Abstract
Bacterial proteins that are abnormally truncated due to incomplete mRNA or the presence of rare codons are extended by an SsrA tag during ribosome rescue in a trans-translation process important for maintaining protein quality. In Escherichia coli, the SsrA-tagged proteins become the target of the Tsp, Lon, FtsH, ClpXP, and ClpAP proteases. Here we show that degradation of model SsrA-tagged proteins in Streptococcus pneumoniae depends primarily or exclusively on ClpXP in vivo. In addition, we show the E. coli SsrA tag is also a target of S. pneumoniae ClpXP in vivo, even though the N-terminal portions of the tags differ significantly between the two species, suggesting there may be no adaptor protein for SsrA in S. pneumoniae.
Efficient use of translational machinery is disrupted by stalled ribosomes. Causes of such stalling include the presence of a rare codon, broken message, scarcity of tRNA, and others (21). To rescue stalled ribosomes and to relieve the stress caused by accumulation of stalled ribosomes and incompletely synthesized proteins, a unique RNA known as transfer message RNA (tmRNA) is recruited to the stalled ribosome (12). The tmRNA, also known as the ssrA RNA or 10S RNA, has dual roles, in which it acts both as a tRNA and as an mRNA (12). The polypeptide chain is provided with alanine on the stalled ribosome by charged tmRNA; then, the ribosome resumes protein synthesis, now directed by a small internal open reading frame of the tmRNA (12, 26). The result is addition of a small SsrA peptide tag at the C terminus and eventual release of the newly synthesized, but likely defective, polypeptide (12, 26). The SsrA peptide tag added to the incomplete polypeptide serves as a recognition signal for proteolysis. This mechanism, designated trans-translation, maintains the quality of proteins by eliminating many aberrant ones. ssrA genes are widely conserved among all bacteria and most encode an SsrA sequence of approximately 10 amino acids (11).
In Escherichia coli, five proteases (Tsp, FtsH, Lon, ClpXP, and ClpAP) recognize the SsrA sequence and can participate to some extent in degrading polypeptide chains attached to it (3, 6, 9, 24), but ClpXP is thought to be the major protease responsible for the degradation of SsrA-tagged proteins in vivo under normal conditions in two gram-negative bacterial species (E. coli and Caulobacter crescentus) and in the gram-positive Bacillus subtilis (2, 6, 27). An adaptor protein, SspB, in E. coli and its ortholog (SspBα) in C. crescentus promote degradation of SsrA-tagged proteins by ClpXP, while no similar adaptors are known in gram-positive bacteria (2, 7, 14, 15). For most other bacterial species, SsrA-tagged proteins are speculated to be substrates of ClpXP (10). However, ClpX is not conserved in all bacteria, while FtsH is, and for bacteria lacking ClpX, FtsH or Lon is considered to be the likely protease for SsrA-tagged proteins (8, 10). The mycoplasmas, for example, which lack ClpX and ClpP orthologs, possess a Lon protease with exceptional activity on SsrA-tagged substrates (8).
An SsrA tagging system is active in Streptococcus pneumoniae (pneumococcus), but the specific proteases responsible for the recognition and degradation of its SsrA-tagged proteins are unknown (18). The pneumococcal SsrA sequence (AKNNTSYALAA) resembles the one in E. coli (AANDENYALAA) in length and also in the sequence recognized directly by ClpX (underlined) (5). There are fewer cellular proteases in S. pneumoniae than in E. coli or in B. subtilis. As it has just five putative clp genes (clpC, clpE, clpL, clpP, and clpX) and carries ftsH, encoding a well-characterized membrane-bound protease, S. pneumoniae lacks genes for proteases of the Lon, Tsp, ClpA, and ClpYQ classes (20). Thus, ClpXP and FtsH are both good candidates for SsrA-specific proteases; however, it should be noted that while degradation by ClpXP in vivo requires the presence of the adaptor protein SspB in E. coli (4), no apparent pneumococcal homologs of SspB are known.
To identify pneumococcal cytoplasmic proteases that are involved in degrading SsrA-tagged proteins in vivo, we constructed synthetic genes for model SsrA-tagged proteins and determined the effects of individual protease mutations on the level of the tagged proteins. The results showed that proteins tagged with SsrA at their C terminus are strongly stabilized by mutations of clpX and clpP but not by mutations affecting any other Clp protease or FtsH and suggest that ClpXP recognition of SsrA tags may not require an adaptor protein.
GFP-SsrA is unstable in S. pneumoniae.
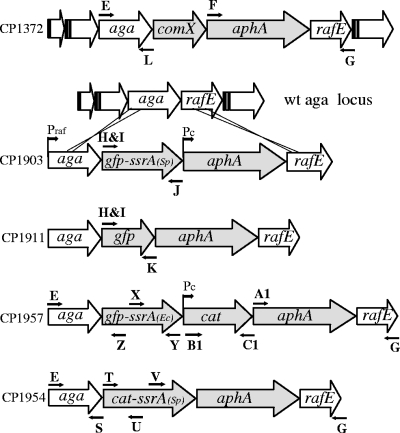
To determine the fate of GFP-SsrA in vivo, we constructed strains in which genes encoding GFP-SsrA or green fluorescent protein (GFP) were inserted in the chromosome of S. pneumoniae downstream of the native site of the gene aga, creating a transcriptional fusion dependent on the raffinose-inducible aga promoter (Table 1; Fig. 1). The resulting expression cassette, aga-gfp-ssrA-aphA, also includes a constitutive promoter for the kanamycin resistance marker, aphA. To ensure maximal expression of GFP-SsrA, a synthetic ribosome binding site, 5′-AGGAGGTA, was positioned 6 bases upstream of the start codon of GFP. This sequence is found at pneumococcal ribosomal protein genes and is predicted to be a strong ribosome binding site (23). The constructs were confirmed by PCR analysis using primers binding outside and within the insertions and by sequencing each entire insert (Table 2). As aga encodes the enzyme α-galactosidase, which hydrolyzes p-nitrophenyl-d-galactopyranoside to the yellow product p-nitrophenol, the expression of the synthetic operon can be readily assayed spectrophotometrically (22).
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| CP1250 | hex- malM511 str-1 bgl-1; Rx derivative, low β-galactosidase background | 19 |
| CP1343 | CP1250, but ΔclpL::erm; Ermr | 17 |
| CP1344 | CP1250, but ΔclpC::tet; Tetr | 17 |
| CP1359 | CP1250, but ΔclpP::tet; Tetr | 17 |
| CP1361 | CP1250, but ΔclpX::erm slx; Ermr | 17 |
| CP1363 | CP1250, but ΔftsH::erm; Ermr | 17 |
| CP1851 | CP1250, but ΔclpE::erm; Ermr | 25 |
| CP1903 | CP1250, but aga::gfp-ssrA::Pckan; Kanr | CP1250 × aga-gfp-ssrASp-kan-rafE |
| CP1904 | CP1903, but ΔclpP::tet; Kanr Tetr | CP1359 × CP1903 |
| CP1905 | CP1903, but ΔclpX::erm slx; Kanr Ermr | CP1361 × CP1903 |
| CP1906 | CP1903, but ΔclpC::tet; Kanr Tetr | CP1344 × CP1903 |
| CP1907 | CP1903, but ΔclpE::erm; Kanr Ermr | CP1851 × CP1903 |
| CP1903 | CP1903, but ΔclpL::erm; Kanr Ermr | CP1343 × CP1903 |
| CP1910 | CP1903, but ΔftsH::erm; Kanr Ermr | CP1363 × CP1903 |
| CP1911 | CP1250, but aga:: gfp::Pckan; Kanr | CP1250 × aga-gfp-kan-rafE |
| CP1912 | CP1911, but ΔclpP::tet; Kanr Tetr | CP1359 × CP1911 |
| CP1913 | CP1911, but ΔclpX::erm slx; Kanr Ermr | CP1361 × CP1911 |
| CP1914 | CP1911, but ΔclpC::tet; Kanr Tetr | CP1344 × CP1911 |
| CP1915 | CP1911, but ΔclpE::erm; Kanr Ermr | CP1851 × CP1911 |
| CP1916 | CP1911, but ΔclpL::erm; Kanr Ermr | CP1343 × CP1911 |
| CP1954 | CP1250, but aga::cat-ssrA::Pckan; Cmr Kanr | CP1250 × aga-cat-ssrA-kan-rafE |
| CP1957 | CP2000, but aga::gfp-ssrAEc::CAT::Pckan; Cmr Kanr | CP1250 × aga-gfp-ssrAEc-kan-rafE |
| CP1958 | CP1957, but ΔclpP::tet; Kanr Cmr Tetr | CP1359 × CP1957 |
| CP1959 | CP1957, but ΔclpX::erm slx; Kanr Ermr | CP1361 × CP1957 |
| CP1969 | CP1954, but ΔclpX::erm slx; Kanr Ermr Cmr | CP1359 × CP1954 |
| CP1971 | CP1954, but ΔclpP::tet; Kanr Cmr Tetr | CP1359 × CP1954 |
| CP2025 | CP1250, but ΔnagA::erm; Ermr | CP1250 × adh-erm-spr1868 |
FIG. 1.
Organization of the aga locus and construction of cassettes for regulated synthesis of GFP- and SrA-tagged proteins. Fragments containing a truncated aga gene (′aga) or the aphA (kanamycin resistance) gene and a truncated rafE gene (rafE′) were amplified with primer pairs E-L and F-G, respectively, from the CP1372 DNA template. gfp-mut2 was first cloned into pMAL-c2X after amplification from plasmid pKL147, kindly provided by Adam Driks (Loyola University), using primers A and B. ssrA was amplified from CP1250 with primers C and D and inserted downstream of gfp-mut2. A heterologous fragment with gfp-ssrA was amplified in two steps: one to add a synthetic ribosome binding site (primers H and J) using plasmid pMAL-c2X (gfp-ssrA) as a template and the second to add a restriction site to the first product (primers I and J). After digestion at restriction sites incorporated by the PCR primers, the second product was ligated with the targeting aga and aphArafE fragments as indicated. After transformation into CP1250 with Kan selection, the structure of the new synthetic locus in CP1903 was verified by sequencing. Filled arrows, genes manipulated during the construction; open arrows, conserved genes of the raffinose locus and its flanking genes. Promoters: Praf, aga promoter; Pc, artificial constitutive promoter derived from amiA. CP1911, CP1957, and CP1954 were constructed similarly but by using primer pairs H-K/I-K, E-Z/X-Y/B1-C1/A1-G, and E-S/T-U/V-G.
TABLE 2.
Oligonucleotides used in this study
| Primer | Lab name | Sequencea | Location |
|---|---|---|---|
| A | SA 47 | 5′-ggttgccggcATGAGTAAAGGAGAAGAACT | gfp |
| B | SA 48 | 5′-ggttggatccTTTGTATAGTTCATCCATGC | gfp |
| C | SA 51 | 5′-ggttgtcgacGCAAAAAATAACACTTCTTA | ssrASp |
| D | SA 52 | 5′-ggttaagcttTGGAGCCGGTGGGAGTCGAA | ssrA |
| E | SA 81 | 5′-ATATTCTCTTTGAGTCCTGCTCTGG | aga |
| F | SA 85 | 5′-ggttgccggcGGATCCGTTTGATTTTTAATGG | aphA |
| G | SA 86 | 5′-caacgatatcTTTGACTAACTGT | rafE |
| H | SA 90 | 5′-aggaggtaaatctaATGAGTAAAGGAGAAGAAC | gfp |
| I | SA 92 | 5′-ggttggtaccAGGAGGTAAATCTAATGAGT | gfp |
| J | SA 93 | 5′-ggttgccggcCAAACACCTGCCAACATATT | gfp-ssrASp |
| K | SA 94 | 5′-ggttgccggcCGACTCTAGAGGATCCTTATTT | gfp |
| L | SA 96 | 5′-ggttggtaccTCATAGTTTTCTAAAAATATACT | aga |
| M | SA 98 | 5′-ggttgccggcTTTGTATAGTTCATCCATGC | gfp |
| N | SA 100 | 5′-TGGCGAAGTTTACTCAGGTG | aga |
| O | SA 101 | 5′-TTTCCCGTTCCACATCATAGG | aphA |
| P | SA 102 | 5′-GCATGGCACTCTTGAAAAAGTC | gfp |
| Q | SA 103 | 5′-GTAACAGCTGCTGGGATTACAC | gfp |
| R | SA 104 | 5′-ATATAATGGTTCGGGGAAATTG | cat |
| S | SA 105 | 5′-ggttggatcCTCATAGTTTTCTAAAAATATACTG | aga |
| T | SA 106 | 5′-ggttggatccGCTTGATGAAAATTTGTTTGAT | cat |
| U | SA 107 | 5′-ggttctcgagTAAAAGCCAGTCATTAGGCC | cat |
| V | SA 108 | 5′-ggttctcgagGTAACAGCTGCTGGGATTAC | gfp |
| W | SA 109 | 5′-AGGAGTCCAAATACCAGAGAATG | cat |
| X | SA 120 | 5′-ggttggatccGCAAACGACGAAAACTACGC | ssrAEc |
| Y | SA 121 | 5′-ggttgtcgacCGTCCGAAATTCCTACATCC | ssrAEc |
| Z | SA 122 | 5′-ACTCTAGAGGATCCTTTGTATAG | gfp |
| A1 | SA 123 | 5′-ggtttctagaCGGTGGATCCGTTTGATTTT | aphA |
| B1 | SA 124 | 5′-ggttgtcgacCGGTATCGATAAGCTTGATGA | cat |
| C1 | SA 125 | 5′-ggtttctagaGGTTAGTGACATTAGAAAACCG | cat |
| D1 | SA 130 | 5′-GATCTGTCAATGGTTCAGATAC | spc |
| E1 | SA 131 | 5′-AAAGATATTGCGGGAAATGC | spc |
| F1 | SA 153 | 5′-ggttctcgagCAAAATTTGTTTGATTTGTATCT | erm |
| G1 | SA 154 | 5′-ggttggatccGTCGGCAGCGACTCATAG | erm |
| H1 | SA 157 | 5′-ggccggatccAGAGAGAAAGCAGAAGTTAGAG | adhA |
| I1 | SA 158 | 5′-GCCAGACAAGGAAAGAAATATG | adhA |
| J1 | SA 159 | 5′-TAATTGGAGTTTGGCAGTTG | SPR1868 |
| K1 | SA 160 | 5′-ggccctcgagTAGGCATAATGTTAACCTCCTT | SPR1868 |
Restriction nuclease recognition sites are underlined and nucleotides matching templates are in uppercase letters.
For induction of the aga locus with raffinose, cultures were grown in medium containing 0.1% raffinose and expression of the compound aga locus was monitored by determining α-galactosidase activity. Lysates of the wild type and of strains carrying GFP-SsrA or GFP constructs all displayed 10-fold induction of aga expression compared to controls without raffinose, indicating comparable expression levels for both gfp constructs (data not shown). Strains CP1903 and CP1911, expressing the GFP-SsrA or GFP genes, respectively, were examined for accumulation of GFP by a Western blotting assay using anti-GFP antibody. GFP-SsrA was not detectable, whereas untagged GFP was readily detected as a band at a position corresponding to the size of purified GFP (Fig. 2). We conclude that GFP-SsrA is degraded in vivo in the wild-type background but the untagged version of GFP is more stable, implying that the SsrA tag renders GFP a substrate of cytoplasmic proteases.
FIG. 2.
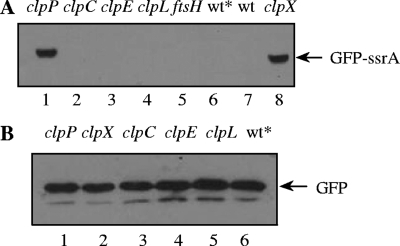
Stabilization of GFP-SsrA in clpX and clpP mutants. (A) Strains expressing gfp-ssrA in clpP (CP1904), clpC (CP1906), clpE (CP1907), clpL (CP1908), ftsH (CP1910), or clpX (CP1905) mutant backgrounds were harvested, lysed, and loaded in lanes 1 to 5 and 8 of a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. CP1903, expressing gfp-ssrA in the wild-type background (wt*), and the wild-type with no gfp insertion at the aga locus (wt) were loaded in lanes 6 and 7, respectively. (B) Strains expressing untagged gfp in clpP (CP1912), clpX (CP1913), clpC (CP1914), clpE (CP1915), or clpL (CP1916) mutant backgrounds or in the wild-type background (wt*; CP1911) were harvested, lysed, and loaded in lanes 1 to 6. Strains expressing gfp-ssrA or untagged gfp in protease-proficient backgrounds are represented as wt*. Cultures were grown to an optical density at 550 nm of 0.25 in a casein hydrolysate yeast extract medium (13) supplemented with raffinose (1 g/liter) for maximal induction of the aga locus. Cell pellets were resuspended and heated in a 1/100 volume of lysis buffer (100 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate, 0.2% bromophenol blue, 20% glycerol, and 200 mM dithiothreitol) for determination of GFP or GFP-SsrA by Western blotting with anti-GFP antibody (Roche). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to polyvinylidene difluoride membranes, the membranes were probed with a mouse anti-GFP primary antibody (1:1,000; Roche) and with an anti-mouse immunoglobulin G secondary antibody conjugated to horseradish peroxidase (1:10,000; GE). Detection was performed with an enhanced chemiluminescence substrate (ECL Plus; GE) and Hyblot CL film (Denville Scientific) with exposures between 10 and 600 s.
GFP-SsrA is stable in ClpP- and ClpX-deficient cells but unstable in all other protease mutants.
Since S. pneumoniae carries genes for ClpX, ClpP, and other Clp protease subunits, we asked whether ClpXP or another protease might participate in the degradation of SsrA-tagged GFP in pneumococcus by determining the accumulation of GFP-SsrA in clpX, clpP, clpC, clpE, clpL, and ftsH mutant strains. Although clpX was reported to be essential in S. pneumoniae (20), P. Luo obtained a deletion of clpX in the presence of an unlinked suppressor of this lethality (slx) (16; A. Piotrowski, personal communication), allowing the examination of clpX mutants reported here.
The level of untagged GFP was not elevated in any of these protease-deficient backgrounds, suggesting that GFP itself is not a substrate of any of the corresponding proteases (Fig. 2). GFP-SsrA was detected in clpX and clpP mutants but not in the wild-type background or in the other mutant strains (Fig. 2). The parallel effects of clpP and clpX mutations strongly suggest that the observed stabilizing effect of these mutations on GFP-SsrA arises from disruption of ClpXP protease activity and not from any polar effects of the two mutations. We conclude that ClpXP is the major pneumococcal Clp protease that degrades SsrA-tagged GFP. However, the possibility that FtsH or another protease has some minor role in degradation of SsrA-tagged proteins cannot be ruled out, as ClpX is present in the other protease mutants and could mask minor additional activities.
GFP tagged with E. coli SsrA is degraded in S. pneumoniae.
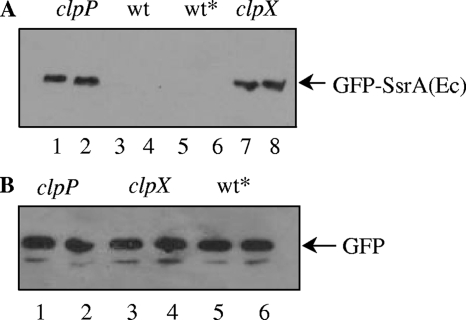
The sequence of the E. coli SsrA tag (SsrAEc), AANDENYALAA, is similar to that of S. pneumoniae (SsrASp), AKNNTSYALAA. The first 7 amino acids, AANDENY of the E. coli SsrA tag contain the binding site for its adaptor protein, SspB, while the terminal 3 amino acids (LAA) interact with ClpX (5). In vitro, SspB enhances the degradation rate of proteins tagged with SsrA by at least 10-fold and is required for their degradation by ClpXP in vivo (4). The three terminal residues of the pneumococcal SsrA tag are identical to those of the E. coli tag; the tags only differ in the portion specific for the adaptor protein. We reasoned that if S. pneumoniae requires an adaptor protein for ClpXP specific for AKNNTSY, then proteins tagged with SsrAEc would not be degraded, as the AANDENY portion of the tag recognized by the E. coli adaptor protein is considerably different in sequence and overall charge. To seek an indication of a pneumococcal adaptor with a different specificity for SsrA, we constructed a strain containing gfp fused to the ssrAEc tag and determined the levels of GFP-SsrAEc and GFP in vivo by Western blotting. Surprisingly, GFP-SsrAEc was completely absent from wild-type cells, while untagged GFP was readily detected in parallel controls, as expected. Suspecting that ClpXP may be responsible for the absence of GFP-SsrAEc, as it is for GFP-SsrASp, we transformed the SsrAEc construct into clpX and clpP mutants. GFP-SsrAEc was readily detectable in the clpX and clpP mutants at levels comparable to GFP-SsrASp in the same backgrounds (Fig. 3), strongly suggesting that ClpXP degrades GFP tagged with SsrAEc, just as it degrades GFP-SsrASp. This result implies that ClpXP likely does not require an adaptor to degrade SsrA-tagged protein in S. pneumoniae.
FIG. 3.
Targeting of E. coli SsrA by ClpXP in S. pneumoniae. (A) Strains expressing gfp-ssrAEc in clpP (CP1958) and clpX (CP1959) null mutant backgrounds were harvested, lysed, and loaded in lanes 1 and 2 and 7 and 8, respectively. The wild type with no gfp insertion at the aga locus (CP1250) and a strain expressing gfp-ssrAEc in the wild-type background (wt*; CP1957) were loaded in lanes 3 and 4 and lanes 5 and 6, respectively. (B) Strains expressing gfp in clpP (CP1912), clpX (CP1913), or wt (wt*; CP1911) backgrounds were loaded in lanes 1 and 2, 3 and 4, or 5 and 6, respectively. Cultures were induced with raffinose and prepared for Western analysis as described for Fig. 2, but all the samples were loaded in duplicate.
Degradation of SsrA-tagged CAT by ClpXP.
To test the generality of SsrA targeting by ClpXP and to seek an adaptor for SsrA by an independent method, we constructed strains expressing chloramphenicol (Cm) acetyltransferase (CAT) fused to a terminal SsrA tag. If CAT-SsrA were also targeted by ClpXP, Cm resistance could depend on loss of clpX, clpP, or loss of an adaptor for ClpXP. In the new cassette, cat-ssrA was expressed constitutively and was linked to a Kan (aphA) marker to allow selection independent of accumulation of CAT-SsrA. The structure of the new cassette (cat-ssrA-aphA) was confirmed by sequencing. In a clpX or clpP mutant background, the synthetic cat-ssrA gene conferred resistance to Cm comparable to that provided by the untagged CAT gene (Table 3), while the clpX+ clpP+ CAT-SsrA strain CP1954 was as sensitive to Cm as its wild-type parent, CP1250. Thus, CAT-SsrA was stabilized in clpX and clpP mutants, indicating that ClpXP degrades a second SsrA-tagged protein.
TABLE 3.
Stabilization of CAT-SsrA by clpP or clpX mutations
| Strain | CAT allele | Genotype
|
Cm MICa (μg/ml) | |
|---|---|---|---|---|
| clpX | clpP | |||
| CP1250 | + | + | 8 | |
| CP1359 | + | − | 4 | |
| CP1954 | cat-ssrA | + | + | 8 |
| CP1972 | cat-ssrA | + | − | 32 |
| CP1969 | cat-ssrA | − | + | 32 |
| CPM7 | pEVP3 cat | + | + | 32 |
The lowest level of Cm in a twofold dilution series at which colonies failed to appear during 20 h at 37°C.
Transformation of the Cm-sensitive cat-ssrA strain with a complex random spc insertion library (1) failed to reveal any insertions outside of clpX and clpP that stabilized the model SsrA-tagged CAT (data not shown). While this result is consistent with the conclusion that pneumococcal ClpXP acts without an adaptor to recognize SsrA, other explanations are also possible, including incomplete saturation by the mariner transposition reactions or essentiality of the putative adaptor. Degradation of SsrA-tagged proteins in vivo by ClpXP without the participation of an adaptor protein may not be exceptional; while this study was under way it was reported that degradation of SsrA-tagged proteins in B. subtilis similarly appears to lack an adaptor for ClpXP (7). Perhaps the amino-proximal portion of the gram-positive SsrA tag acts primarily as a spacer between its C-terminal ClpX-interacting portion and the body of the protein linked to it.
In summary, ClpXP appears to be the primary protease of S. pneumoniae that degrades proteins tagged with SsrA signals of S. pneumoniae or E. coli and it appears to act without the participation of an adaptor protein.
Nucleotide sequence accession numbers.
Nucleotide sequences of the new cassettes reported here have been deposited as GenBank accession numbers FJ495555 to FJ495558.
Acknowledgments
We thank Timothy E. Sommerville and Sume M. Joseph for assistance with the construction of gfp-ssrA and cat-ssrA strains and Peter Burghout for supplying the mariner T7 library.
This material is based upon work supported in part by the National Science Foundation under grant no. MCB 0543187.
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Bijlsma, J. J., P. Burghout, T. G. Kloosterman, H. J. Bootsma, A. de Jong, P. W. Hermans, and O. P. Kuipers. 2007. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl. Environ. Microbiol. 731514-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chien, P., B. S. Perchuk, M. T. Laub, R. T. Sauer, and T. A. Baker. 2007. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc. Natl. Acad. Sci. USA 1046590-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy, J. S., L. L. Aung, and A. W. Karzai. 2007. Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 1896564-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell, C. M., A. D. Grossman, and R. T. Sauer. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 571750-1761. [DOI] [PubMed] [Google Scholar]
- 5.Flynn, J. M., I. Levchenko, M. Seidel, S. H. Wickner, R. T. Sauer, and T. A. Baker. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA 9810584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 121338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith, K. L., and A. D. Grossman. 2008. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 701012-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gur, E., and R. T. Sauer. 2008. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proc. Natl. Acad. Sci. USA 10516113-16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman, C., D. Thevenet, P. Bouloc, G. C. Walker, and R. D'Ari. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 121348-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7449-455. [DOI] [PubMed] [Google Scholar]
- 11.Keiler, K. C., L. Shapiro, and K. P. Williams. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. USA 977778-77783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271990-993. [DOI] [PubMed] [Google Scholar]
- 13.Lee, M. S., B. A. Dougherty, A. C. Madeo, and D. A. Morrison. 1999. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl. Environ. Microbiol. 651883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessner, F. H., B. J. Venters, and K. C. Keiler. 2007. Proteolytic adaptor for transfer-messenger RNA-tagged proteins from α-proteobacteria. J. Bacteriol. 189272-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 2892354-2356. [DOI] [PubMed] [Google Scholar]
- 16.Luo, P. 2003. Ph.D. thesis. University of Illinois at Chicago, Chicago.
- 17.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molnos, J., R. Lange, and K. E. Amrein. 2000. An improved vector system for insertional gene inactivation inspired by the tmRNA-tagging system of S. pneumoniae. J. Microbiol. Methods 42197-201. [DOI] [PubMed] [Google Scholar]
- 19.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21853-862. [DOI] [PubMed] [Google Scholar]
- 20.Robertson, G. T., W. L. Ng, R. Gilmour, and M. E. Winkler. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 1852961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche, E. D., and R. T. Sauer. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 184579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenow, C., M. Maniar, and J. Trias. 1999. Regulation of the alpha-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 91189-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250144-155. [DOI] [PubMed] [Google Scholar]
- 24.Spiers, A., H. K. Lamb, S. Cocklin, K. A. Wheeler, J. Budworth, A. L. Dodds, M. J. Pallen, D. J. Maskell, I. G. Charles, and A. R. Hawkins. 2002. PDZ domains facilitate binding of high temperature requirement protease A (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of the small stable RNA A (ssrA)-encoded peptide. J. Biol. Chem. 27739443-39449. [DOI] [PubMed] [Google Scholar]
- 25.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 1873052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu, G. F., G. E. Reid, J. G. Zhang, R. L. Moritz, and R. J. Simpson. 1995. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 2709322-9326. [DOI] [PubMed] [Google Scholar]
- 27.Wiegert, T., and W. Schumann. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 1833885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]