M., Logan, and E. Soo. 2006. Functional characterisation of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 28118489-18498. [DOI] [PubMed] [Google Scholar]
- 26.Merino, S., X. Rubirés, A. Aguilar, and J. M. Tomás. 1996. The O:34-antigen lipopolysaccharide as an adhesin in Aeromonas hydrophila. FEMS Microbiol. Lett. 13997-101. [DOI] [PubMed] [Google Scholar]
- 27.Merkx-Jacques, A., R. K. Obhi, G. Bethune, and C. Creuzenet. 2004. The Helicobacter pylori flaA1 and wbpB genes control lipopolysaccharide and flagellum synthesis and function. J. Bacteriol. 1862253-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Namdari, H., and E. J. Bottone. 1990. Microbiologic and clinical evidence supporting the role of Aeromonas caviae as a pediatric enteric pathogen. J. Clin. Microbiol. 28837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okazaki, R., T. Okazaki, J. L. Strominger, and A. M. Michelson. 1962. Thymidine diphosphate-4-keto-6-deoxy-d-glucose, an intermediate in thymidine diphosphate l-rhamnose synthesis in Escherichia coli strains. J. Biol. Chem. 2373014-3026. [PubMed] [Google Scholar]
- 32.Paulsen, I. T., A. M. Beness, and M. H. Saier. 1997. Computer based analysis of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 1432685-2699. [DOI] [PubMed] [Google Scholar]
- 33.Power, P. M., and M. P. Jennings. 2003. The genetics of glycosylation in Gram-negative bacteria. FEMS Microbiol. Lett. 218211-222. [DOI] [PubMed] [Google Scholar]
- 34.Rabaan, A. A., I. Gryllos, J. M Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 694257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27493-497. [Google Scholar]
- 36.Schirm, M., I. C. Schoenhofen, S. M. Logan, K. C. Waldron, and P. Thibault. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem. 777774-7782. [DOI] [PubMed] [Google Scholar]
- 37.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 481579-1592. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11554-561. [DOI] [PubMed] [Google Scholar]
- 39.Schoenhofen, I. C., D. J. McNally, J.-R. Brisson, and S. M. Logan. 2006. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 168C-14C. [DOI] [PubMed] [Google Scholar]
- 40.Schoenhofen, I. C., D. J. McNally, E. Vinogradov, D. Whitfield, N. M. Youn, S. Dick, W. W. Wakarchuk, J.-R. Brisson, and S. M. Logan. 2006. Functional characterisation of the dehydratase/aminotransferase pairs from Helicobacter and Campylobacter. J. Biol. Chem. 281723-732. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer,. H. P., and R. Chuanchuen. 2001. Small broad-host-range lacZ operon fusion vector with low background activity. BioTechniques 311258-1262. [DOI] [PubMed] [Google Scholar]
- 42.Seshadri, R., S. Joseph, A. Chopra, J. Shaw, J. Graf, D. Haft, M. Wu, R. Madupu, M. J. Rosovitz, Q. Ren, L. Talon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. J. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: the makings of a pathogen. J. Bacteriol. 1888272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thibault, P., S. M. Logan, J. F. Kelly, J.-R Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 27634862-34870. [DOI] [PubMed] [Google Scholar]
- 44.Thornley, J. P., J. G. Shaw, I. A. Gryllos, and A. Eley. 1997. Virulence properties of clinically significant Aeromonas species. Rev. Med. Microbiol. 861-72. [Google Scholar]
- 45.Vimr, E. R., and F. A. Troy. 1985. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate lyase. J. Bacteriol. 164854-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilcox, M. H., A. M. Cook, A. Eley, and R. C. Spencer. 1992. Aeromonas spp. as a potential cause of diarrhoea in children. J. Clin. Pathol. 45959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]