Abstract
The Mycobacterium tuberculosis cmk gene, predicted to encode a CMP kinase (CMK), was cloned and expressed, and its product was purified to homogeneity. Steady-state kinetics confirmed that M. tuberculosis CMK is a monomer that preferentially phosphorylates CMP and dCMP by a sequential mechanism. A plausible role for CMK is discussed.
Nucleoside monophosphate (NMP) kinases are key enzymes in the metabolism of ribo- and deoxyribonucleoside triphosphates, catalyzing the reversible phosphoryl transfer from a nucleoside triphosphate (usually ATP) to a specific NMP (25). The resulting nucleoside diphosphates are subsequently phosphorylated to generate nucleoside triphosphates, precursors of nucleic acids. Bacterial CMP kinase (CMK), which is part of pyrimidine nucleotide interconversion pathways, catalyzes the transfer of a phosphate group from ATP to either CMP or dCMP to form CDP or dCDP and ADP, respectively (1). In Mycobacterium tuberculosis, the causative agent of human tuberculosis, a putative cmk (Rv1712) gene has been identified in the genome of the H37Rv strain by sequence similarity (8).
In addition, it has been proposed that the cmk gene product is essential for the optimal in vitro growth of M. tuberculosis based on transposon site hybridization studies (15). However, this approach is a large-scale screening methodology, and hence, cmk gene manipulation experiments must be carried out to firmly support the hypothesis of its essentiality in M. tuberculosis. The first step should thus be demonstration that the Rv1712 locus indeed encodes a protein having CMK activity in M. tuberculosis. Here, we report heterologous recombinant protein expression, purification to homogeneity, N-terminal amino acid sequencing, electrospray ionization-mass spectrometry (ESI-MS) analysis, and size exclusion chromatography of a functional cmk-encoded M. tuberculosis CMK (MtCMK). Steady-state kinetics showed that MtCMK preferentially phosphorylates CMP and dCMP, and double-reciprocal plots indicated a sequential mechanism for MtCMK with ternary complex formation. The availability of the MtCMK protein in large quantities will allow further functional and structural efforts to be undertaken in order to provide a framework on which to base the design of chemical compounds that inhibit the enzyme activity.
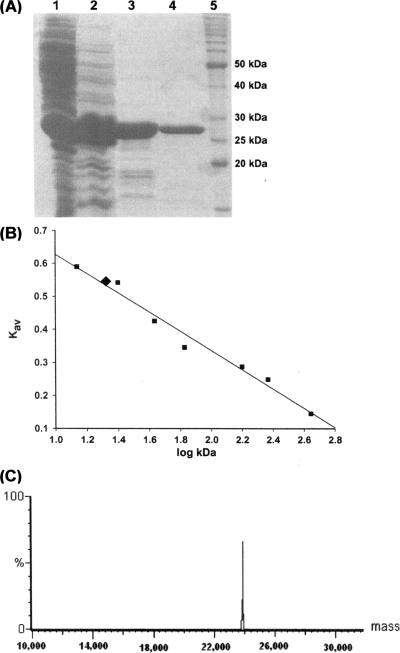
A 693-bp fragment consistent with the size expected for the predicted cmk coding sequence was PCR amplified from M. tuberculosis H37Rv genomic DNA (see Fig. S1A in the supplemental material). For directional cloning, a forward primer (5′-GGCATATGAGTCGCCTAAGCGCAGCGGTAGT-3′) and a reverse primer (5′-GTGGATCCTCACCGCACTGCCTCACTTCGC-3′) were designed to contain, respectively, NdeI and BamHI restriction sites (underlined). The PCR fragment was purified and ligated into the pET-23a(+) expression vector (Novagen), and its sequence was determined by automated DNA sequencing. The resulting pET-23a(+)::cmk plasmid was introduced into Escherichia coli BL21(DE3) by electroporation and selected on LB plates containing 50 μg/ml ampicillin. Three liters of LB broth containing 50 μg/ml ampicillin was inoculated with a single colony of MtCMK-expressing E. coli and incubated for 9 h (after cells had reached an optical density at 600 nm of 0.4) at 37°C at 180 rpm. No isopropyl-β-d-thiogalactopyranoside was added to the culture because this was the best experimental condition we found for expression of recombinant MtCMK in soluble form in E. coli (see Fig. S1B in the supplemental material). Approximately 10 g of cells was resuspended in 60 ml of 50 mM Tris-HCl buffer (pH 7.5) containing 0.2 mg/ml lysozyme and disrupted by sonication on ice (10 times for 10 s at an amplitude of 60% in a Vibra-Cell ultrasonic processor), and debris was removed by centrifugation at 48,000 × g for 30 min. The supernatant was incubated with 1% (wt/vol) streptomycin sulfate and centrifuged at 48,000 × g for 30 min. The resulting supernatant was purified to homogeneity by a three-step protocol consisting of an anionic-exchange column, a gel filtration column, and a strong anionic-exchange resin, yielding 5 mg of recombinant protein per liter of cell culture (Fig. 1A). The preparation was stored at −80°C with no apparent loss of activity, and recombinant MtCMK was stable for up to 30 days at 4°C. Analytical gel filtration chromatography revealed a single peak of approximately 21 kDa (Fig. 1B), indicating that MtCMK is a monomer in solution. The first 16 N-terminal amino acid residues of MtCMK were determined by the Edman degradation method, unambiguously identifying the homogeneous recombinant protein as MtCMK and confirming removal of the N-terminal methionine. ESI-MS analysis revealed just one peak at the expected mass for the MtCMK subunit (23,930.56 Da; Fig. 1C), which is in agreement with removal of the N-terminal methionine (predicted molecular mass including methionine, 24,061.12 Da).
FIG. 1.
Purification, determination of oligomeric state by gel filtration, and ESI-MS analysis of MtCMK. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of fractions obtained during the purification of MtCMK. Lanes: 1, crude extract after dialysis; 2 through 4, MtCMK after elution on Q-Sepharose Fast Flow, Sephacryl S200, and Mono Q HR columns, respectively; 5, protein molecular size standards (50, 40, 30, 25, and 20 kDa; Invitrogen). (B) The linear relationship between the average elution position (Kav) and the logarithm of the molecular masses (log kDa) of protein standards (squares) allowed the determination of 21 kDa as the molecular mass of native MtCMK (diamond). (C) ESI-MS analysis revealing a single peak at 23,930.56 Da.
To prove the correct in silico assignment to the M. tuberculosis Rv1712 locus, the biological activity of recombinant MtCMK was probed by steady-state kinetics. MtCMK activity was assayed in the forward direction by coupling the ADP product formation to the pyruvate kinase and lactate dehydrogenase reactions by following the protocol described by others (2, 12). The true steady-state kinetic constants (Table 1) and substrate specificity were determined for CMP, dCMP, and UMP (see Fig. S2, S3, and S4, respectively, in the supplemental material). The double-reciprocal plots showed intersecting patterns for all of the substrates tested, consistent with ternary complex formation and a sequential mechanism. Double-reciprocal plots intersect to the left of the y axis and thus rule out a rapid equilibrium ordered mechanism. Similar intersecting initial velocity patterns have been reported for E. coli (6). The kcat/Km ratio is an apparent second-order rate constant that determines the specificity for competing substrates (9). Accordingly, the results presented here demonstrate that MtCMK preferentially phosphorylates CMP and dCMP and that UMP is a poor substrate.
TABLE 1.
True steady-state kinetic parameters of MtCMK with three NMPs and ATP as a phosphate donor
| Phosphate acceptor | Mean Km (μM) ± SE | Mean Vmax (U/mg) ± SE | Mean kcat (s−1) ± SE | kcat/Km ratio (M−1 s−1) |
|---|---|---|---|---|
| CMP | 120 ± 9 | 131 ± 4 | 52 ± 2 | 1.1 × 106 |
| dCMP | 165 ± 7 | 75 ± 2 | 30 ± 1 | 0.2 × 106 |
| UMP | 13,854 ± 771 | 32 ± 1 | 12.2 ± 0.4 | 0.88 × 103 |
A promising target for drug development should be essential for the survival of a pathogen and absent from its host. Alternatively, the target may play an important role in adaptation of the pathogen to a particular physiological state of the host. There are two major pathways for pyrimidine nucleotide synthesis: the de novo pathway and the salvage pathway. Since de novo pyrimidine ribonucleotide synthesis requires higher energy levels, cells use the salvage pathway to reutilize pyrimidine bases and nucleosides derived from preformed nucleotides (10).
Hydrophilic agents traverse the mycobacterial cell wall slowly because of mycobacterial porin inefficiency in the permeation of solutes and the low concentration of porins. Lipophilic agents are retarded by the lipid bilayer, which is of unusually low fluidity (17). Internalization of chemical compounds having a negative net charge into cells is hampered by the net negative charge of the mycobacterial cell wall (4). Therefore, mono-, di-, or triphosphate nucleosides are not likely to enter the mycobacterial cell unless there is a transport system to carry out this process. Notwithstanding, no transporters for bases, nucleosides, or nucleotides of nucleic acids could be identified in M. tuberculosis (3). It is thus likely that M. tuberculosis has to recycle bases, nucleosides, and/or nucleotides to survive in the hostile environment offered by host macrophages. In general, pyrimidine bases and nucleosides, which are the transportable precursors of nucleotides, are not available as exogenous nutrients to most bacteria. It has been estimated that thymidine and uridine, for instance, may be available to mycobacteria growing in the host at a concentration range of 0.5 to 5.5 μM (22). It was described that Mycobacterium leprae, the causative agent of leprosy, is able to incorporate exogenously supplied pyrimidines as bases or nucleosides, but not as a nucleotide, into its nucleic acids (22). Besides M. leprae, it was reported that other pathogenic mycobacteria (such as Mycobacterium avium and Mycobacterium microti) cannot take up uridine nucleotides directly, but they are able to utilize the pyrimidines by hydrolyzing them to uridine and then taking up the latter (23). Exogenous bases are usually transported into the cell by specific membrane proteins, such as cytosine, uracil, and xanthine permeases. However, none of these have been described in the genome of M. tuberculosis or in those of other relevant pathogenic mycobacteria.
Although CMP is not produced in the de novo pathway, it might accumulate either from CTP during the synthesis of phospholipids or from the hydrolytic cleavage of mRNA. Therefore, the physiological role of CMK is also to recycle CMP to CDP, which is either rapidly phosphorylated by the unspecific nucleoside diphosphate kinase NdkA to CTP or reduced to dCDP (6). In bacteria, CDP (as well as ADP, UDP, or GDP) can also result from the phosphorolytic cleavage of mRNA by polynucleotide phosphorylase (7, 14). It therefore seems worthwhile to look for a possible link between these two CDP-producing enzymes, i.e., CMK and polynucleotide phosphorylase. The cmk gene is located in the M. tuberculosis chromosome between engA (probable GTP-binding protein) and Rv1711 (probable RNA pseudouridylate synthase). The EngA family is thought to act as a cellular messenger by forming interactions with the ribosome, with overexpression of EngA in E. coli restoring the growth of mutants null for an rRNA methyltransferase which modifies the 23S rRNA in intact 50S ribosomal subunits (24). The 16S and 23S rRNAs of E. coli contain 11 pseudouridines, which are synthesized by seven pseudouridine synthases (19). Although speculative, the location of the cmk gene in the M. tuberculosis chromosome may support its role in recycling of nucleotides derived from RNA degradation.
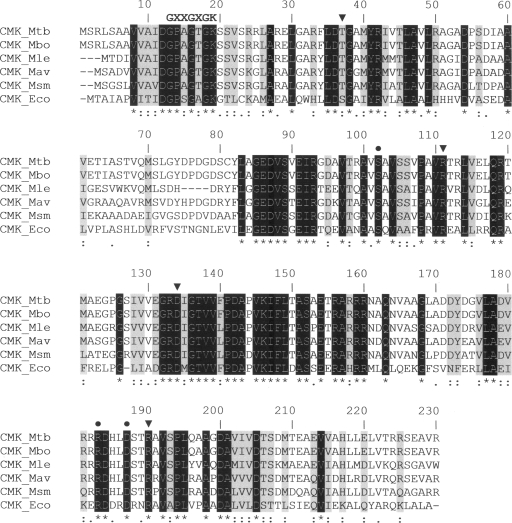
NMP kinases usually exhibit a high degree of sequence identity at the amino acid level, despite variations observed in their substrate specificity (11, 13, 20). NMP kinases are composed of three domains: the CORE, the LID (which closes upon binding of the phosphate donor ATP), and the NMP-binding domain (21). NMP kinases are divided into short and long forms. The latter group consists of adenylate kinases with an insertion of around 27 residues in the LID domain (1). Bacterial CMKs represent a third distinct family of NMP kinases, as they possess a short LID domain but have an insertion of 40 amino acid residues in the NMP-binding domain (5). A multiple sequence alignment of mycobacterial CMKs and E. coli CMK demonstrates that the P-loop sequence is conserved and that amino acid residues interacting with the pyrimidine ring and pentose moiety of CMP are conserved (Fig. 2). These structural differences could be exploited in the development of novel inhibitors targeted more specifically toward M. tuberculosis and other pathogenic mycobacteria. The results presented here provide a solid foundation to carry out genetic studies (16) to demonstrate the role, if any, of cmk in M. tuberculosis survival in the human host.
FIG. 2.
Multiple sequence alignment of mycobacterial CMKs with the known three-dimensional structure of E. coli CMK (CMK_Eco). The putative CMK sequences of M. tuberculosis (Mtb), M. bovis (Mbo), M. leprae (Mle), M. avium (Mav), and M. smegmatis (Msm) were aligned with CMK_Eco by using the default settings of the CLUSTALW program (18). Identical conserved residues are shown in white on a black background and are also indicated by asterisks below the alignment. Strongly similar and weakly similar residues are identified by colons and periods, respectively. Strongly similar residues are also shaded in gray. Triangles and circles above the alignment indicate residues that interact with the pyrimidine ring of CMP (Ser36, Arg110, Asp132, and Arg188 in CMK_Eco) and those that are key in pentose recognition (Ser101, Arg181, and Asp185 in CMK_Eco), respectively, as determined by site-directed mutagenesis and crystallographic studies (1, 13). The conserved fingerprint sequence GXXGXGK (where X stands for any amino acid) of NMP kinases, which forms the phosphate-binding loop (P loop), is indicated at the N-terminal portion of CMKs (6).
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Millennium Initiative Program and the National Institute of Science and Technology on Tuberculosis, MCT-CNPq, Ministry of Health-Department of Science and Technology (Brazil) to D.S.S. and L.A.B. D.S.S. (304051/1975-06), L.A.B. (520182/99-5), and M.S.P. (500079/90-0) are National Council for Scientific and Technological Development of Brazil (CNPq) research career award recipients. C.T. was supported by a studentship from FARMASA.
Footnotes
Published ahead of print on 30 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bertrand, T., P. Briozzo, L. Assairi, A. Ofiteru, N. Bucurenci, H. Munier-Lehmann, B. Golinelli-Pimpaneau, O. Bârzu, and A. M. Gilles. 2002. Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme. J. Mol. Biol. 3151099-1110. [DOI] [PubMed] [Google Scholar]
- 2.Blondin, C., L. Serina, L. Wiesmüller, A. M. Gilles, and O. Bârzu. 1994. Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal. Biochem. 220219-221. [DOI] [PubMed] [Google Scholar]
- 3.Braibant, M., P. Gilot, and J. Content. 2000. The ATP cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24449-467. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 5.Briozzo, P., B. Golinelli-Pimpaneau, A. M. Gilles, J. F. Gaucher, S. Burlacu-Miron, H. Sakamoto, J. Janin, and O. Bârzu. 1998. Structures of Escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure 61517-1527. [DOI] [PubMed] [Google Scholar]
- 6.Bucurenci, N., H. Sakamoto, P. Briozzo, N. Palibroda, L. Serina, R. S. Sarfati, G. Labesse, G. Briand, A. Danchin, O. Bârzu, and A. M. Gilles. 1996. CMP kinase from Escherichia coli is structurally related to other nucleoside monophosphate kinases. J. Biol. Chem. 2712856-2862. [DOI] [PubMed] [Google Scholar]
- 7.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76889-900. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 9.Fersht, A. 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman & Co., New York, NY.
- 10.Islam, M. R., H. Kim, S. W. Kang, J. S. Kim, Y. M. Jeong, H. J. Hwang, S. Y. Lee, J. C. Woo, and S. G. Kim. 2007. Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Mol. Biol. 63465-477. [DOI] [PubMed] [Google Scholar]
- 11.Liou, J. Y., G. E. Dutschman, W. Lam, Z. Jiang, and Y. C. Cheng. 2002. Characterization of human UMP/CMP kinase and its phosphorylation of d- and l-form deoxycytidine analogue monophosphates. Cancer Res. 621624-1631. [PubMed] [Google Scholar]
- 12.Millar, G., A. Lewendon, M. G. Hunter, and J. R. Coggins. 1986. The cloning and expression of the aroL gene from Escherichia coli K12. Purification and complete amino acid sequence of shikimate kinase II, the aroL-gene product. Biochem. J. 237427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofiteru, A., N. Bucurenci, E. Alexov, T. Bertrand, P. Briozzo, H. Munier-Lehmann, and A. M. Gilles. 2007. Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase. FEBS J. 2743363-3373. [DOI] [PubMed] [Google Scholar]
- 14.Py, B., H. Causton, E. A. Mudd, and C. F. Higgins. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14717-729. [DOI] [PubMed] [Google Scholar]
- 15.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 16.Schneider, C. Z., T. Parish, L. A. Basso, and D. S. Santos. 2008. The two chorismate mutases from both Mycobacterium tuberculosis and Mycobacterium smegmatis: biochemical analysis and limited regulation of promoter activity by aromatic amino acids. J. Bacteriol. 190122-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder, E. K., O. N. de Souza, D. S. Santos, J. S. Blanchard, and L. A. Basso. 2002. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis. Curr. Pharm. Biotechnol. 3197-225. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidyanathan, P. P., M. P. Deutscher, and A. Malhotra. 2007. RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA 131868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rompay, A. R., M. Johansson, and A. Karlsson. 1999. Phosphorylation of deoxycytidine analog monophosphates by UMP-CMP kinase: molecular characterization of the human enzyme. Mol. Pharmacol. 56562-569. [DOI] [PubMed] [Google Scholar]
- 21.Vonrhein, C., G. J. Schlauderer, and G. E. Schulz. 1995. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure 3483-490. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler, P. R. 1989. Pyrimidine scavenging by Mycobacterium leprae. FEMS Microbiol. Lett. 48179-184. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler, P. R. 1990. Biosynthesis and scavenging of pyrimidines by pathogenic mycobacteria. J. Gen. Microbiol. 136189-201. [DOI] [PubMed] [Google Scholar]
- 24.Xu, L., S. P. Muench, A. Roujeinikova, S. E. Sedelnikova, and D. W. Rice. 2006. Cloning, purification and preliminary crystallographic analysis of the Bacillus subtilis GTPase YphC-GDP complex. Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 62435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan, H., and M. D. Tsai. 1999. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol. 73103-134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.