Abstract
SigH regulates a transcriptional network that responds to heat and oxidative stress in mycobacteria. Seven sigH paralogs are reported to exist in the Mycobacterium smegmatis genome. A comprehensive real-time reverse transcriptase PCR analysis during different stages of growth and upon exposure to various stress conditions and antimycobacterial compounds showed differential expression of sigH paralogs during stationary phase and severalfold increases in the levels of transcription of sigH1, sigH4, sigH5, sigH6, and sigH7 under specific stress conditions.
Sigma factors play a major role in the regulation of bacterial gene expression, and their contents vary considerably in different mycobacterial genomes (18). The recently sequenced Mycobacterium smegmatis genome is predicted to encode 26 sigma factors, which is twice the number present in Mycobacterium tuberculosis (13 sigma factors) (23). M. smegmatis contains orthologs of all M. tuberculosis sigma factors except sigC, sigI, and sigK. There is an enrichment of the sigH subfamily in this species that contains seven paralogs (23). SigH, an extracytoplasmic function (ECF) sigma factor, is a key regulator of a transcriptional network that responds to oxidative and heat stresses in mycobacteria (17). M. smegmatis and M. tuberculosis SigH are highly similar proteins, and their levels are found to be increased by heat shock and oxidative stress in both species. However, an M. tuberculosis sigH mutant is more susceptible to heat and oxidative stress. An M. smegmatis sigH mutant showed survival at 53°C, like the wild type, but increased sensitivity toward cumene hydroperoxide (5). Similar survival instincts of the sigH mutant and the wild type suggest the presence of multiple mechanisms to counter these stresses in this species. Although the regulatory mechanism of sigH expression and sigH's role have been worked out (13, 16, 22), the multiplicity of its paralogous circuit in M. smegmatis remains to be analyzed. Determining the conditions under which the expression of sigH paralogs are induced or repressed is vital to the understanding of their possible function and role in the regulation of gene expression in M. smegmatis in response to different environmental stimuli. We examined the expression of sigH paralogs in M. smegmatis at different stages of growth and upon exposure to various stress conditions, like heat shock, cold shock, nutrient starvation, oxidative stress, and antibiotic stress, using quantitative real-time reverse transcriptase PCR.
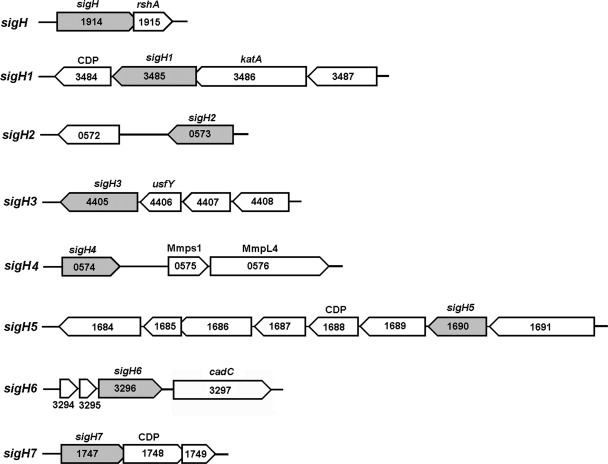
Organization of sigH paralogs in the M. smegmatis genome.
ECF sigma factors are known to exist as operons in several bacterial genomes (8, 24). Of the eight sigH subfamily members, present at different loci in the M. smegmatis genome, six are clustered in putative operons, while sigH2 and sigH4 are monocistronic (Fig. 1). sigH overlaps rshA, a gene encoding sigH's cognate anti-sigma factor. The sigH1 and sigH3 operons include four genes; sigH1 is the third gene in its operon and is followed by a gene encoding a putative transcriptional regulator with cupin domains (MSMEG_3484), while sigH3 is the last gene in its operon and is preceded by usfY (MSMEG_4406) and the MSMEG_4408 gene, both of which are predicted to encode membrane proteins. The monocistronic units sigH2 (MSMEG_0573) and sigH4 (MSMEG_0574) are present on complementary strands; sigH2 is followed by genes encoding a series of hypothetical proteins, and sigH4 is followed by genes encoding two mycobacterial transmembrane proteins, Mmps1 (MSMEG_0575) and MmpL4 (MSMEG_0576). sigH5 is preceded by a transcriptional regulatory protein (MSMEG_1691) in a putative eight-gene operon and followed by an oxoacyl reductase (MSMEG_3484) and a cupin domain protein (MSMEG_3484). sigH6 is the last gene of a putative tricistronic operon; it is preceded by two genes encoding hypothetical proteins and followed by a gene encoding a transcriptional regulator protein (MSMEG_3297) after a gap consisting of a 175-bp intergenic region. sigH7 is the first gene in a putative tricistronic operon and overlaps a gene encoding a cupin domain protein (MSMEG_1748).
FIG. 1.
Genomic organization of sigH paralogs in M. smegmatis. Sigma factors other than sigH2 and sigH4 are arranged in a polycistronic operon. Open reading frames encoding cupin domain proteins (CDP) and putative membrane proteins follow sigH paralogs in the putative operon. Blunt-tipped arrowheads indicate the overlap of open reading frames.
Expression of sigH paralogs during growth and stress conditions.
M. smegmatis strain mc2155 was grown at 37°C in 7H9 medium, and 10-ml cultures were removed at timely intervals of 12 h, 24 h, 48 h, and 72 h and processed for RNA isolation, as described previously (20). For stress experiments, aliquots of exponentially growing cultures were subjected to various treatments for 4 h: cold shock and heat shock (15°C and 53°C, respectively), nutrient starvation (phosphate-buffered saline, pH 7.0), and oxidative stress (10 mM H2O2). For antibiotic treatments, compounds at their high-end critical concentrations, namely, isoniazid (200 μg/ml), rifampin (rifampicin) (200 μg/ml), ethambutol (5 μg/ml), and streptomycin (2 μg/ml) (20), were added to the culture, which was incubated for 4 h and then processed for RNA isolation. About 5.0 μg of DNA-free RNA was reverse transcribed using random hexamers and Moloney murine leukemia virus reverse transcriptase, and cDNA was purified, diluted five times with sterile water, and processed for quantitative real-time PCR using appropriate primers of sigH paralogs (see Table S1 in the supplemental material) and Roche's SYBR green I master kit on a model 480 LightCycler (Roche Diagnostics). Melt curve analysis was performed to ensure the homogeneity of each amplicon. Relative expression levels were determined after normalization with the sigA transcript level using LightCycler 480 II software version 1.5 (Roche, Germany). The average relative expression levels and standard deviations were determined from the data generated from three independent experiments.
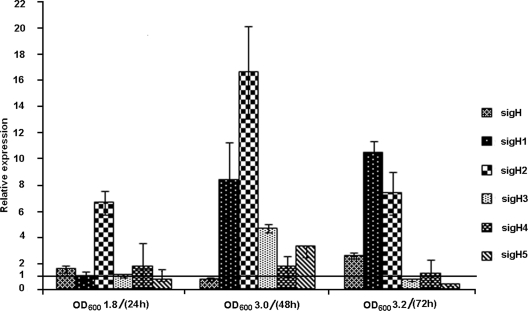
First, we examined the expression of sigH paralogs from the early exponential phase to the late stationary phase of growth. Figure 2 shows the relative expression levels of sigH paralogs at the various phases of growth. The transcript level of sigH increased marginally during late log phase, declined to nearly the control level in stationary phase, and then increased to 2.6-fold during late stationary phase. sigH1 expression remained relatively constant during the early and late log phases but gradually increased to 8.3-fold and 10.4-fold during the stationary and late stationary phases, respectively. sigH2 showed a gradual increase of 6.6-fold during log phase, an increase of 16.6-fold during stationary phase, and then a decrease to 7.3-fold in late stationary phase. sigH3 and sigH5 levels were induced to 4.7-fold and 3.2-fold, respectively, during stationary phase but declined to much lower levels during late stationary phase. sigH4 did not respond to stationary phase. sigH6 and sigH7 transcripts were far below the control levels during these stages. An M. smegmatis sigH mutant displayed survival in the logarithmic and stationary phases of growth similar to that of the wild type (5). The lower level of sigH expression indicates the lesser dependence of M. smegmatis on sigH-mediated gene expression during different stages of growth. Further, it is possible that the parallel expression of sigH paralogs would have compensated for the loss incurred by sigH in the mutant strain and thereby ensured the similar survival rates of the wild type and sigH mutant.
FIG. 2.
Relative levels of expression of sigH paralogs at the late log (optical density at 600 nm [OD600] of 1.8), stationary (OD600 of 3.0), and extended stationary (OD600 of 3.2) phases. Expression levels of sigH paralogs at early log phase (OD600 of 0.5) were considered controls (line at 1 on the y axis). sigH6 and sigH7 transcripts were far below the control level.
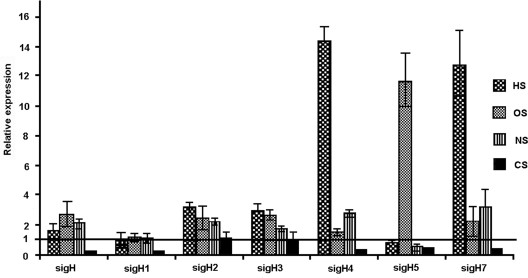
M. smegmatis, as a saprophyte, encounters several environmental stresses in its ecological niche. We examined the expression of sigH paralogs after subjecting M. smegmatis cultures to different stress conditions. Heat stress induced the transcript levels of sigH2 (3.2-fold), sigH3 (2.9-fold), sigH4 (14-fold), and sigH7 (12.6-fold) (Fig. 3). A sudden rise in the transcription of sigH4 and sigH7 upon heat shock suggests a specific role for these sigma factors in regulating the expression of genes that help bacteria overcome heat stress. A low level of sigH induction (1.6-fold) during heat stress possibly is the reason behind the similar survival instincts of the sigH mutant and its wild-type counterpart after heat shock, as reported earlier (5). Transcript levels of sigH1 and sigH5 were relatively comparable to their control levels of expression. Cold shock did not induce the expression of any sigH subfamily genes; moreover, sigH paralogs except sigH2 and sigH3, whose expression levels did not change, showed a decline in their transcript levels with respect to those of the untreated control. The sigH6 transcript was at an almost-negligible level under these conditions.
FIG. 3.
Relative levels of expression of sigH paralogs during heat shock (HS), oxidative stress (OS), cold shock (CS), and nutrient starvation (NS). Note the severalfold increases in sigH4 and sigH7 levels during heat shock and in the sigH5 level during oxidative stress. The sigH1 level remains unchanged, and other genes are variably induced. Expression levels without treatment were taken as controls (line at 1 on the y axis). The sigH6 transcript remains almost undetectable.
Exponentially growing M. smegmatis cultures quickly adapt to a sudden nutrient depletion and can survive for a long period under starvation conditions (21). We simulated nutrient starvation by incubating the M. smegmatis cultures in phosphate-buffered saline. This treatment stimulated levels of sigH (2.1-fold), sigH2 (2.2-fold), sigH3 (1.6-fold), sigH4 (2.8-fold), and sigH7 (3.2-fold) expression, while sigH1 remained unaltered and sigH5 showed a decline compared to its expression in the untreated control (Fig. 3). The sigH6 transcript remained almost undetectable.
M. smegmatis mounts a protective oxidative-stress response (19). An M. smegmatis sigH mutant showed more susceptibility to organic peroxides than to hydrogen peroxides (5). To examine the role of sigH paralogs in hydrogen peroxide-mediated stress, we treated M. smegmatis cultures with H2O2 and monitored the expression of sigH paralogs under treatment conditions. The majority of sigH paralogs responded to oxidative stress, a marked feature of sigH family proteins. We observed increased levels of expression of sigH (2.7-fold), sigH2 (2.5-fold), sigH3 (2.6-fold), and sigH7 (2.2-fold), while sigH1 levels remained almost unchanged and sigH4 levels marginally increased with respect to levels in the untreated control (Fig. 3). A dramatic rise in sigH5 (12-fold) transcript level was particularly noticeable, as its expression did not increase during any other stress conditions applied. It may be recalled that sigH5 showed enhanced expression during stationary phase and that M. smegmatis stationary-phase cells were shown to respond better to hydrogen peroxide stress (21). Interestingly, M. smegmatis sigma factors SigB and SigF, which were also shown to render resistance to peroxide stress, showed increased expression during stationary phase (7, 12). These findings reinforce the notion that bacteria adapt to particular stress conditions by regulating the expression of different subsets of genes, orchestrated by an overlapping network of ECF sigma factors. It is tempting to speculate a role for sigH5 in a transcriptional network that confers protection against hydrogen peroxide stress in M. smegmatis.
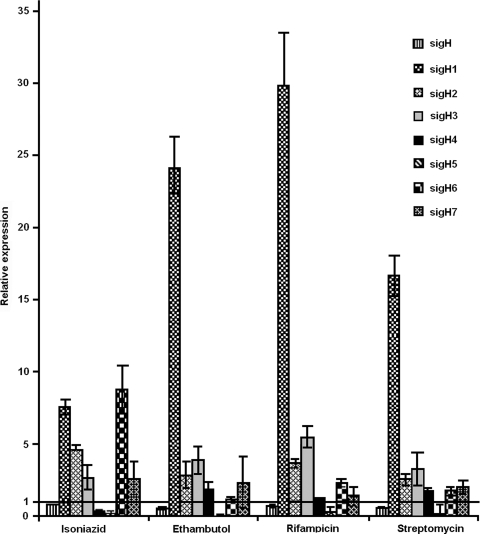
M. smegmatis is naturally less susceptible to some of the primary-line antimycobacterial drugs, like isoniazid and rifampin (10, 14). An interdependence of oxidative-stress response and isoniazid resistance has been reported to occur among mycobacterial species (3, 4). In M. tuberculosis, an extensive transcriptional network responding to oxidative stress was shown to be regulated by SigH (17). To investigate the role of M. smegmatis sigH paralogs in response to various drugs, we examined the expression of these sigma factors after subjecting M. smegmatis cultures to critical concentrations of antimycobacterial drugs. Antibiotic stress stimulated various transcript levels of sigH paralogs, among which an exemplary increase in sigH1 expression in response to all four of the antibiotics isoniazid (7.5-fold), ethambutol (24-fold), rifampin (30-fold), and streptomycin (17-fold) was particularly noticeable (Fig. 4). Earlier, enhanced sigH1 expression was noticed only during the stationary and extended stationary phases, not during other stress conditions applied. Since sigH1 responded markedly to different antibiotics with dissimilar modes of actions, it is difficult to speculate on a role for sigH1 in mounting a unifying mechanism whereby a single transcription factor might confer resistance to multiple drugs. In Bacillus subtilis, another saprophytic soil microbe like M. smegmatis, an ECF sigma factor, SigW, showed enhanced expression in response to several antibiotics with different modes of actions and to toxic peptides (9). SigW was found to regulate the expression of an ∼60-gene regulon, which encodes proteins that inactivate, sequester, or eliminate toxic compounds from the cell (1). It is possible that sigH1 controls the expression of genes which provide intrinsic resistance to a range of antimicrobial compounds and other toxic products that cells accumulate during stationary phase and upon exposure to different kinds of drugs. This needs to be further studied. Another noticeable observation was the maximally induced level of sigH6 (ninefold) in response to isoniazid treatment. It may be recalled that sigH6 transcripts were found to be at negligible levels during different stages of growth and upon exposure to other stress conditions applied in this study. Since sigH6 did not respond to ethambutol, it appears that common cell wall biosynthesis inhibitors do not regulate sigH6 transcription. Notably, isoniazid affects the cell wall biosynthesis process by inhibiting the FAS-II elongation pathway of mycolic acid biosynthesis. It would be of interest to examine the status of sigH6 transcripts in response to other FAS-II pathway inhibitors. Further studies are required to understand the role of SigH6 in helping M. smegmatis overcome isoniazid stress, as it is more tolerant to this antimycobacterial drug.
FIG. 4.
Levels of expression of sigH paralogs upon exposure to antimycobacterial compounds relative to untreated-control expression levels (line at 1 on the y axis). sigH1 is highly induced in response to all antibiotics, while other genes show various levels of expression. The sigH6 transcript is maximally induced in response to isoniazid.
SigH regulates the transcriptional network that responds to oxidative, nitrosative, and heat stresses in mycobacteria (13, 17). An M. smegmatis sigH mutant showed increased sensitivity to oxidative stress by cumene hydroperoxide but no defect in survival at 53°C or oxidative stress due to hydrogen peroxide (5). However, an M. smegmatis sigH sigE double mutant was impaired in survival after both heat shock and oxidative stress by cumene hydroperoxide (5). SigH has been shown to be responsible for the stress-inducible expression of SigE, and both SigH and SigE are required for the full induction of SigB (17). Recently, a sigF mutant of M. smegmatis was found to be significantly impaired in survival after a 50°C heat shock and hydrogen peroxide treatment (7, 15). It is possible that SigF of M. smegmatis either belongs to a SigH-mediated network of transcriptional regulation or provides a separate, additional mechanism of response to these stress conditions. The role of SigF and SigH in the resistance of M. smegmatis to oxidative stress is also highlighted with the recent observation that the promoter of dps, a gene expressed under conditions of oxidative stress and starvation, can be recognized only by RNA polymerase containing one of these sigma factors (2). These findings highlight the regulatory overlap of different sigma factors that work together to allow adaptation to heat and oxidative stress in mycobacteria.
In view of the apparent redundancy of sigma factors mounting overlapping stress responses, we examined the expression of different sigH paralogs in M. smegmatis and observed a diverse and distinctive expression of sigH paralogs during growth and under different stress conditions. The majority of them responded to heat shock and oxidative stresses, a particular feature of the SigH family of sigma factors. Some of them showed a fairly higher-level expression during the stationary and late stationary phases, presumably to impart a better tolerance to these cells against changing physiological states. Overlapping expression of these sigma factors under similar physiological and stress conditions reinstates the earlier reported redundancy of ECF sigma factors. However, an exemplary rise in the expression of sigH4 and sigH7 in response to heat shock, sigH5 in response to H2O2, sigH1 in response to antibiotics, and sigH6 in response to isoniazid treatment suggests that, probably, these sigma factors are specifically regulated by appropriate signaling molecules to orchestrate the regulation of a selected set of genes. ECF sigma factors are cotranscribed with one or more negative regulators and were reported to interact with the products of their neighboring genes residing in the same operon (24). Often, these include a transmembrane protein with an extracytoplasmic sensory domain and an intracellular inhibitory domain functioning as an anti-sigma factor that binds and inhibits its cognate sigma factor (16). SigH activity is transcriptionally regulated at its autoregulated promoter and posttranslationally via interaction with its cognate anti-sigma factor, RshA, whose gene resides in the same operon (13). The presence of cupin domain protein genes downstream of sigH1, sigH5, and sigH7 is noteworthy. Transcriptional regulators with cupin domains were reported to be present in several microbial genomes: M. tuberculosis (Rv3833, AraC family of transcriptional regulators), B. subtilis (ydeC), and Pseudomonas aeruginosa (pae-1, a heat shock regulator) (11). Interestingly, MSMEG_3484, a cupin domain protein gene present downstream of sigH1 in its putative operon also possesses an AraC binding domain and is hypothesized to be a transcriptional regulator. Transcriptional regulators of the AraC family were shown to regulate the expression of several stress-responsive genes in bacteria (6). It would be of interest to study the interactions of the cupin domain proteins and other membrane proteins, whose genes are present in the vicinity of sigH paralogs, and examine their roles as putative anti-sigma factors to their cognate sigma factors. It would help us decode the signaling cascade operating to orchestrate the complex regulatory circuits of heat and oxidative stress through sigH subfamily members. The generation of mutants with single and multiple deletions of sigH paralogs with subsequent transcriptome analysis would enable the identification of the regulon of these sigH family proteins. A more comprehensive study is required to delineate the overlapping network of a regulon mediated by sigH paralogs in M. smegmatis during growth and in response to various stress conditions. It would reveal the biological significance of an unusual expansion of the sigH family of sigma factors in M. smegmatis.
Supplementary Material
Acknowledgments
This work was supported by CSIR network project CMM0018 on Alternative Animal Model. Anirudh K. Singh is the recipient of a CSIR research fellowship.
We are thankful to Preethi Srinivasan (BITS trainee) for her initial technical assistance.
This is communication no. 7682 of the CDRI.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis sigmaW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316443-457. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury, R. P., S. Gupta, and D. Chatterji. 2007. Identification and characterization of the dps promoter of Mycobacterium smegmatis: promoter recognition by stress-specific extracytoplasmic function sigma factors σH and σF. J. Bacteriol. 1898973-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, S. T. 1994. Mycobacterium tuberculosis: drug-resistance mechanisms. Trends Microbiol. 2411-415. [DOI] [PubMed] [Google Scholar]
- 4.Dhandayuthapani, S., Y. Zhang, M. H. Mudd, and V. Deretic. 1996. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J. Bacteriol. 1783641-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes, N. D., Q. L. Wu, D. Kong, X. Puyang, S. Garg, and R. N. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 1814266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frota, C. C., K. G. Papavinasasundaram, E. O. Davis, and M. J. Colston. 2004. The AraC family transcriptional regulator Rv1931c plays a role in the virulence of Mycobacterium tuberculosis. Infect. Immun. 725483-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebhard, S., A. Hümpel, A. D. McLellan, and G. M. Cook. 2008. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 1542786-2795. [DOI] [PubMed] [Google Scholar]
- 8.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 4647-110. [DOI] [PubMed] [Google Scholar]
- 9.Helmann, J. D. 2006. Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds. Sci. Prog. 89243-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 12311-18. [DOI] [PubMed] [Google Scholar]
- 11.Khuri, S., F. T. Bakker, and J. M. Dunwell. 2001. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 18593-605. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee, R., and D. Chatterji. 2005. Evaluation of the role of sigma B in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 338964-972. [DOI] [PubMed] [Google Scholar]
- 13.Park, S. T., C. M. Kang, and R. N. Husson. 2008. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 10513105-13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piddock, L. J., K. J. Williams, and V. Ricci. 2000. Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Antimicrob. Chemother. 45159-165. [DOI] [PubMed] [Google Scholar]
- 15.Provvedi, R., D. Kocincova, V. Dona, D. Euphrasie, M. Daffe, G. Etienne, R. Manganelli, and J. M. Reyrat. 2008. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacterial. 1907859-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raman, S., A. Cascioferro, R. N. Husson, and R. Manganelli. 2008. Mycobacterial sigma factors and surface biology, p. 223-234. In M. Daffe and J. M. Reyrat (ed.), The mycobacterial cell envelope. ASM Press, Washington, DC.
- 17.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 1836119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30926-994. [DOI] [PubMed] [Google Scholar]
- 19.Sherman, D. R., P. J. Sabo, M. J. Hickey, T. M. Arain, G. G. Mahairas, Y. Yuan, C. E. Barry III, and C. K. Stover. 1995. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc. Natl. Acad. Sci. USA 926625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh, A. K., and B. N. Singh. 2008. Conservation of sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr. Microbiol. 56574-580. [DOI] [PubMed] [Google Scholar]
- 21.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50949-959. [DOI] [PubMed] [Google Scholar]
- 23.Waagmeester, A., J. Thompson, and J. M. Reyrat. 2005. Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 13505-509. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150591-599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.