Abstract
We have previously described a novel conjugal DNA transfer process that occurs in Mycobacterium smegmatis. To identify donor genes required for transfer, we have performed a transposon mutagenesis screen; we report here that LpqM, a putative lipoprotein-metalloproteinase, is essential for efficient DNA transfer. Bioinformatic analyses predict that LpqM contains a signal peptide necessary for the protein's targeting to the cell envelope and a metal ion binding motif, the likely catalytic site for protease activity. Using targeted mutagenesis, we demonstrate that each of these motifs is necessary for DNA transfer and that LpqM is located in the cell envelope. The requirement for transfer is specific to the donor strain; an lpqM knockout mutant in the recipient is still proficient in transfer assays. The activity of LpqM is conserved among mycobacteria; homologues from both Mycobacterium tuberculosis and Mycobacterium avium can complement lpqM donor mutants, suggesting that the homologues recognize and process similar proteins. Lipoproteins constitute a significant proportion of the mycobacterial cell wall, but despite their abundance, very few have been assigned an activity. We discuss the potential role of LpqM in DNA transfer and the implications of the conservation of LpqM activity in M. tuberculosis.
In previous work, we have described a novel conjugation system in Mycobacterium smegmatis (15). Although this process meets the criteria of conjugation (successful transfer requires prolonged cell-cell contact and is DNase resistant), the mechanism of transfer is unique (27-29). Transfer is chromosomally encoded, and despite exhaustive bioinformatics searches, we have yet to identify any genes encoding obvious transfer functions. A comprehensive transposon mutagenesis screen of the donor strain failed to identify transfer-defective mutants. Instead, the screen identified hyperconjugative mutants that mapped to a large, 30-kb locus, esx-1 (7). esx-1 encodes a secretory apparatus, ESX-1, and we hypothesized that the secretion of proteins by ESX-1 negatively regulates transfer, either because the secreted proteins physically block transfer or because they act as intercellular quorum sensors. In a more recent study of the M. smegmatis recipient strain, we showed that a functional ESX-1 apparatus is essential for recipient activity (4). Recipient activity required secretion of at least two esx-1-encoded proteins, EsxA and EsxB. We have therefore proposed that in the recipient, as in the donor, ESX-1 is secreting proteins that regulate, rather than mediate, DNA transfer. The ESX-1 apparatus is highly conserved, and esx-1-encoded mutants of Mycobacterium tuberculosis, Mycobacterium bovis, and Mycobacterium marinum are attenuated (reviewed in references 1 and 6). Proteins known to be secreted by ESX-1 from M. tuberculosis include EsxA and EsxB (formerly known as ESAT-6 and CFP10, respectively), EspA, and Rv3881 (10, 12, 17, 25). Further underscoring the functional involvement of ESX-1 in both conjugation and virulence, our screen for transfer-defective recipient mutants also identified homologues of EspA (MSMEG5168a) and Rv3881 (MSMEG0076) (4). However, the mechanisms by which these proteins function, both in M. tuberculosis virulence and in regulation of M. smegmatis conjugation, are unknown.
In this study, we have reexamined donor contributions to DNA transfer by exploiting a hyperconjugative donor mutant strain. This strategy, which increased the sensitivity of the assay, has allowed us to isolate donor-defective insertions for the first time. One insertion mapped to a gene encoding a putative metallo-lipoprotein, LpqM. We describe the genetic characterization of this protein and discuss its potential role as an extracellular protease in DNA transfer.
MATERIALS AND METHODS
Bacterial strains and media.
The M. smegmatis donor strains used were derivatives of strains mc2155 (24) and MKD158 (resistant to hygromycin [Hygr]) (7) and mc2155 Δesx-1. The mc2155 Δesx-1 strain contains a replacement of the esx-1 genes Msmeg_0056 to Msmeg_0082 with a gene encoding Hygr, which was constructed by J. Wang and J. Flint by allele replacement. Briefly, 2-kb segments of DNA upstream of msmeg_0056 and downstream of msmeg_0082 were PCR amplified and cloned into the mycobacterial plasmid pPR23, along with a cassette encoding hygromycin resistance; the two amplified segments were cloned such that they flanked the Hygr gene in an appropriate orientation for allele replacement. pPR23 is temperature sensitive for replication, and it encodes the counter-selectable marker, sacB, and gentamicin resistance (Gmr) (16). Hygr allele replacements were selected for at the nonpermissive temperature in the presence of sucrose. Sucrose-resistant Hygr Gms recombinants were purified, and the precise replacement of the esx-1 region with the Hygr gene was confirmed by Southern analysis and PCR. The recipient strain used was MKD8, which is resistant to streptomycin (Smr) (15). M. smegmatis was grown at 37°C in Trypticase soy broth or Sauton's medium (0.5 g/liter KH2PO4, 0.5 g/liter MgSO4, 4.0 g/liter l-asparagine, 6% glycerol, 0.05 g/liter ferric ammonium citrate, 2.0 g/liter citric acid, and 100 μl 1% ZnSO4). The media were supplemented with antibiotics at the following concentrations: 100 μg/ml hygromycin, 10 μg/ml kanamycin, and 200 μg/ml streptomycin.
Escherichia coli DH5α was used throughout the study for routine molecular genetic techniques. Cell cultures were grown in LB medium at 37°C. When appropriate, the medium was supplemented with antibiotics at the following concentrations: 75 μg/ml hygromycin, 50 μg/ml kanamycin, and 15 μg/ml gentamicin.
Transposon mutagenesis.
A mutant transposon insertion library was made in the mc2155 Δesx-1 strain, using a temperature-sensitive phage to deliver a mariner transposon encoding kanamycin resistance (Kmr) (2, 22). Individual insertions were mapped as previously described (7). Briefly, chromosomal BamHI fragments encoding Kmr were cloned, and the site of insertion was determined by DNA sequence analysis using primers that annealed to the Kmr gene. The chromosomal locations of the insertions and the gene annotations are based on the April 2007 version of the M. smegmatis chromosomal sequence in The Institute for Genomic Research database (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?database=gms). Transposon insertions were transduced from the mc2155 Δesx-1 strain into MKD158, using the generalized transducing phage Bxz1 as described previously (13).
Microtiter mating screen.
A screen for donor mutants that are defective in conjugation was carried out as described previously (7). The Hygr marker replacing the esx-1 locus was used to monitor transfer from the donor strain into the recipient (MKD8). Transconjugants were selected on Trypticase soy agar medium containing hygromycin and streptomycin. DNA transfer is temperature sensitive, and thus, all crosses are carried out at 30°C (15).
Mating assay.
The effect of insertion mutations on donor activity was determined using a quantitative mating assay to measure the conjugation frequency (the number of transconjugants per donor) (15). In all crosses, transconjugants were selected on medium containing hygromycin and streptomycin. Transfer frequencies are the averages of at least three crosses, which were performed in parallel with wild-type donor and recipient controls.
Site-directed mutagenesis.
The lpqM gene was amplified by PCR from genomic DNA and cloned into pUC19. Mutations were created by PCR, using Pfu polymerase (Stratagene). Primer sequences used for mutagenesis are available on request. Mutations were confirmed by DNA sequence analysis. The lpqM gene knockout mutant, MKD158 ΔlpqM, was generated by allele exchange. Briefly, 800-bp segments of DNA upstream and downstream of lpqM were amplified and cloned on either side of a cassette encoding kanamycin resistance in pPR23 (16). Allele replacements were then selected at the nonpermissive temperature in the presence of sucrose. Kmr Gms recombinants were confirmed by PCR.
Complementation studies.
The M. smegmatis lpqM gene with 150 bp of upstream sequence was amplified from mc2155 genomic DNA and cloned into pPR23 (16). This multicopy, nonintegrating plasmid (pPR23MslpqM) was transformed into mc2155 ΔlpqM, and complementation was determined in a mating-out assay. All strains harboring pPR23 derivatives for lpqM complementation were grown at 30°C. The lpqM homologues from M. tuberculosis and Mycobacterium avium were constructed in a similar manner, except that each gene was expressed from the M. smegmatis lpqM promoter region. An epitope-tagged M. smegmatis lpqM derivative was constructed in pUAB200 (23). A DNA duplex encoding the c-Myc epitope (amino acid sequence, EEQKLISEDL) was ligated in-frame with the C terminus of lpqM to create an lpqM-c-Myc gene fusion. All constructs were confirmed by DNA sequencing.
Subcellular fractionation.
Crude cell wall, membrane, and cytosolic fractions were prepared by differential centrifugation as described previously (8, 9, 19). mc2155 was transformed with pUAB200-lqpM-c-Myc. Six hundred milliliters of overnight cultures grown at 37°C were pelleted by centrifugation at 7,000 rpm for 10 min at 4°C. Cells were resuspended in phosphate-buffered saline containing a protease inhibitor cocktail (Roche) and were then lysed by sonication (five 15-s pulses with a Branson microtip) to generate a total cell lysate. Insoluble material from the total cell lysate, including unbroken cells, was removed by centrifugation (15,000 rpm for 10 min at 4°C). The clarified supernatant fraction was further separated by ultracentrifugation at 200,000 × g for 2.5 h at 4°C (Beckman NVT90 rotor) to separate the membrane fractions from the soluble cytoplasmic fraction. The membrane fraction and insoluble pellets were resuspended in phosphate-buffered saline containing 1% Triton X-100 before analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. GroEL was used to monitor the fractionation procedure. GroEL is an abundant cytoplasmic protein that should be enriched into the soluble, cytosolic fractions during the procedure.
Western blotting.
Protein fractions were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to a polyvinylidene difluoride membrane (Amersham Hybond-P; GE Healthcare) by electrophoresis at 35 eV overnight at 4°C. Unless otherwise stated, all the remaining procedures were carried out at room temperature. The membrane was blocked for 1 h with 5% nonfat milk in TBS-T buffer (50 mM Tris [pH 7.5], 150 mM NaCl, and 0.1% Tween 20). c-Myc antibody (Invitrogen) was used at a 1:5,000 dilution, and GroEL antibody (Stressgen) was used at a dilution of 1:5,000 in TBS-T buffer. Secondary antibody and detection reagents were from the Amersham ECL Western Breeze kit (GE Healthcare) and were used according to the manufacturer's instructions.
RESULTS
LpqM is required for conjugation in M. smegmatis.
In a previous genetic screen for conjugative donor mutants, only hyperconjugative mutants were isolated, despite the fact that we had expected to isolate both hyper- and hypoconjugative mutant strains (7). The majority of the hyperconjugative mutations mapped to the esx-1 locus of M. smegmatis; we showed that this locus negatively regulates DNA transfer. A likely explanation for our inability to identify transfer-defective mutants was that the assay was suboptimal such that the background level of false-positive mutations was too high. To circumvent this issue, we took advantage of the hyperconjugative phenotype of esx-1 mutants. A defined deletion of the genes msmeg_0056 to msmeg_0082 was created within esx-1 by allele exchange. We chose to generate a deletion, as this could not revert and would also avoid the isolation of insertions in the esx-1 region, resulting in a hyperconjugative phenotype. As expected, the deletion elevated the DNA transfer frequency (>10-fold), resulting in confluent growth of transconjugant mating spots in the microtiter mating assay and thereby increasing the sensitivity of the screen. The Δesx-1 strain was subjected to mariner::km transposon mutagenesis as previously described (7). Nine transfer-defective mutants were isolated from a screen of 18,000 mutants, and these had insertions in msmeg_0033, msmeg_1435, msmeg_1455, msmeg_3080, msmeg_4913, and msmeg_6128. Notably, four independent insertions were isolated in msmeg_4913, and quantitative transfer assays indicated that these were the most deleterious mutations, reducing transfer by over 160-fold and often down to undetectable levels (Table 1, rows 1 and 2).
TABLE 1.
DNA transfer frequenciesa
| Cross | Relevant genotype | No. of transconjugants per donor |
|---|---|---|
| 1 | Δesx-1 | 1.4 (± 0.1) × 10−4 |
| 2 | Δesx-1 lpqM::Tn | 8.8 (± 0.8) × 10−7 |
| 3 | Δesx-1 ΔdinG | 8.9 (± 3.4) × 10−5 |
| 4 | Δesx-1 lpqM::Tn pPR23MslpqM | 1.7 (± 0.7) × 10−5 |
| 5 | Δesx-1 ΔlpqM | 8.5 (± 0.8) × 10−7 |
| 6 | Δesx-1 ΔlpqM pPR23MslpqM | 7.5 (± 1.5) × 10−5 |
| 7 | MKD158 (wild-type) | 5.1 (± 0.3) × 10−5 |
| 8 | lpqM::Tn | 1.3 (± 0.7) × 10−8 |
| 9 | ΔlpqM | 1.9 (± 0.8) × 10−8 |
| 10 | ΔlpqM pPR23MslpqM | 8.4 (± 1.4) × 10−6 |
| 11 | ΔlpqM pPR23MtblpqM | 5.2 (± 0.7) × 10−7 |
| 12 | ΔlpqM pPR23MalpqM | 8.0 (± 0.5) × 10−7 |
| 13 | ΔlpqM pPR23Δ22-lpqM | 7.8 (± 0.2) × 10−8 |
| 14 | ΔlpqM pPR23C22A-lpqM | 3.2 (± 0.5) × 10−8 |
| 15 | ΔlpqM pPR23YT-lpqM | 6.0 (± 0.6) × 10−8 |
Each frequency is the average of at least three crosses, using approximately equal numbers of donor and recipient cells (2 × 108).
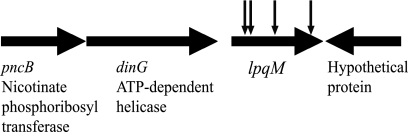
msmeg_4913 encodes a putative lipoprotein protease, LpqM, and is the third gene in a 3-gene operon (Fig. 1). We note that although the gene preceding lpqM encodes a putative DNA helicase, DinG, this helicase is not required for transfer. A precise deletion of dinG had no effect on DNA transfer frequencies (Table 1, row 3). To confirm that the transfer defect was a result of the transposon insertion, the lpqM gene was amplified and cloned into the multicopy, nonintegrating plasmid pPR23, creating pPR23MslpqM. This plasmid complemented the transfer defect, although only to 10% of wild-type levels (Table 1, row 4). A precise lpqM deletion was also generated by allele exchange, resulting in a defect in transfer similar to the defect caused by the transposon insertion in lpqM. Again, the transfer defect was rescued by complementation with pPR23MslpqM (Table 1, rows 5 and 6). The effect of the deletion establishes the requirement for LpqM in DNA transfer and rules out potential dominant-negative effects created by the transposon insertion.
FIG. 1.
Schematic map of the lpqM region of the M. smegmatis genome. The map is based on the genome sequence available from TIGR (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?database=gms). The vertical arrows indicate the sites of insertion of the mariner::km transposon. The segment of DNA cloned for complementation studies included the 150-bp region upstream of lpqM, which encompasses the 92-bp intergenic region between dinG and lpqM.
LpqM affects transfer independently of ESX-1 function and is donor specific.
The lpqM mutants described above were isolated in a Δesx-1 hyperconjugative background. Possibly, the requirement for LpqM in transfer was dependent on the lack of a functional ESX-1 apparatus. To rule out this scenario, we transduced one of the original Kmr transposon insertions into the wild-type donor strain, MKD158. In addition, a precise deletion of lpqM was generated in MKD158, using the same allele exchange vector as had been used for the Δesx-1 strain. Both of these lpqM mutant strains were found to be defective in DNA transfer compared to the parent MKD158 (Table 1, compare rows 7 to 9). As expected, we observed lower frequencies of transfer in the wild-type background than in the Δesx-1 strain. The transfer frequencies of both MKD158 lpqM mutant derivatives were elevated >100-fold when lpqM was expressed in trans (Table 1, compare rows 9 and 10). These data confirm that the LpqM requirement is independent of ESX-1 function. For further confirmation, we have determined that secretion of EsxB by ESX-1 occurs at wild-type levels in an lpqM mutant (data not shown).
Using the same allele exchange vector, we generated a deletion of lpqM in the recipient strain, MKD8 (15). DNA transfer from a wild-type donor into the ΔlpqM recipient derivative was unaffected, thereby establishing that the essential role of LpqM in transfer is specific to the donor strain.
LpqM is functionally conserved across species.
Bioinformatic analyses of LpqM indicate that it has homologues in other mycobacteria, including M. tuberculosis (Rv0419 [48.3% amino acid identity]) and M. avium (MAV4736 [47.2% amino acid identity]) (Fig. 2). There is also an LpqM paralogue within M. smegmatis (MSMEG4893 [50% amino acid identity]), but its inability to complement any lpqM mutation suggests that it encodes a protein that performs different functions or is a pseudogene. The M. avium and M. tuberculosis lpqM homologues were individually cloned into pPR23 and were then tested for their ability to complement the ΔlpqM strain. In each case, transfer activity was partially restored, indicating that functional conservation of LpqM activity exists among these mycobacterial species (Table 1, compare rows 9 to 12). Complementation does not restore LpqM activity to the wild-type levels, and we believe this is a result of low expression levels of lpqM from the complementing vector. The cloned DNA included a 92-bp putative promoter region immediately upstream of lpqM, but it is possible that in vivo transcription is normally initiated from a stronger promoter transcribing the entire operon (Fig. 1). The important finding is that in each case the overall level of transfer is elevated above that of the ΔlpqM mutant.
FIG. 2.
The M. smegmatis LpqM protein has homologues in other mycobacteria. The predicted amino acid sequences of LpqM from M. tuberculosis (Rv0419), M. bovis (Mb0427), and M. avium (Mav_4736) are aligned with the sequence from M. smegmatis (MSMEG913). Although the correct initiation codon for M. tuberculosis LpqM and M. bovis LpqM has not been experimentally determined, it has been assigned to the first methionine (M) codon. However, it is possible that the translation of these two homologues begins at valine (V) 19; in that case, the alignment with M. smegmatis LpqM and M. avium LpqM would be even better. The black bar over the first 23 amino acids of the M. smegmatis LpqM protein sequence is the predicted signal peptide and lipobox (LAACS). Residues 162 to 166 are also indicated by a black bar; they represent the conserved zinc metal binding site (HEXXH).
Transfer activity requires the conserved signal peptide motif and membrane targeting.
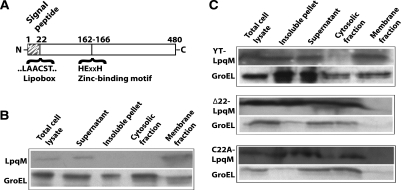
Computational analyses of the LpqM protein predict it to contain an N-terminal signal peptide motif (amino acids 1 to 22) (Fig. 2 and 3A). The signal peptide is followed by an absolutely conserved cysteine residue, embedded within a larger “lipobox” motif (LAACS) that is known to be important for outer membrane localization of lipoproteins (30, 31). Briefly, the signal peptide is hypothesized to target the protein to the cell wall. Following translocation across the cytoplasmic membrane, the lipobox cysteine is covalently linked to a diacylglycerol moiety by a membrane-bound diacylglycerol transferase. A signal peptidase then cleaves off the signal peptide, and the diacylglycerol moiety is used to anchor the lipoprotein in the cell wall. We constructed a series of pPR23lpqM mutant plasmid derivatives in order to confirm the significance of these conserved amino acids and to examine their impact on transfer. The plasmids were introduced into the ΔlpqM donor strain, and then each mutant derivative was assessed for its ability to complement the deletion of lpqM. Both deletion of LpqM residues 2 to 22 (LpqMΔ22) and substitution of Cys22 by Ala [LpqM(C22A)] reduced the ability of the plasmid to complement the defect in DNA transfer, indicating that the signal peptide and membrane anchor are necessary for efficient DNA transfer (Table 1, compare rows 10, 13, and 14).
FIG. 3.
LpqM is a membrane-associated protein and requires an intact signal peptide and lipobox for membrane localization. (A) Schematic representation of the LpqM protein indicating the locations of the key amino acid motifs that have been subjected to mutagenesis. (B) The wild-type LpqM protein was present in the membrane fraction. The cellular fractionation of LpqM, which used an LpqM-c-Myc derivative, was followed by Western blot analysis employing c-Myc antibodies (see Materials and Methods). Following sonication, the total cell lysate was clarified by centrifugation into the insoluble pellet and supernatant fractions. The supernatant was further fractionated by ultracentrifugation into cytosolic and membrane fractions. As predicted, LpqM was present in the total cell lysate, the lysate supernatant, and the membrane fractions, but not the cytosolic fraction. The soluble GroEL protein is found predominantly in the cytoplasm. Note that the insoluble pellet is from the initial, clarifying spin of the total cell lysate and includes both insoluble proteins and unbroken cells; therefore, this fraction is often contaminated with both LpqM and GroEL. (C) Removal of the signal peptide, i.e., deletion of residues 2 to 22 (Δ22), or a C22A substitution of the essential lipobox cysteine prevents LpqM targeting to the membrane fraction, and instead, it accumulates in the cytosol. However, mutation of the zinc binding catalytic site in the LpqM(YT) derivative does not disrupt membrane targeting.
The cellular location of each mutant protein was assessed to determine the effect of deleting the signaling functions. To facilitate these studies, a c-Myc epitope tag was fused to the C terminus of LpqM. This derivative had wild-type activity, as evidenced by its ability to complement the ΔlpqM mutation (data not shown). Importantly, the c-Myc tag allowed the use of epitope-specific antibodies to determine the location of the fusion protein. Cells expressing the fusion protein were harvested, and the cellular components were fractionated and then subjected to Western blot analysis. As predicted, wild-type LpqM is primarily associated with the membrane fraction and is absent from the cytosolic fraction (Fig. 3B). In contrast, c-Myc-tagged versions of LpqM(C22A) and LpqMΔ22 are absent from the membrane fraction and are retained in the cytosolic fraction, confirming the function of the predicted signal peptide (Fig. 3C). The overall levels of both signal peptide mutant proteins are similar to wild-type LpqM levels, indicating that the absence of the mutant proteins from the membrane fraction is not due to protein turnover, but rather to the mutation.
LpqM contains a zinc ion binding site essential for DNA transfer.
Consistent with its predicted role as a metalloproteinase, LpqM contains a conserved zinc ion binding motif (HEXXH) (Fig. 2 and 3A). The side chains of the two histidine residues in the motif coordinate the binding of the zinc metal ion, while the glutamate residue is thought to play a catalytic role (14). To address whether the predicted HEXXH motif is indeed an important component of the catalytic site and necessary for transfer, we mutated the motif to YTXXH, so as to disrupt both the zinc coordination and catalysis. The LpqM(YT) derivative could not rescue DNA transfer from the ΔlpqM donor strain, indicating that zinc binding, and therefore protease activities, is important for transfer (Table 1, row 15). Importantly, LpqM(YT) was expressed at wild-type levels and was found to be associated with the membrane, indicating that the transfer defect was not a consequence of the misfolding or improper targeting of the protein (Fig. 3C).
DISCUSSION
LpqM is the first mycobacterial protein that has been shown to be essential for donor conjugal transfer activity. The requirement for protease catalytic activity was demonstrated by disruption of the zinc metal binding motif, which abolished transfer, despite appropriate expression of LpqM and its localization to the membrane. The requirement of a membrane-associated protease in DNA transfer is consistent with previous models that we have proposed concerning the regulation of conjugation (4, 7). Briefly, we have observed that proteins secreted from both the donor and recipient by the ESX-1 secretory apparatus regulate transfer, and we have suggested that the secreted proteins act as quorum sensors, or as sex pheromones. Thus, through the secretion of different sets of proteins, the donor and recipient cells can respond, if conditions for genetic exchange are appropriate. A possible role for LpqM in conjugation would be to process extracellular proteins into a form able to activate or repress transfer. In this scenario, we envisage LpqM processing either donor or recipient proteins or proteins from both cell types. LpqM would process donor proteins translocated across the cytoplasmic membrane before their release into the surrounding milieu, while recipient proteins would be translocated from the exterior across the outer membrane, before being processed and imported into the donor cell. If recipient proteins were substrates of LpqM, the recipient ESX-1 apparatus could secrete some of these. However, if LpqM processes donor proteins, these would have to be secreted via an apparatus other than ESX-1, given that we have demonstrated that the LpqM requirement is independent of ESX-1 function. The identification of LpqM substrates in future studies will help to refine this hypothesis.
The ability of LpqM homologues to complement the M. smegmatis lpqM mutant indicates that LpqM protease activity is conserved across several mycobacterial species. However, the complementation does not necessarily indicate that conjugation occurs in M. tuberculosis or M. avium; it indicates only that the LpqM protein recognizes and processes similar substrates in each species. It is well established that M. tuberculosis secretes proteins that are essential for its virulence, but the targets of these proteins are unknown (3, 20, 26). The ability of the M. tuberculosis protein to complement the M. smegmatis lpqM mutant suggests that at least some of the proteins processed by the M. tuberculosis LpqM protein are of mycobacterial origin and not from the cell it has infected. Again, the identification of LpqM substrates will allow us to test this model further.
The cell envelope of mycobacteria is a thick, lipid-rich, complex structure that forms an important permeability barrier around the cell; it is thought to play a critical role in the biology and pathogenicity of mycobacteria (5). Although the subject of much research, only recently has the envelope been recognized to be composed of multilayered structures that include an outer membrane (11). Thus, the cell envelope is not dissimilar to those of gram-negative bacteria, with two membranes sandwiching a periplasmic space. Determination of the composition of these distinct layers, the functions of the layers, and the process by which molecules (including DNA) are translocated through them is obviously an important goal in the ongoing development of antimycobacterial drugs.
Lipoproteins constitute a major component of the cell envelope in mycobacteria; bioinformatic studies project that the M. tuberculosis genome encodes 99 lipoproteins representing 2.5% of the proteome (26). These lipoproteins are thought to be translocated across the cytoplasmic membrane and anchored to structures within the periplasm, although some could instead be anchored to the outer membrane. They are predicted to participate in diverse cellular functions, including transport, cell wall metabolism, cell adhesion, signaling, and protein degradation, and thus, some lipoproteins will play a significant role in virulence. However, only a few of the potential lipoproteins have been shown to be located in the membrane and had their putative activities confirmed. A role for lipoproteins in virulence has been implicated by showing that lspA mutants of M. tuberculosis are attenuated (21). lspA is the lipoprotein signal peptidase responsible for removal of the signal peptide following transacetylation and translocation. The peptidase cleavage generates the mature form of the lipoprotein that is anchored in the membrane. This lspA mutant illustrates that lipoprotein processing is important in M. tuberculosis virulence. A lipoprotein encoded by the M. tuberculosis gene Rv2224 is also essential for virulence, and it is a protease, as our data suggest for LpqM (18). Rv2224 cleaves GroEL2 and releases it into the culture medium. The directed release of proteins processed by Rv2224 into the culture supernatant is similar to the one that we have proposed for the processing and release of donor proteins by LpqM. Regardless of the mechanism, our data now add conjugation to the list of functional roles attributed to mycobacterial lipoproteins.
Acknowledgments
We thank Todd Gray and members of the Derbyshire laboratory for comments on the manuscript and Erin DeConno for help with figures. The Wadsworth Center Applied Genome Technology Core facility performed the DNA sequence analyses.
K.P. was a Summer REU Fellow and was an Emerging Infectious Diseases Fellow, funded by the CDC and administered by the Association of Public Health Laboratories. K.T.N. was supported by funding from an NIAID training grant (1T32AI055429) and K.M.D. by grants AI042308 and AI042308-08S1.
Footnotes
Published ahead of print on 20 February 2009.
REFERENCES
- 1.Abdallah, A. M., N. C. Gey van Pittius, P. A. Champion, J. Cox, J. Luirink, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, and W. Bitter. 2007. Type VII secretion-mycobacteria show the way. Nat. Rev. Microbiol. 5883-891. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. J. Jacobs. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 9410961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunstein, M., and J. T. Belisle. 2000. Genetics of protein secretion, p. 203-220. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- 4.Coros, A., B. Callahan, E. Battaglioli, and K. M. Derbyshire. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69794-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daffe, M., and J. M. Reyrat. 2008. The mycobacterial cell. ASM Press, Washington, DC.
- 6.DiGiuseppe Champion, P. A., and J. S. Cox. 2007. Protein secretion systems in mycobacteria. Cell. Microbiol. 91376-1384. [DOI] [PubMed] [Google Scholar]
- 7.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 10112598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbons, H. S., F. Wolschendorf, M. Abshire, M. Niederweis, and M. Braunstein. 2007. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J. Bacteriol. 1895090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu, S., J. Chen, K. M. Dobos, E. M. Bradbury, J. T. Belisle, and X. Chen. 2003. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 21284-1296. [DOI] [PubMed] [Google Scholar]
- 10.Guinn, K. M., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 1053963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 10012420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, S., J. Kriakov, C. Vilcheze, Z. Dai, G. F. Hatfull, and W. R. Jacobs, Jr. 2004. Bxz1, a new generalized transducing phage for mycobacteria. FEMS Microbiol. Lett. 241271-276. [DOI] [PubMed] [Google Scholar]
- 14.Lipscomb, W. N., and N. Strater. 1996. Recent advances in zinc enzymology. Chem. Rev. 962375-2434. [DOI] [PubMed] [Google Scholar]
- 15.Parsons, L. M., C. S. Jankowski, and K. M. Derbyshire. 1998. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol. Microbiol. 28571-582. [DOI] [PubMed] [Google Scholar]
- 16.Pelicic, V., M. Jackson, J.-M. Reyrat, W. R. Jacobs, and B. Gicquel. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 9410955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9533-539. [DOI] [PubMed] [Google Scholar]
- 18.Rengarajan, J., E. Murphy, A. Park, C. L. Krone, E. C. Hett, B. R. Bloom, L. H. Glimcher, and E. J. Rubin. 2008. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc. Natl. Acad. Sci. USA 105264-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezwan, M., M. A. Laneelle, P. Sander, and M. Daffe. 2007. Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 6832-39. [DOI] [PubMed] [Google Scholar]
- 20.Rigel, N. W., and M. Braunstein. 2008. A new twist on an old pathway-accessory Sec systems. Mol. Microbiol. 69291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sander, P., M. Rezwan, B. Walker, S. K. Rampini, R. M. Kroppenstedt, S. Ehlers, C. Keller, J. R. Keeble, M. Hagemeier, M. J. Colston, B. Springer, and E. C. Bottger. 2004. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 521543-1552. [DOI] [PubMed] [Google Scholar]
- 22.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 9812712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh, A., D. Mai, A. Kumar, and A. J. Steyn. 2006. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. USA 10311346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 25.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 10013001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutcliffe, I. C., and D. J. Harrington. 2004. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 28645-659. [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., and K. M. Derbyshire. 2004. Plasmid DNA transfer in Mycobacterium smegmatis involves novel DNA rearrangements in the recipient, which can be exploited for molecular genetic studies. Mol. Microbiol. 531233-1241. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J., P. K. Karnati, C. M. Takacs, J. C. Kowalski, and K. M. Derbyshire. 2005. Chromosomal DNA transfer in Mycobacterium smegmatis is mechanistically different from classical Hfr chromosomal DNA transfer. Mol. Microbiol. 58280-288. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J., L. M. Parsons, and K. M. Derbyshire. 2003. Unconventional conjugal DNA transfer in mycobacteria. Nat. Genet. 3480-84. [DOI] [PubMed] [Google Scholar]
- 30.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 31.Wu, H. C., and M. Tokunaga. 1986. Biogenesis of lipoproteins in bacteria. Curr. Top. Microbiol. Immunol. 125127-157. [DOI] [PubMed] [Google Scholar]