Abstract
Three mutants deficient in hydrogen/formate uptake were obtained through screening of a transposon mutant library containing 5,760 mutants of Desulfovibrio desulfuricans G20. Mutations were in the genes encoding the type I tetraheme cytochrome c3 (cycA), Fe hydrogenase (hydB), and molybdopterin oxidoreductase (mopB). Mutations did not decrease the ability of cells to produce H2 or formate during growth. Complementation of the cycA and mopB mutants with a plasmid carrying the intact cycA and/or mopB gene and the putative promoter from the parental strain allowed the recovery of H2 uptake ability, showing that these specific genes are involved in H2 oxidation. The mop operon encodes a periplasm-facing transmembrane protein complex which may shuttle electrons from periplasmic cytochrome c3 to the menaquinone pool. Electrons can then be used for sulfate reduction in the cytoplasm.
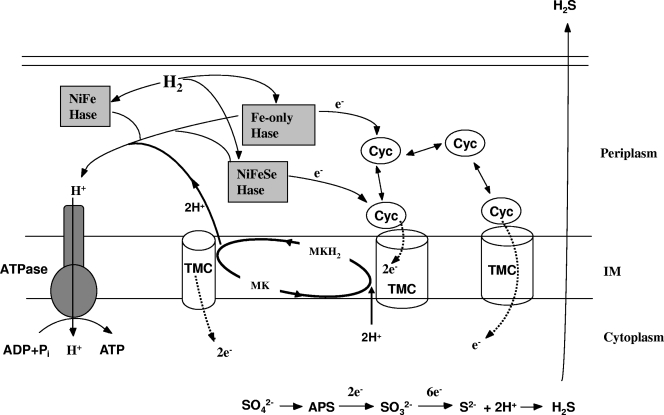
Members of the genus Desulfovibrio are gram-negative sulfate-reducing bacteria that derive energy from the dissimilatory reduction of sulfate coupled to the oxidation of dihydrogen (H2) or organic substrates (1, 35, 38). During the oxidation of organic compounds or when exogenous H2 is available, H2 is thought to diffuse into the periplasm, where it can be oxidized by periplasmic hydrogenases (1, 24) (Fig. 1). The electrons generated from H2 oxidation in the periplasm are transferred into the cytochrome network (14) for delivery to inner membrane electron conduit proteins. The latter process may involve transfer through the menaquinone pool, allowing cells to couple H2 oxidation to the generation of a proton motive force (PMF) (Fig. 1). Eventually, electrons are transferred to cytoplasmic proteins for the reduction of sulfate.
FIG. 1.
Proposed conceptual model for the electron transfer pathway of H2 oxidation by Desulfovibrio. Three classes of periplasmic hydrogenases (NiFe, NiFeSe, and Fe-only hydrogenases [Hase]) may be involved in H2 metabolism. The electrons (e−) generated from H2 oxidation in the periplasm are transferred to the cytochrome c3 (Cyc) network for delivery to transmembrane electron carriers (TMC) and then possibly into the menaquinone (MK) pool, creating reduced menaquinone (MKH2) within the inner membrane (IM) or directly across the inner membrane for sulfate reduction in the cytoplasm. The Mop complex described here may be one of these TMC. The protons generated from H2 oxidation in the periplasm drive ATP synthesis (by the ATP synthase), as they contribute directly to a proton gradient. The protons imported through ATP synthase are also used in the sulfate reduction pathway. H2 is either supplied externally or produced in the cytoplasm through organic substrate degradation.
Hydrogenases in Desulfovibrio species catalyze both H2 oxidation and production reactions. Three classes of periplasmic hydrogenases, NiFe hydrogenase, NiFeSe hydrogenase, and Fe-only hydrogenase, are present in Desulfovibrio (20, 31, 36, 37). Cytochrome c3 has been suggested previously to be the physiological electron carrier for hydrogenases in Desulfovibrio vulgaris (14). Conducting electrons across the cytoplasmic membrane from periplasmically oxidized H2, ultimately reducing sulfate (38), requires membrane proteins or transmembrane complexes, and several, including the Hmc and Tmc protein complexes, have been proposed as possible electron transfer channels (23, 27, 28, 34). However, physiological evidence supporting a role for these complexes in transferring electrons from H2 is still lacking (23, 25, 38).
While many proteins that are thought to be involved in H2 metabolism and bioenergetic pathways within Desulfovibrio species have been studied, the connections between them and the complete electron transfer pathways remain to be resolved (38). In order to identify novel genes involved in H2 metabolism in D. desulfuricans G20, we screened a mutant library constructed in a previous study (13). The library was screened individually for those mutants deficient in syntrophic growth with Methanospirillum hungatei on lactate as an electron donor. We reasoned that as H2 is the proposed electron carrier among syntrophic partners, a defect in syntrophic growth would likely include the inability to produce or consume H2. The screening resulted in the identification of three mutants unable or slow to grow on H2. The mutations included changes in the Fe-only hydrogenase and cytochrome c3, whose functions in H2 oxidation are reasonably well understood (14, 35, 38). This result demonstrated the efficacy of the screening strategy and provided a proof of principle for the process. Furthermore, we obtained a mutant with a mutation in a putative transmembrane protein complex, a molybdopterin oxidoreductase (MopABCD), which may be a potential electron conduit protein for H2 oxidation (Fig. 1).
MATERIALS AND METHODS
Medium and culture growth.
A mineral salts (MS) medium was used to grow D. desulfuricans G20. Each liter of MS medium contained 50 ml of solution A (KH2PO4, 8 g/liter; NH4Cl, 20 g/liter; MgCl2·6H2O, 4 g/liter; CaCl2·2H2O, 1 g/liter), 50 ml of solution B (K2HPO4, 8 g/liter), 10 ml of trace metal solution (nitrilotriacetic acid, 12.8 g/liter; FeSO4·7H2O, 0.42 g/liter; MnCl2·4H2O, 0.1 g/liter; CoCl2·6H2O, 0.17 g/liter; CuCl2·2H2O, 0.02 g/liter; ZnSO4·7H2O, 0.21 g/liter; Na2MoO4·2H2O, 0.01 g/liter), 10 ml of Wolfe's vitamin solution [biotin, 2 mg/liter; folic acid, 2 mg/liter; pyridoxine HCl, 10 mg/liter; thiamine HCl, 5 mg/liter; riboflavin, 5 mg/liter; nicotinic acid, 5 mg/liter; calcium d-(+)-pantothenate, 5 mg/liter; cyanocobalamin, 0.1 mg/liter; p-aminobenzoic acid, 5 mg/liter; thioctic acid, 5 mg/liter], 3.52 g of NaHCO3, 1 g of yeast extract, and 1 ml of resazurin (0.1%). The medium was reduced with a solution of cysteine and Na2S (at 1.25% each) by adding 0.05 ml to 5 ml of medium. Strain G20 was grown in serum tubes (23 ml) containing 5 ml of MS medium, and growth was monitored using a spectrophotometer at 600 nm (to determine the optical density at 600 nm [OD600]). When lactate (50 mM) or formate (50 mM) was used as the electron donor, the headspace was N2/CO2 (80/20, vol/vol). For studies with H2 as the electron donor, the headspace was flushed with H2/CO2 (80/20, vol/vol). Acetate (10 mM) was included in cultures with formate or H2 as the sole electron donor. Sulfate at 50 mM was added as the electron acceptor unless otherwise indicated.
Mutant screening.
The mutant library of D. desulfuricans G20 with 5,760 mutants was constructed using a mini-Tn10 transposon-bearing plasmid, which was shown to mutagenize D. desulfuricans G20 efficiently and randomly. This library therefore provides about 1.5-fold coverage of the 3,775 candidate protein-encoding genes found in the G20 genome. The detailed procedures for the construction and validation of this mutant library have been described previously (13).
For the screening of potential mutations related to H2 metabolism, syntrophic cocultures were established by the inoculation of early-stationary-phase cultures of M. hungatei JF-1 (ATCC 27890; 1 ml at an OD600 of 0.7) and individual G20 mutants (0.1 ml at an OD600 of 0.7) into 5 ml of MS-lactate medium in a serum tube (23 ml). The OD600 was routinely measured to monitor growth. Syntrophic cocultures containing the G20 parental strain reached a maximum OD within 4 to 5 days. Mutants that grew significantly more slowly than the parental strain (reaching a maximum OD600 2 or more days later) or did not grow were identified as potential targets, after which their growth in H2-sulfate medium was further tested.
Identification of the insertion site.
To identify the transposon insertion site within the chromosomes of mutants, the region within the genome surrounding the transposon insertion site was amplified using two rounds of arbitrarily primed PCR as described previously (8). Primer sequences used in this study are given in the supplemental material. To determine which gene was interrupted, sequences obtained from the PCR products were compared to sequences in the NCBI database and to the D. desulfuricans G20 genome sequence in the GenBank database (accession no. NC_007519) by using BLASTN. Protein sequence analysis was done and transmembrane helix predictions were made using programs available at http://us.expasy.org/.
Complementation experiments.
The entire mop, hyd, and cyc operons, as well as 100 to 500 bases of flanking DNA (see the supplemental material), were each amplified from the parental strain genomic DNA by using phosphorylated gene-specific primers (see the supplemental material) and Phusion high-fidelity DNA polymerase (Finnzymes Oy, Finland) to obtain blunt-end PCR products. The PCR products included, for mop, the Dde_2932 to Dde_2935 loci (cloned fragment position in the genome, 2912471 to 2917382; 4,912 bp); for hyd, the Dde_0081 and Dde_0082 loci (75350 to 77578; 2,229 bp); and for cycA, the Dde_3182 locus (3169617 to 3170418; 802 bp). Entire operons, including promoter regions, were amplified, as an upstream insertion may have the effect of inactivating the entire downstream part of the operon. The blunt-end PCR products were then purified and ligated into plasmid pMO719 by using T4 DNA ligase. pMO719 was constructed by cloning the pBG1 cassette (which contains a Desulfovibrio replicon) (39) into the EcoRI sites of pCR8/GW/TOPO (Invitrogen) with spectinomycin as a selective marker (K. L. Keller et al., unpublished data). Before ligation, pMO719 was digested with EcoRV and dephosphorylated using Antarctic phosphatase (New England Biolabs, Ipswich, MA). Escherichia coli GC5 competent cells (GeneChoice Inc, Frederick, MD) were transformed with the ligated products. Plasmids containing the correct inserts were subsequently isolated, and D. desulfuricans G20 was transformed with the plasmids by electroporation as described previously (19). For the identification of plasmids with the correct insertions, selected colonies grown on Luria-Bertani agar plates (for E. coli) or lactate-sulfate agar plates (for D. desulfuricans G20) (13) (both with 800 μg/ml spectinomycin) were picked and grown in Luria-Bertani or MS liquid medium (both with 400 μg/ml spectinomycin) overnight. Plasmids were extracted and digested with PvuII. pMO719 was used as a negative control. The digested products were separated on 1% agarose gels, and band patterns were compared. The putative positive constructs were further verified by sequencing the region flanking the insertion.
Enzyme assays.
Whole-cell suspensions were prepared anaerobically by washing cells twice in anoxic 50 mM Tris-HCl, pH 8.0 (containing 2 mM dithiothreitol), subjecting the cells to centrifugation at 8,000 × g for 10 min, and finally resuspending the pellet in the same buffer. Hydrogenase and formate dehydrogenase activities were routinely assayed by photometric measurement of the reduction at 578 nm of 1 mM methyl viologen (MV; ɛ578 = 9.7 mM−1 cm−1) or 1 mM benzyl viologen (BV; ɛ578 = 9.2 mM−1 cm−1) at 25°C with hydrogen or formate (10 mM) as the electron donor, respectively. For the hydrogenase assay, absorbance at 578 nm was monitored in anaerobic cuvettes filled with an oxygen scavenging system (0.5 U/ml glucose oxidase, 250 U/ml catalase, and 2.5 mM glucose) in 50 mM Tris-HCl, pH 8.0. Cuvettes were flushed with H2 for 1 min. For formate dehydrogenase, the assay was conducted in 50 mM Tris-HCl, pH 8.0 (containing 2 mM dithiothreitol), in rubber-stoppered glass cuvettes flushed with nitrogen for 1 min. The reaction was started by the addition of cells, and the kinetics of reduction was recorded. Controls without an electron donor in the assay mixture were conducted simultaneously. The specific activity was defined as the micromoles of MV or BV reduced per minute per milligram of protein.
Chemical analyses.
For the analysis of the protein concentration, 0.2 ml of the cell suspension was added to 0.2 ml of 1 N NaOH and the mixture was incubated at 37°C for 4 h. The protein concentration was measured using the Bradford protein assay (5) with bovine serum albumin as the standard.
Formate, lactate, and acetate in the culture were measured with a high-pressure liquid chromatography system equipped with an Aminex HPX-87H column (Bio-Rad) and a refractive index detector. H2 in the headspace was measured with a reduced gas analyzer (Trace Analytical, Inc.), and the level of H2 is expressed as the total amount of H2 produced, in micromoles, and corrected with respect to the volume of the culture.
RESULTS AND DISCUSSION
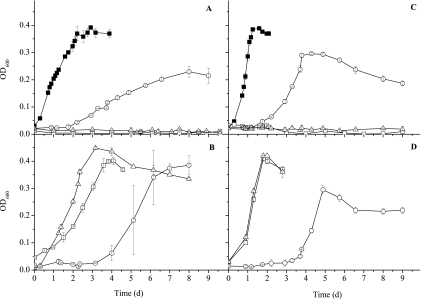
Through our screening process, three mutants deficient in growth in the presence of H2-sulfate were obtained. These mutants had transposon insertions in genes annotated as encoding the tetraheme cytochrome c3 (Dde_3182; cycA), the iron hydrogenase small subunit (Dde_0082; hydB), and the molybdopterin oxidoreductase molybdopterin-binding subunit (Dde_2933; mopB). The three mutants exhibited growth rates similar to that of the parental strain when lactate was the electron donor (data not shown). When formate or H2 was used as the electron donor and sulfate was the electron acceptor, the parental strain reached a maximum OD600 of 0.3 to 0.4 within 2 to 4 days. However, the mopB and cycA mutants were unable to grow on formate or H2 during the 10-day incubation period, and the hydB mutant grew more slowly than the parental strain (Fig. 2). With pyruvate as the electron donor, the three mutants grew more slowly and reached a lower maximum OD600 than the parental strain (Table 1).
FIG. 2.
Growth curves comparing the D. desulfuricans G20 parent (▪), mopB mutant (□), cycA mutant (▵), and hydB mutant (○) grown in MS medium containing 50 mM sulfate with 110-kPa partial H2 pressure in the headspace (A and B) or 50 mM formate (C and D). Panels A and C show data for transposon mutants and the parental strain, and panels B and D show data for complemented mutants. All the values are means of two measurements with average deviations.
TABLE 1.
Comparison of the growth rates and motilities of transposon mutants with those of the corresponding complemented mutants
| Characteristic | Phenotypea of:
|
||||||
|---|---|---|---|---|---|---|---|
| Mutants
|
Parental strain | Complemented mutants
|
|||||
| cycA::Tn10 | mopB::Tn10 | hydB::Tn10 | cycA::Tn10 (pXZL3182) | mopB::Tn10 (pXZL2933) | hydB::Tn10 (pXZL0082) | ||
| Growth on: | |||||||
| Lactate-SO42− | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| H2SO42− | − | − | + | +++ | +++ | +++ | ++ |
| Formate-SO42− | − | − | + | +++ | +++ | +++ | + |
| Pyruvate-SO42− | + | + | ++ | +++ | +++ | +++ | ++ |
| Syntrophy with JF-1 | − | − | ++ | +++ | +++ | +++ | ++ |
| Motility | − | + | + | +++ | ++ | ++ | ++ |
+++, ++, +, and − indicate growth or motility relative to that of the parental strain, with +++ being equivalent to the level of the parental strain.
To determine whether the observed phenotypes resulted from the elimination of the specific gene in which the insertion was detected or from the impact on surrounding genes, the mutants were individually complemented to be certain that the phenotype could be restored. After the cloning of the entire complementary operons, as well as 100 to 500 bases of flanking DNA, the complemented mopB and cycA mutants recovered the ability to growth on H2 and formate. Growth rates of the complemented strains were similar to that of parental strain (Fig. 2), confirming that the identified genes are indeed involved in H2- and formate-dependent growth. Lag times were somewhat longer for the complemented mutants growing on H2 than for the parental strain, and the reason for this finding has not been determined. The hydB mutant was able to grow to some extent on H2 and formate, and complemented hydB mutants grew better on H2 but not better on formate than noncomplemented mutants (Fig. 2). The lag time observed during the growth of the hydB mutant or complemented mutants was not reduced during subsequent transfer of the cultures. Complemented mopB and cycA mutants completely recovered the ability to grow syntrophically with M. hungatei, and pyruvate-grown cultures reached an OD600 similar to that of the parental strain culture (Table 1). The OD600 of the complemented hydB mutant culture grown on pyruvate was, however, lower than that of the parental strain culture. The three mutants were all much less motile than the parental strain based on microscopic observations. All complemented mutants recovered motility to some degree, especially the cycA mutant; however, complemented mutants still were less motile than the parental strain. The hydB mutant may have been difficult to complement due to the fact that the Fe-only hydrogenase requires maturation machinery and the correct temporal, spatial, and stoichiometric production of the subunits together with the maturation proteins (4).
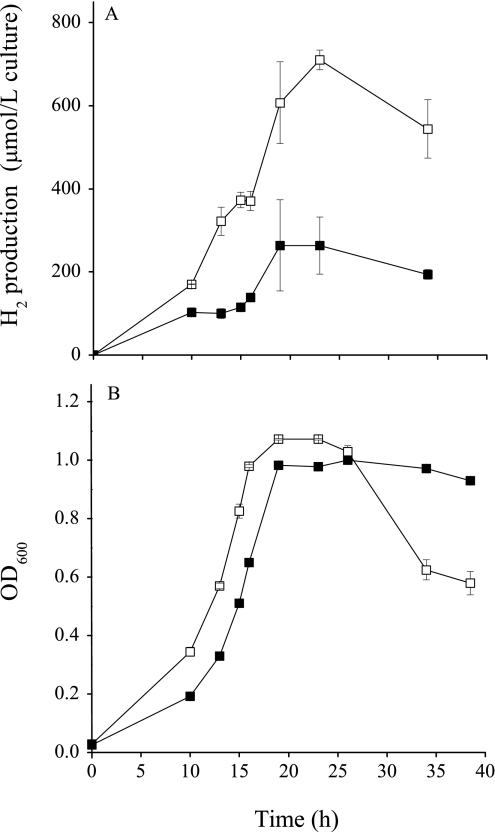
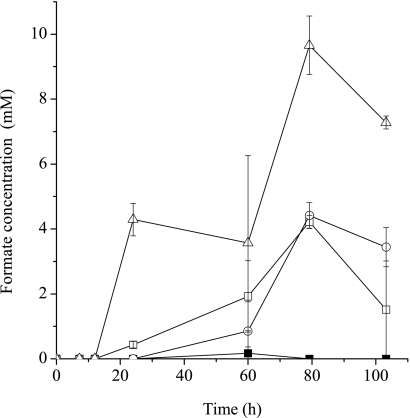
H2 levels in cultures grown with lactate, pyruvate, or formate in the presence of sulfate were monitored. Although the mutants and the parental strain grew similarly on lactate, results consistently showed that H2 accumulated to significantly higher levels in mutant cultures than in the parental strain culture (Fig. 3). H2 was not detected in the 7-day-old parental strain culture but accumulated in mutant cultures with all three above-listed substrates (date not shown). The rapid decline in the OD600 of the mopB mutant culture after 25 h of growth (Fig. 3) may be due to the inability of the mopB mutant to reuse H2 present from lactate oxidation to sustain biomass levels. Formate was also produced during growth with lactate and accumulated to much higher levels during the growth of mutants than during the growth of the parental strain (Fig. 4). The accumulation of formate and H2 in all of the mutant cultures was observed, regardless of whether the mutants were grown under donor- or acceptor-limited conditions (data not shown). We speculate that high H2 and formate levels occurred as these compounds were produced during the oxidation of lactate in all strains; however, the mutants were unable to reconsume these compounds for sulfate reduction during the later stages of growth. The inoculation of 0.1 ml of a culture (OD600 = 0.8) of mutant or parental strain cells into 5 ml of medium with lactate in the absence of sulfate produced equivalent levels of H2 (Fig. 5), suggesting that sulfate-dependent growth (and H2 consumption) was important in maintaining lower levels of H2.
FIG. 3.
Total H2 produced by the cultures (A) and growth (B) of the G20 parental strain (▪) and the mopB mutant (□) in MS medium with 50 mM lactate and 50 mM sulfate.
FIG. 4.
Formate concentrations during the growth of the G20 parent (▪), mopB mutant (□), cycA mutant (▵), and hydB mutant (○) in MS medium containing 50 mM lactate and 20 mM sulfate.
FIG. 5.
Total H2 produced in the absence of sulfate by cultures of the G20 parent (▪), mopB mutant (□), cycA mutant (▵), and hydB mutant (○) incubated in MS medium containing 50 mM lactate.
The data presented above suggest roles for type I tetraheme cytochrome c3, Fe hydrogenase, and molybdopterin oxidoreductase in H2 oxidation. The fact that the hydB mutant can grow at all indicates that an alternative route for H2 oxidation exists. A mutant with a transposon mutation in the NiFe hydrogenase small subunit (encoded by hydA; Dde_2134) was also obtained (21). This mutant grew normally with H2-sulfate, further confirming, as has been observed previously (6), that the role of NiFe hydrogenase in H2 oxidation is not clear. Here, it was observed that G20 mutants lacking the Fe hydrogenase accumulated H2 with lactate or formate as the electron donor (data not shown) and grew slowly with H2 as the electron donor. The poor growth of hyd mutants of Desulfovibrio on H2, as well as H2 accumulation during growth on lactate, has been demonstrated previously (21, 30). The combined data support the assertion that the Fe hydrogenase acts as an uptake hydrogenase for Desulfovibrio. Recent experiments suggest that the Fe hydrogenase is most important when H2 is present at high concentrations or when cells are growing on lactate and that in the presence of low-level intracellular H2, the lower-activity, higher-affinity NiFeSe hydrogenase is used (6).
The mopB and cycA mutants exhibit similar phenotypes of no detectable growth on H2 and formate. One possibility is that the mopB and cycA proteins act together to form an electron transfer pathway in which electrons from formate or H2 oxidation are moved to some electron transfer intermediate or directly to another set of respiratory proteins involved in sulfate reduction (Fig. 1). The type I tetraheme periplasmic cytochrome c3 is perhaps the most well studied protein in Desulfovibrio (32), and yet much remains to be learned regarding its function. In Desulfovibrio, the pool of periplasmic cytochrome c3 has been suggested to act as an electron acceptor for periplasmic hydrogenases and formate dehydrogenases (14, 18, 23). Studies done with cell-free preparations have shown cytochrome c3 to interact with and to mediate electron transfer from the Fe hydrogenase to a transmembrane high-molecular-mass cytochrome c (Hmc) (26). A nuclear magnetic resonance study has also described the interaction of the Fe hydrogenase with cytochrome c3 (11), providing further evidence for the possible interaction of these two proteins. However, in vivo studies of the interaction of cytochrome c3 with other proteins are still lacking, and as a result, cellular interactions are not fully understood. In addition, the expression levels of the hmc operon during H2-dependent growth were unexpectedly low (38), and deletions of the hmc operon demonstrated that this transmembrane complex is not essential for H2 uptake (10). These data cast doubt on the importance of this Hmc complex in electron transfer from the cytochrome c3 pool during H2-dependent sulfate reduction.
In G20, a four-gene operon (locus Dde_2932 to locus Dde_2935) encodes the bis-molybdopterin guanine dinucleotide (bis-MGD)-containing oxidoreductase described here as MopABCD. Bis-MGD proteins constitute a diverse class of redox proteins, which have been further divided into subfamilies based on evolutionary as well as biochemical properties. The D. desulfuricans G20 Mop protein complex belongs within the dimethyl sulfoxide reductase family, of which many representatives have three subunits. These include a molybdopterin-binding subunit, an Fe-S cluster-containing subunit, and an integral membrane component that is thought to interact directly with menaquinone or menaquinone derivatives (3, 9, 16). The mop operon was compared to existing Desulfovibrio genomic data and appears to be conserved in all the available genomes of Desulfovibrio (7). The Mop complex is definitely produced at reasonable levels, as proteomic analyses have detected MopB and MopC in cell extracts of D. vulgaris (41) and MopC in D. desulfuricans G20 (22). Bis-MGD proteins with the highest degrees of similarity to G20 Mop include the Mop in D. vulgaris strain Hildenborough, polysulfide reductase (NrfD) in “Desulfococcus oleovorans” Hxd3 and “Solibacter usitatus” Ellin6076, and nitrate reductase in Geobacter uraniireducens Rf4. Orthologous polysulfide reductases and nitrate reductases whose crystal structures have been studied have a transmembrane subunit which may interact with menaquinone during electron transfer (16, 17).
The cellular location of the MopABCD complex of strain G20 was determined in silico using PSORT-B (12) and CELLO II (40). Both analyses indicated that the first three encoded proteins (the Dde_2932 to Dde_2934 proteins) are most likely located in the periplasm and that the fourth protein, encoded by the Dde_2935 gene, is a transmembrane subunit. In MopB, a Tat motif, RRXFXK, starting at position 5 was observed, indicating the likely use of the twin arginine system for translocation to the periplasm.
MopA has been annotated as a hypothetical protein (209 amino acids [aa]; pI 5.44; molecular mass, 23.8 kDa). Six heme c-binding motifs (CXXCH) are found in the amino acid sequence (see Fig. S1 in the supplemental material). This subunit, which has a c-type heme-containing domain, is most likely a member of the cytochrome c family, which may play a role in interaction with hydrogenase or other redox proteins during electron transfer. MopB is a putative molybdopterin-binding subunit (689 aa; pI 7.22; molecular mass, 73.2 kDa) in which the conserved domain is similar to a group of related uncharacterized putative hydrogenase-like homologs of molybdopterin-binding proteins (see Fig. S2 in the supplemental material). MopC is a typical iron-sulfur cluster-binding subunit (255 aa; pI 7.43; molecular mass, 28.8 kDa) in which the region of positions 3 to 244 is annotated as Fe-S cluster-containing hydrogenase component 1 (COG0437). The amino acid sequence contains three motifs for binding iron-sulfur clusters and one heme c-binding motif (see Fig. S3 in the supplemental material). MopD is a transmembrane protein (418 aa; pI 7.10; molecular mass, 47 kDa) with 10 predicted transmembrane helices (TM1 to TM10) (see Fig. S4 in the supplemental material). The conserved domain of aa 30 to 416 is annotated as a hydrogenase 2 cytochrome b type (COG5557), which possibly contains heme b capable of interacting with quinone/menaquinone. Pairs of conserved histidines in transmembrane helices of NarI from E. coli and Hmc5 from D. vulgaris are involved in heme b binding (2). In the G20 MopD, histidine 182 and H199 are found in the fourth transmembrane helix, and these residues are also conserved in many other bacterial Mop proteins. H137 is also conserved; however, it is not likely within a transmembrane helix. The predicted positions of these histidines are not necessarily suggestive of their involvement in heme binding (29), and therefore, the existence of cytochrome b is still ambiguous. MopD is homologous to many polysulfide reductases (Psr). Some Psr proteins do not contain any hemes in the membrane-bound subunit, unlike many other heme b-containing membrane-bound bis-MGD enzymes. However, these Psrs still bind quinone or its analogs (17). Quinone-binding sites are difficult to identify at the sequence level due to the limited number of quinone-binding structures available and the diversity of quinone-binding proteins (33). The recent crystallization of polysulfide reductase (corresponding to locus TTC0153 to locus TTC0155) of Thermus thermophilus showed a quinone-binding pocket in the PsrC subunit (TTC0153) responsible for binding quinone or quinone derivatives (17). MopD in G20 showed high levels of identity to MopD (DVU0692) of D. vulgaris (80%) and polysulfide reductase NrfD (Dole_2549) of D. oleovorans Hxd3 (54%) but a low level of identity (12.9%) to PsrC (TTC0153) of T. thermophilus.
The function of the MopABCD complex is largely unknown. As the mop mutant is defective in H2 and formate uptake, it is likely that the protein acts either as a hydrogenase/formate dehydrogenase or as an electron transfer chain component in the H2 oxidation pathway. We are not familiar with any studies that have documented a molybdoprotein that can catalyze H2 oxidation or a single protein that can carry out both formate and H2 oxidation. To address the question of whether this complex catalyzes either of these processes, we assayed hydrogenase and formate dehydrogenase activities in intact cells by using MV or BV as the electron acceptor. The mop mutant was shown to have hydrogenase activity similar to that of the parental strain and higher formate dehydrogenase activity than the parental strain (Table 2). This finding suggests that MopABCD may not function as a hydrogenase or formate dehydrogenase but that, rather, defects in H2 and formate oxidation are more likely to be due to electron transfer chain interruption.
TABLE 2.
Specific activities of formate dehydrogenase and hydrogenase in the washed cells of the parental strain and the mopB mutant grown on lactate-sulfate mediuma
| Strain | Sp act (μmol min−1·mg−1) of:
|
|||
|---|---|---|---|---|
| Formate dehydrogenase
|
Hydrogenase
|
|||
| MV | BV | MV | BV | |
| Parent | 0.69 ± 0.19 | 0.35 ± 0.21 | 9.6 ± 0.8 | 9.4 ± 1.8 |
| mopB mutant | 1.49 ± 0.28 | 0.64 ± 0.17 | 10.3 ± 2.0 | 10.0 ± 2.2 |
MV or BV was used as the electron acceptor in the assays. Activity was expressed as the mean ± the standard deviation (n = 3) in micromoles of MV or BV per minute per milligram of protein.
The best-studied systems with components similar to molybdopterin oxidoreductase include the formate dehydrogenase/nitrate reductase of E. coli (16) and the polysulfide reductase of Wolinella succinogenes (9). In both cases, formate or H2 is oxidized by periplasmic components of protein complexes and electrons are then transferred through a cytochrome b-containing subunit to menaquinone or methyl-menaquinone. The electrons from the reduced menaquinone are then taken up by a similar complex facing the cytoplasm, resulting in the reduction of nitrate or a complex facing the periplasm for polysulfide reduction. By coupling electron transfer to the reduction/oxidation of a quinone derivative, cells are able to use these enzymes to generate a PMF (3, 15, 16). Although we have not addressed the ability of mutants to generate a PMF, the poor levels of motility of all three mutants relative to that of the parental strain suggest that the mutations have impaired this ability. This type of coupled system which uses Mop for one or both of the redox reactions appears to be an ideal candidate for H2 or formate oxidation coupled to the reduction of sulfate. Thus, if the G20 Mop is similar to the well-characterized proteins described above, it will likely transfer electrons from reduced components in the periplasm, most likely from the tetraheme cytochrome c3, via the hexaheme cytochrome c of the Mop complex to the menaquinone pool. A redox loop mechanism will be established if a cytoplasm-facing, menaquinone-interacting membrane-bound complex exists which, coupled to H2 oxidation, transfers electrons from the menaquinone pool to the cytoplasmic side for sulfate reduction. Two complexes, Qmo and Dsr, apparently conserved in all sulfate reducers, have been suggested as possible conduits for transferring electrons from the menaquinone pool to adenosine phosphosulfate and sulfite, respectively (28, 29).
This redox loop mechanism does not exclude other transmembrane complexes that may function as possible alternative electron transfer channels and does not address the fate of electrons that do not pass through H2. Further purification, crystallization, and cytochrome c3-Mop interaction studies will help in providing direct evidence of the electron transfer process.
Supplementary Material
Acknowledgments
We thank Todd Kitten of Virginia Commonwealth University for suggestions on the arbitrary PCR method. Zhiguo Fang and Helong Jiang provided help with high-pressure liquid chromatography measurements. We also thank the anonymous reviewers for the valuable comments and suggestions.
The research was funded by the U.S. Department of Energy (DOE) Hydrogen Initiative. Contributions of K.L.K. were funded by the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the Office of Science, Office of Biological and Environmental Research, Genomics Program: GTL through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and DOE.
Footnotes
Published ahead of print on 20 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Badziong, W., and R. K. Thauer. 1980. Vectorial electron transport in Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate as sole energy source. Arch. Microbiol. 125167-174. [Google Scholar]
- 2.Berks, B. C., M. D. Page, D. J. Richardson, A. Reilly, A. Cavill, F. Outen, and S. J. Ferguson. 1995. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol. Microbiol. 15319-331. [DOI] [PubMed] [Google Scholar]
- 3.Biel, S., J. Simon, R. Gross, T. Ruiz, M. Ruitenberg, and A. Kroger. 2002. Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur. J. Biochem. 2691974-1983. [DOI] [PubMed] [Google Scholar]
- 4.Böck, A., P. King, M. Blokesch, and M. Posewitz. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 511-71. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey, S. A., H. S. Park, J. K. Voordouw, Z. He, J. Zhou, and G. Voordouw. 2007. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 1896159-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caffrey, S. M., H. S. Park, J. Been, P. Gordon, C. W. Sensen, and G. Voordouw. 2008. Gene expression by the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough grown on an iron electrode under cathodic protection conditions. Appl. Environ. Microbiol. 742404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, S., J. C. Noe, S. Paik, and T. Kitten. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 6389-94. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, W., and O. Klimmek. 2002. The function of methyl-menaquinone-6 and polysulfide reductase membrane anchor (PsrC) in polysulfide respiration of Wolinella succinogenes. Eur. J. Biochem. 2691086-1095. [DOI] [PubMed] [Google Scholar]
- 10.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174143-151. [DOI] [PubMed] [Google Scholar]
- 11.ElAntak, L., X. Morelli, O. Bornet, C. Hatchikian, M. Czjzek, A. Dolla, and F. Guerlesquin. 2003. The cytochrome c3-[Fe]-hydrogenase electron-transfer complex: structural model by NMR restrained docking. FEBS Lett. 5481-4. [DOI] [PubMed] [Google Scholar]
- 12.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. L. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 313613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groh, J. L., Q. Luo, J. D. Ballard, and L. R. Krumholz. 2005. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 717064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. W. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22554-559. [DOI] [PubMed] [Google Scholar]
- 15.Jormakka, M., B. Byrne, and S. Iwata. 2003. Proton motive force generation by a redox loop mechanism. FEBS Lett. 54525-30. [DOI] [PubMed] [Google Scholar]
- 16.Jormakka, M., S. Törnroth, B. Byrne, and S. Iwata. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 2951863-1868. [DOI] [PubMed] [Google Scholar]
- 17.Jormakka, M., K. Yokoyama, T. Yano, M. Tamakoshi, S. Akimoto, T. Shimamura, P. Curmi, and S. Iwata. 2008. Molecular mechanism of energy conservation in polysulfide respiration. Nat. Struct. Mol. Biol. 15730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeGall, J., and G. Fauque. 1988. Dissimilatory reduction of sulfur compounds p. 587-639. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, New York, NY.
- 19.Li, X., and L. R. Krumholz. 2007. Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 1893705-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lissolo, T., E. S. Choi, J. Legall, and J. H. D. Peck. 1986. The presence of multiple intrinsic membrane nickel-containing hydrogenases in Desulfovibrio vulgaris (Hildenborough). Biochem. Biophys. Res. Commun. 139701-708. [DOI] [PubMed] [Google Scholar]
- 21.Luo, Q., J. L. Groh, J. D. Ballard, and L. R. Krumholz. 2007. Identification of genes that confer sediment fitness to Desulfovibrio desulfuricans G20. Appl. Environ. Microbiol. 736305-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo, Q., K. K. Hixson, S. J. Callister, M. S. Lipton, B. E. L. Morris, and L. R. Krumholz. 2007. Proteome analysis of Desulfovibrio desulfuricans G20 mutants using the accurate mass and time (AMT) tag approach. J. Proteome Res. 63042-3053. [DOI] [PubMed] [Google Scholar]
- 23.Matias, P. M., I. A. C. Pereira, C. M. Soares, and M. A. Carrondo. 2005. Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol. 89292-329. [DOI] [PubMed] [Google Scholar]
- 24.Odom, J. M., and H. D. Peck. 1981. Hydrogen cycling as a general mechanism for energy coupling in the sulfate reducing bacteria, Desulfovibrio sp. FEMS Microbiol. Lett. 1247-50. [Google Scholar]
- 25.Pereira, I. A. C. 2007. Respiratory membrane complexes of Desulfovibrio, p. 24-35. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 26.Pereira, I. A. C., C. V. Romao, A. V. Xavier, J. LeGall, and M. Teixeira. 1998. Electron transfer between hydrogenases and mono- and multiheme cytochromes in Desulfovibrio ssp. J. Biol. Inorg. Chem. 3494-498. [Google Scholar]
- 27.Pereira, P. M., M. Teixeira, A. V. Xavier, R. O. Louro, and I. A. Pereira. 2006. The Tmc complex from Desulfovibrio vulgaris Hildenborough is involved in transmembrane electron transfer from periplasmic hydrogen oxidation. Biochemistry (Moscow) 4510359-10367. [DOI] [PubMed] [Google Scholar]
- 28.Pires, R. H., A. I. Lourenco, F. Morais, M. Teixeira, A. V. Xavier, L. M. Saraiva, and I. A. C. Pereira. 2003. A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim. Biophys. Acta 160567-82. [DOI] [PubMed] [Google Scholar]
- 29.Pires, R. H., S. S. Venceslau, F. Morais, M. Teixeira, A. V. Xavier, and I. A. C. Pereira. 2006. Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex—a membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry (Moscow) 45249-262. [DOI] [PubMed] [Google Scholar]
- 30.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prickril1, B. C., S. He, C. Li, N. Menon, E. Choi, A. E. Przybyla, D. V. DerVartanian, J. H. D. Peck, G. Fauque, J. LeGall, M. Teixeira, I. Moura, J. J. G. Moura, D. Patil, and B. H. Huynh. 1987. Identification of three classes of hydrogenase in the genus, Desulfovibrio. Biochem. Biophys. Res. Commun. 149369-377. [DOI] [PubMed] [Google Scholar]
- 32.Rapp-Giles, B. J., L. Casalot, R. S. English, J. A. Ringbauer, A. Dolla, and J. D. Wall. 2000. Cytochrome c3 mutants of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 66671-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rich, P., and N. Fisher. 1999. Generic features of quinone-binding sites. Biochem. Soc. Trans. 27561-565. [DOI] [PubMed] [Google Scholar]
- 34.Rossi, M., W. B. R. Pollock, M. W. Reij, R. G. Keon, R. Fu, and G. Voordouw. 1993. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J. Bacteriol. 1754699-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voordouw, G. 1995. The genus Desulfovibrio: the centennial. Appl. Environ. Microbiol. 612813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voordouw, G., and S. Brenner. 1985. Nucleotide sequence of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough). Eur. J. Biochem. 148515-520. [DOI] [PubMed] [Google Scholar]
- 37.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 563748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wall, J. D., A. P. Arkin, N. C. Balci, and B. Rapp-Giles. 2007. Genetics and genomics of sulfate respiration in Desulfovibrio, p. 1-12. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 39.Wall, J. D., B. J. Rapp-Giles, and M. Rousset. 1993. Characterization of a small plasmid from Desulfovibrio desulfuricans and its use for shuttle vector construction. J. Bacteriol. 1754121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, C. S., Y. C. Chen, C. H. Lu, and J. K. Hwang. 2006. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 64643-651. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, W. W., M. A. Gritsenko, R. J. Moore, D. E. Culley, L. Nie, K. Petritis, E. F. Strittmatter, D. G. Camp, R. D. Smith, and F. J. Brockman. 2006. A proteomic view of Desulfovibrio vulgaris metabolism as determined by liquid chromatography coupled with tandem mass spectrometry. Proteomics 64286-4299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.