Abstract
Although BBA74 initially was described as a 28-kDa virulence-associated outer-membrane-spanning protein with porin-like function, subsequent studies revealed that it is periplasmic and downregulated in mammalian host-adapted spirochetes. To further elucidate the role of this protein in the Borrelia burgdorferi tick-mammal cycle, we conducted a thorough examination of its expression profile in comparison with the profiles of three well-characterized, differentially expressed borrelial genes (ospA, ospC, and ospE) and their proteins. In vitro, transcripts for bba74 were expressed at 23°C and further enhanced by a temperature shift (37°C), whereas BBA74 protein diminished at elevated temperatures; in contrast, neither transcript nor protein was expressed by spirochetes grown in dialysis membrane chambers (DMCs). Primer extension of wild-type B. burgdorferi grown in vitro, in conjunction with expression analysis of DMC-cultivated wild-type and rpoS mutant spirochetes, revealed that, like ospA, bba74 is transcribed by σ70 and is subject to RpoS-mediated repression within the mammalian host. A series of experiments utilizing wild-type and rpoS mutant spirochetes was conducted to determine the transcriptional and translational profiles of bba74 during the tick-mouse cycle. Results from these studies revealed (i) that bba74 is transcribed by σ70 exclusively during the larval and nymphal blood meals and (ii) that transcription of bba74 is bracketed by RpoS-independent and -dependent forms of repression that are induced by arthropod- and mammalian host-specific signals, respectively. Although loss of BBA74 does not impair the ability of B. burgdorferi to complete its infectious life cycle, the temporal compartmentalization of this gene's transcription suggests that BBA74 facilitates fitness of the spirochete within a narrow window of its tick phase. A reexamination of the paradigm for reciprocal regulation of ospA and ospC, performed herein, revealed that the heterogeneous expression of OspA and OspC displayed by spirochete populations during the nymphal blood meal results from the intricate sequence of transcriptional and translational changes that ensue as B. burgdorferi transitions between its arthropod vector and mammalian host.
Lyme disease, the most common arthropod-borne infection in the United States, is caused by the spirochete Borrelia burgdorferi (87). In nature, B. burgdorferi has an obligate biannual enzootic cycle involving small mammalian host reservoirs, typically Peromyscus leucopus, and an Ixodes tick vector (51, 96). To successfully complete this life cycle, B. burgdorferi must adapt to and propagate within two markedly different physiologic milieus (67, 75, 96). A number of investigators have reported that manipulation of parameters, such as temperature, pH, DNA supercoiling, cell density, and partial O2 pressure, during in vitro cultivation can trigger changes in gene and protein expression resembling those that occur when spirochetes adapt to their mammalian host (4, 20, 21, 44, 45, 65, 74, 77, 78, 89, 102). It also is now evident, however, that spirochetes must be exposed to as yet unidentified mammalian host-specific signals to induce the full range of transcriptional and translational changes that occur during infection (1, 13, 17, 19, 39, 97). Gene regulation during the tick phase of the enzootic cycle has come to be recognized as equally important to the spirochete's survival strategy (24, 29, 32, 53, 67, 73, 74, 91, 92, 96). In this regard, recent evidence obtained from expression profiling of p66 and ospD points to the existence of arthropod-specific signals that modulate B. burgdorferi gene expression at various times during the tick phases of the cycle (24, 91).
Studies of differential gene expression in B. burgdorferi have given rise to several regulatory paradigms (90). The two most extensively investigated are the reciprocal regulation of outer surface protein A (ospA) and ospC as spirochetes alternate between the arthropod vector and mammalian host (25, 29, 41, 61, 64, 68, 76, 77, 106) and the temperature- and blood meal-dependent induction of the ospE, ospF, and elp (erp) genes (3, 6, 28, 39, 58, 88). An important advance in our understanding of the molecular mechanisms underlying these paradigms was the discovery from the genomic sequence that Lyme disease spirochetes coordinate their physiologic adaptations and pathogenic programs with just three sigma factors: the housekeeping sigma factor, σ70 (RpoD), and the alternate sigma factors RpoN and RpoS (33). Seminal studies by Norgard and coworkers (42, 83, 103) demonstrated that expression of RpoS is regulated by RpoN in concert with the response regulator protein Rrp2. Expression of RpoS, in turn, is essential for the induction of the established virulence factors ospC, dbpBA, and bbk32 as well as numerous other genes thought to be required to establish mammalian infection (18, 19, 42, 59, 79, 80, 100). Analysis of rpoS and RpoS-dependent genes within infected ticks has demonstrated that induction of the RpoS regulon begins during the nymphal blood meal prior to spirochete transmission (i.e., the RpoS-ON state) (19, 35, 36, 39, 41, 76, 77). RpoS is also required for repression of ospA and other tick-phase genes (e.g., bba62 and lp6.6) in response to mammalian host-specific signals (17, 19) although it is not known if RpoS-dependent repression occurs during nymphal tick feeding or only after spirochetes have transited to their murine host. Because ospA is transcribed by σ70 (84), the repression mechanism most likely involves blockage of transcription by an RpoS-dependent trans-acting factor; we along with others have proposed that the poly(T) tract upstream of the ospA promoter also contributes to repression (13, 17, 19, 84). Downregulation of ospA may also be influenced by environmental factors, such as pH (102), and or DNA topology (4). Expression of genes downregulated by RpoS within the mammalian host is thought to resume once spirochetes are acquired by naïve larvae (i.e., the RpoS-OFF state) (19).
Although the presence of homologous upstream regions and highly similar expression profiles initially suggested that the ospE, ospF, and elp genes are controlled by a common regulatory mechanism (2, 28, 58, 89), expression of ospF has been shown to be RpoS dependent while the ospE and elp paralogs are transcribed by σ70 (17, 18, 26, 27). Promoter mapping studies revealed that sequences in the −10 regions of the ospE, ospF, and elp promoters play a critical role in determining recognition by σ70 or RpoS (27). Transcription of ospE and elp paralogs by σ70 would make the corresponding proteins available at points within the enzootic cycle when RpoS is not present. By analogy with regulatory mechanisms identified in other bacteria (14, 37), differential expression of σ70-dependent genes in B. burgdorferi presumably involves trans-acting factors that interact directly or indirectly with RNA polymerase to enhance or diminish the efficiency of gene transcription. Along these lines, Babb et al. (5) have reported that the Erp-binding factor, chromosomal (EbfC), a borrelial YbaB ortholog (55), binds to a region upstream of the ospE paralogs although it is unclear what, if any, effect EbfC binding has on transcription of these genes.
BBA74 initially was described as a 28-kDa virulence-associated outer-membrane-spanning protein with porin-like function (81, 82). More recently, we demonstrated that recombinant BBA74 lacks the physical properties typical of porins and that the native protein is located in the periplasmic space of B. burgdorferi (63). Bioinformatics analysis, however, has been unable to elucidate a possible function for BBA74 as it has no known orthologs. A clue that BBA74 functions within the arthropod vector was provided by microarray studies of spirochetes cultivated within dialysis membrane chambers (DMCs), which showed that bba74 is downregulated in response to mammalian host-specific signals (13, 19). To extend these results, we performed a more thorough characterization of bba74 in comparison to the paradigmatic genes ospC, ospA, and ospE. These analyses revealed that bba74 is transcribed by σ70 exclusively during the larval and nymphal blood meals and that this novel expression pattern is a result of RpoS-independent and -dependent forms of repression that are induced by arthropod host- and mammalian host-specific signals, respectively. Although loss of BBA74 does not impair the ability of Borrelia to complete its infectious life cycle (8, 36, 91), the complex regulation of this gene is consistent with the notion that BBA74 facilitates fitness of the spirochete within a narrow window of its tick phase. Our analysis of bba74 expression during the enzootic cycle also provided an opportunity to reexamine the ospA-ospC regulatory paradigm (75, 90, 96). Contrary to the prediction of strictly reciprocal expression of these genes and their corresponding proteins, spirochetes in the midguts of fed nymphs express large amounts of OspA as well as OspC. Our findings suggest that the heterogeneous expression of OspA and OspC by spirochete populations within fed nymphs results from the intricate sequence of transcriptional and translational changes that ensue as spirochetes transition from the RpoS-OFF to RpoS-ON state.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. burgdorferi strains B31-MI (33), B31-A3 (36), 297 wild-type clone CE162, and the rpoS mutant CE174 (18) were grown in Barbour-Stoenner-Kelly H (BSK-H) medium supplemented with 6% rabbit serum (7). Standard temperature shift experiments were performed as previously described (19). Mammalian host-adapted spirochetes were obtained by cultivation in DMCs implanted into the peritoneal cavities of rats as previously described (1, 19).
DNA sequence analysis.
The bba74 coding sequence was PCR amplified from strains B31-MI and B31-A3 using the gene-specific PCR primers pS73F and p74comR (see Table S1 in the supplemental material), and the resulting amplicons were sequenced in the Davis Sequencing Facility (CA) using primer p74comF (see Table S1 in the supplemental material). The bba74 upstream regions from strains B31-A3 and 297 were PCR amplified using primers pSP73F and pSP74R (see Table S1 in the supplemental material), and the resulting amplicons were sequenced using these same primers (Genewiz, Inc., South Plainfield, NJ).
Mouse infection experiments.
All animal experiments were performed according to protocols approved by the New York Medical College and University of Connecticut Health Center Animal Care and Use Committees. Four-week-old female C3H/HeJ mice were infected with either B31-MI or B31-A3 strains by intradermal inoculation in doses of 1 × 102, 1 × 103, or 1 × 104 spirochetes. Ear punch biopsies were collected 14 days after inoculation, and mice were sacrificed by CO2 asphyxiation at 21 days postinoculation. Various tissue samples (joints, hearts, and urinary bladders) were collected for culture and RNA isolation (see below). To culture B. burgdorferi, ear punch and tissue samples were transferred to 4 ml of BSK-H medium (Sigma-Aldrich, St. Louis, MO) containing an antibiotic mixture of fosfomycin (2 mg/ml), rifampin (5 mg/ml), and amphotericin B (250 μg/ml) (Sigma-Aldrich). All cultures were maintained at 34°C and examined for the presence of spirochetes every 5 to 7 days by dark-field microscopy beginning 5 days after inoculation. The 50% infectious dose (ID50) values were calculated using the algorithm provided by the National Center for Biotechnology Information (85).
Preparation of B. burgdorferi-infected Ixodes scapularis ticks.
To generate B. burgdorferi-infected ticks, approximately 300 to 400 pathogen-free I. scapularis larvae (Oklahoma State University, Stillwater, OK) were placed on infected C3H/HeJ mice 2 to 3 weeks after inoculation by syringe, allowed to feed to repletion, and collected over water. Fed larvae were stored over a supersaturated K2SO4 solution in an environmental incubator maintained at 22°C with a 16:8-h light-dark photoperiod until they had molted to the nymphal stage. To obtain fed nymphs, naive mice were each infested with 10 to 12 B. burgdorferi-infected flat I. scapularis nymphs confined within a capsule placed on their shaved backs. Nymphs were allowed to feed to engorgement (∼72 h postattachment) and were then removed using forceps.
Immersion-fed larvae were generated according to the method described by Policastro and Schwan (72). Briefly, cultures of wild-type and rpoS mutant isolates were grown to late-logarithmic density in BSK-H medium at 33°C, gently pelleted at 4,000 × g for 10 min, and then resuspended in fresh medium to a final density of 1 × 108 spirochetes per ml. The resulting spirochete suspension was mixed gently with 200 to 300 naïve flat I. scapularis larvae for 1.5 h at room temperature. Following incubation, immersion-fed larvae were washed twice with cold phosphate-buffered saline (PBS) and left to rest for 24 h over a solution of supersaturated K2SO4 before being fed on naïve C3H/HeJ mice as described above.
SDS-PAGE and immunoblot analysis.
Spirochetes were harvested by centrifugation at 7,000 × g and washed three times with PBS (pH 7.4) at 4°C. Each pellet was resuspended in 100 μl of 50 mM Tris-HCl (pH 8.0) containing 0.3% sodium dodecyl sulfate (SDS) and 10 mM dithiothreitol (DTT). Total protein lysates were separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) and either silver stained (62) or transferred to nitrocellulose membranes (GE-Healthcare, Milwaukee, WI). Membranes were blotted with antiserum directed against BBA74 (diluted 1:1,000) (63), OspC (1:5,000) (23), OspA (1:30,000) (23), OspE (1:1,000) (1), and FlaB (1:10,000) (17) in conjunction with horseradish peroxidase-conjugated goat anti-rat or anti-rabbit immunoglobulin G (IgG) (1:30,000) (Southern Biotechnology Associates, Birmingham, AL). Chemiluminescent detection was performed using Supersignal West Pico Chemiluminescent substrate (Pierce, Rockford, IL). For serological experiments, total-protein lysates (B31-MI) and 5 ng of purified recombinant BBA74 (63), OspA (23), OspC (23), or OspE (1) was separated on 12.5% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (GE-Healthcare). The membranes were probed with immune serum (1:1,000) obtained from either needle-inoculated (1 × 104 spirochetes per mouse) or tick-inoculated mice collected at 3 weeks postinoculation. Horseradish peroxidase-conjugated goat anti-mouse IgG (1:20,000) (Southern Biotechnology Associates) was used as the secondary antibody.
RNA isolation.
RNAs were isolated from in vitro and DMC-cultivated organisms using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Tissues from infected mice were homogenized in TRIzol reagent using silicon carbide beads (Beadbeater; Biocore, MD) immediately following isolation. Pools of 150 flat or 30 fed B. burgdorferi-infected nymphs were homogenized in TRIzol using a glass homogenizer and centrifuged at 500 × g for 2 min to remove tick debris. Contaminating genomic DNA was removed from isolated RNA samples by treatment with 10 U of RNase-free TurboDNA free (Ambion, Austin, TX), followed by phenol-chloroform extraction and ethanol precipitation. DNA-free RNAs were stored at −80°C.
Primer extension.
A 25-mer oligonucleotide primer (A74ext) (see Table S1 in the supplemental material) located 80 bp downstream of the predicted bba74 ATG start codon (plasmid lp54 coordinates 51718 to 51742) was end labeled with [γ-33P]ATP using T4 polynucleotide kinase. Ten picomoles of primer was incubated with 7,500 Ci/mmole of [γ-33P]ATP in the presence of 10 units of polynucleotide kinase in 1× polynucleotide kinase buffer (70 mM Tris-HCl, 10 mM MgCl2, and 5 mM DTT, pH 7.6). The reaction mixture was incubated at 37°C for 1 h and terminated by the addition of 0.5 ml of TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.6). The end-labeled primer was phenol-chloroform extracted, ethanol precipitated, and resuspended in 100 μl of TE buffer. Approximately 40 μg of DNase I-treated RNA isolated from B31-MI was mixed with 2.5 × 107 cpm of labeled probe and ethanol precipitated. The pellet was resuspended in 8 μl of TE buffer containing 1.25 μmol of KCl, and annealing was carried out at 55°C for 15 min. Ten units of Superscript II reverse transcriptase (RT) (Invitrogen) was added to the reaction mixture, and the mixture was incubated at 42°C for 2 h in RT buffer (10 mM MgCl2, a 1.6 mM concentration of the deoxynucleoside triphosphates [dNTPs], and 0.1 M DTT). The samples were treated with RNase A (5 U) at room temperature for 15 min, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and ethanol precipitated. The pellets were resuspended in 20 μl of 95% formamide, 1 mM EDTA, 0.25% bromophenol blue, and 50% glycerol. The transcriptional start site was identified by comparing the primer extension product to the region upstream of bba74 sequenced using the dideoxy chain termination method (USB Corp., Cleveland, OH) as per the manufacturer's instructions. Briefly, the bba74 locus, including 500 bp of upstream region, was PCR amplified using primers pS73F and p74comR (see Table S1 in the supplemental material) and cloned into the pGEM-T vector for sequencing reactions. Heat-denatured template was annealed at 55°C with primer A74ext in 200 mM Tris-HCl (pH 7.5), 100 mM MgCl2, and 250 mM NaCl. DNA sequencing reactions were carried out by adding labeling master mix (0.1 M DTT, 1,500 Ci/mmole [γ-33P]ATP, 10 U DNA polymerase, and 7.5 μM dNTPs) containing appropriate dideoxy NTPs to heat-denatured template. Reaction mixtures were incubated at 37°C for 5 min and quenched by the addition of 4 μl of stop buffer (20 mM EDTA, 95% formamide, 0.05% bromophenol blue, and 0.05% xylene cyanol). The sequencing reaction and primer extension products were denatured by heating at 75°C for 2 min and immediately resolved on a 6% acrylamide (19:1, acrylamide-bisacrylamide) sequencing gel containing 8 M urea. Gels were dried and exposed to Kodak XAR (Rochester, NY) film overnight at room temperature.
qRT-PCR.
Quantitative RT-PCR (qRT-PCR) was performed using Superscript III RT (Invitrogen, Carlsbad, CA) in reaction mixtures containing 2 μg of total RNA, 10 mM of each dNTP, and 250 ng of random hexamer primers in first-strand buffer (250 mM Tris-HCl, 375 mM KCl, and 15 mM MgCl2,) and 0.1 M DTT. Reaction mixtures were incubated at 42°C for 2 h. The resulting cDNA was amplified in an iCycler thermal cycler (Bio-Rad, Hercules, CA) using the gene-specific primer pairs listed in Table S1 in the supplemental material. Amplification of cDNAs was carried out using 1× iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions with the annealing temperature and concentration of magnesium chloride optimized for each primer pair. Control reactions (no RT and without template) were included for each assay. Amplicons corresponding to each gene target were cloned into pCR2.1-TOPO (Invitrogen), and purified recombinant plasmid DNAs were diluted (107 to 102 copies/μl) to generate standard curves. Transcript copy numbers were calculated using the iCycler postrun analysis software based on internal standard curves and normalized against copies of flaB present in the same cDNA.
Indirect immunofluorescence.
Infected flat nymphs were individually crushed in 10 μl of CMRL medium (US Biological, Swampscott, MA), smeared on poly-l-lysine-coated glass slides (Sigma-Aldrich) and air dried. Pools of five fed nymphs or 10 larvae were eviscerated in 0.5 ml of cold CMRL medium, and the midguts were pelleted by centrifugation at 500 × g for 5 min at 4°C. The pellets were washed twice with 0.5 ml of ice-cold CMRL medium and resuspended in 0.05 ml of CMRL medium; 10-μl aliquots of washed midgut contents were smeared onto Poly-prep slides (Sigma-Aldrich) and air dried. All slides were fixed by immersion in ice-cold acetone for 10 min at −20°C. The slides were incubated overnight in blocking buffer (PBS supplemented with 10% naïve goat serum [NGS] and 0.2% bovine serum albumin) at 4°C. The slides were gently washed twice in PBS supplemented with 1% NGS, air dried, and incubated with rabbit anti-FlaB antiserum (1:200) in combination with rat antiserum directed against BBA74 (1:200), OspA (1:400), OspC (1:200), or OspE (1:200) in PBS with 10% NGS for 1 h at room temperature. After incubation, the slides were washed twice in PBS supplemented with 1% NGS and incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG (1:750) and Alexa Fluor 594-labeled goat anti-rat IgG (1:750) antibodies (Molecular Probes, Invitrogen) in PBS with 10% NGS for 1 h in the dark. The slides were washed twice with PBS with 1% NGS, followed by two washes with PBS alone and two washes with ultrapure water. Slides were air dried in the dark, and the labeled smears were mounted in VectaShield medium containing 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Each double-labeling experiment was performed in triplicate using three independently prepared pools of fed midguts or whole engorged larvae. For each antigen, the percentage of labeling compared to FlaB was evaluated by three individuals, two of whom were blinded; each person counted a minimum of 1,000 FlaB-labeled organisms per antigen tested. The mean percentage ± standard deviation for each antigen was calculated based on the number of FlaB-labeled organisms.
Statistical analysis.
To determine the statistical significance of differences observed in immunofluorescence assay (IFA) and qRT-PCR studies, percent labeling and normalized transcript copy number values, respectively, were compared within Prism, version 5.0 (GraphPad Software, San Diego, CA), using an unpaired t test with two-tailed P values and a 95% confidence interval.
RESULTS
Transcription and translation of bba74 correlate in spirochetes cultivated in DMCs but not in vitro.
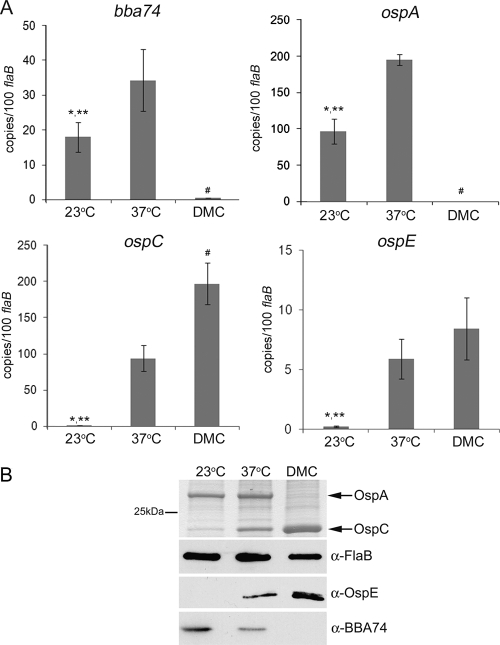
At the outset, we compared the transcriptional profiles of bba74, ospA, ospC, and ospE in spirochetes cultivated in vitro (at 23°C and following a temperature shift to 37°C) and within DMCs. Expression of bba74 increased nearly threefold (P = 0.016) in response to elevated temperature but downshifted markedly in DMC-grown organisms (13, 19, 74); this expression pattern is very similar to that observed for ospA (Fig. 1A). Both ospC and ospE were expressed negligibly at 23°C but were induced strongly upon a temperature shift to 37°C; transcript levels for ospC also showed a further twofold enhancement (P = 0.004) during DMC cultivation (Fig. 1A). Expression of OspA, OspC, and OspE in vitro and within DMCs correlated well with the qRT-PCR results for each respective gene (Fig. 1B). As expected, given the lack of bba74 transcript, BBA74 was undetectable in spirochetes grown within DMCs. In contrast, and despite the increase in transcript copy numbers observed following temperature shift, production of BBA74 decreased in vitro at the higher temperature. In each case, the same expression profile was observed in both strains 297 and B31-MI (data not shown) (18, 19, 39). These experiments, therefore, revealed a dichotomy between the in vitro transcriptional and translational behavior of bba74 that does not occur within DMCs.
FIG. 1.
Transcription and translation of bba74 correlate in vivo but not in vitro. (A) qRT-PCR analyses of bba74, ospA, ospC, and ospE. Values represent the average copy number (± standard deviation) for each gene normalized per 100 copies of flaB. Statistical significance was determined using an unpaired t test; average values are considered significantly different when P is ≤0.05. The single asterisk (*), double asterisks (**), and pound symbol (#) indicate significantly different values for 23°C versus 37°C, 23°C versus DMCs, and 37°C versus DMCs, respectively. (B) B. burgdorferi B31-MI whole-cell lysates were separated by SDS-PAGE and then silver stained to assess expression of OspA and OspC or immunoblotted using specific antisera directed against OspE and BBA74. Reactivity against FlaB was used to confirm that equivalent amounts of lysate were loaded per lane. α, anti.
bba74 is transcribed by σ70 but is subject to RpoS-mediated repression during mammalian host adaptation.
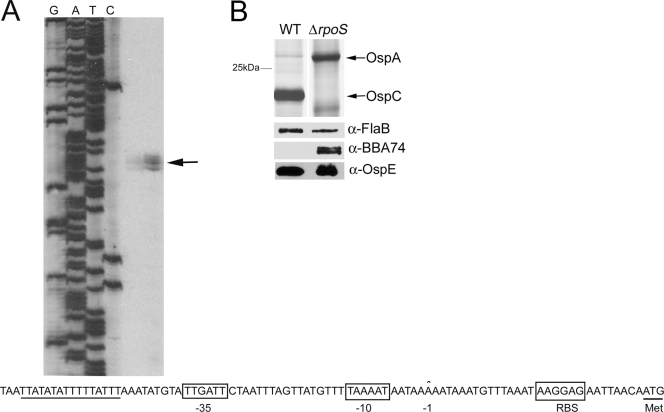
Primer extension analysis was used to identify the promoter elements upstream of the bba74 coding sequence. As shown in Fig. 2A, a transcriptional start site was identified 29 bp upstream of the ATG start codon. Inspection of the upstream DNA revealed excellent matches for the consensus −10 and −35 σ70 recognition motifs (38). Previously, we demonstrated by microarray and qRT-PCR analysis that repression of bba74 in vivo requires RpoS (19). Here, we confirmed these findings at the protein level by immunoblot analysis using lysates prepared from CE174, a well-characterized rpoS mutant (18, 26), and CE162, its strain 297 wild-type parent (Fig. 2B). To ensure the applicability to the B31 strain background of bba74 and BBA74 expression studies performed using the strain 297 rpoS mutant, we compared 500 bp of bba74 upstream DNA sequence from B31-MI with that of strain 297; the resulting alignments revealed only two nucleotide differences between these isolates (data not shown), neither of which was located within the promoter itself. Also note that bba74 was strongly transcribed at 23°C (Fig. 1A), a growth condition under which rpoS and RpoS-dependent genes are not expressed (18, 42). Taken together, these results indicate that bba74 is transcribed exclusively by σ70 and that downregulation of this gene requires both mammalian host-specific signals and RpoS.
FIG. 2.
bba74 is transcribed by σ70 but requires both mammalian host signals and RpoS for downregulation in vivo. (A) The transcriptional start site for bba74 identified by primer extension analysis is indicated by an arrow. Putative −10 and −35 σ70 factor recognition motifs (boxed) as well as a poly(T) tract (underlined) are indicated below. RBS, ribosomal binding site. (B) RpoS-deficient spirochetes fail to downregulate BBA74 during cultivation within DMCs. Whole-cell lysates prepared from wild-type (WT) and rpoS mutant (ΔrpoS) strain 297 isolates were separated by SDS-PAGE and then silver stained to assess expression of OspA and OspC or immunoblotted using specific antisera directed against OspE and BBA74. Reactivity against FlaB was used to confirm that equivalent amounts of lysate were loaded per lane. α, anti.
bba74 is induced during tick feeding but is not expressed during murine infection.
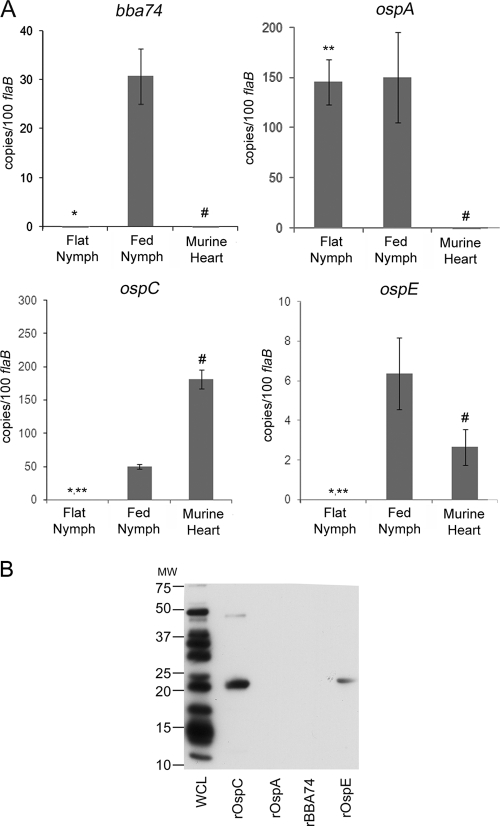
To extend the above findings, we next used qRT-PCR to examine expression of bba74 and BBA74 within flat and fed nymphs at drop-off (72 to 96 h postattachment) and in mouse hearts approximately 17 days postattachment. The data in Fig. 3A show strong induction of bba74 during tick feeding, whereas virtually no transcripts for bba74 were detected in either flat nymphs or infected mice. This expression profile differed from that of ospA, which was expressed intensely in flat nymphs and continued to be expressed at high levels during feeding but was not detected in infected mouse hearts. ospC was induced upon nymphal feeding, and its expression increased further during acute murine infection. ospE expression was induced during feeding, albeit at approximately sevenfold-lower transcript copy numbers than ospC, and continued to be expressed within infected heart tissue. Antibody responses in mice during acute infection were examined to corroborate the qRT-PCR results at the protein level. As shown in Fig. 3B, tick-inoculated mice produced a strong antibody response to OspC and a weaker response to OspE while antibodies directed against BBA74 were not detected. Thus, the transcriptional profile of bba74 is unique in that, like ospE and ospC, it is induced by the mammalian host signals encountered during tick feeding, but, like ospA, it is not expressed during infection.
FIG. 3.
bba74 is expressed during nymphal feeding but not during murine infection. (A) qRT-PCR analysis of bba74, ospA, ospC, and ospE in flat and fed nymphal ticks and in murine hearts. RNAs used to generate cDNAs were obtained from pools of 150 flat or 30 fed B31-MI-infected nymphs at 72 h postattachment and from hearts isolated from three tick-infected C3H/HeJ mice at 17 days postattachment. Values represent the average copy number for each gene (± standard deviation) normalized per 100 copies of flaB. Statistical significance was determined using an unpaired t test; average values are considered significantly different when P is ≤0.05. The single asterisk (*), double asterisks (**), and pound symbol (#) indicate significantly different values for flat versus fed nymphs, flat nymphs versus murine tissue, and fed nymphs versus murine tissue, respectively. (B) Serological responses in mice 3 weeks following infestation with B31-MI-infected I. scapularis nymphs were assessed by immunoblot analysis using B31-MI whole-cell lysates (WCL) or purified recombinant (r) antigens. MW, molecular weight in thousands.
Immunofluorescence microscopy was used to assess expression of BBA74 by individual spirochetes colonizing flat nymphs and within engorged nymphs and larvae. The cumulative results of three independent experiments are presented in Table 1. BBA74 was barely detected in flat ticks but was expressed by more than two-thirds of the spirochetes in both fed nymphal and larval midguts; this result closely resembles the transcriptional pattern observed in flat and fed nymphs (Fig. 3A). OspC was virtually undetectable in flat nymphs and fed larvae but was strongly induced in engorged nymphs; it is important to note, however, that this antigen was detected in less than half of the spirochetes in fed nymphs. OspA, in contrast, was expressed by nearly all of the spirochetes in flat nymphal midguts, by the vast majority of organisms in fed larvae, and by approximately two-thirds of the spirochetes in engorged nymphs. The decreased labeling for OspA in fed compared to flat nymphal midguts was statistically significant (P = 0.035). The labeling pattern for BBA74 most closely resembled that of OspE, which was expressed by very few spirochetes in flat nymphs but was highly expressed in both fed midgut environments; the difference in labeling between fed nymphs and larvae was not significant (P = 0.09). As expected, the expression patterns for all four antigens in strain 297 were highly similar to expression in B31-MI (18, 19, 39; also data not shown). The strong parallel between BBA74 and OspE during all tick stages (Table 1) stands in striking contrast to the differences in their expression levels observed at 23°C in vitro (Fig. 1).
TABLE 1.
Immunofluorescent labeling of B. burgdorferi organisms within infected Ixodes ticks
| Antigen | Percentage of antigen-labeled spirochetes in population of FlaB-labeled spirochetes ina:
|
||
|---|---|---|---|
| Flat nymphs | Fed nymphsb | Fed larvaec | |
| BBA74 | 1.77 ± 0.6 | 78.72 ± 7.18d | 63.4 ± 8.37d |
| OspA | 92.28 ± 6.69e | 67.75 ± 11.96d,e | 89.12 ± 9.21d |
| OspE | 3.21 ± 0.46 | 81.18 ± 7.58d | 69.84 ± 4.43d |
| OspC | ND | 43.68 ± 5.30 | ND |
A minimum of 1,000 FlaB-labeled organisms were counted for each double-labeling experiment. Values presented are the average of three biologically independent experiments ± standard deviations. ND, no labeled spirochetes detected.
Fed nymphs were collected at 72 h postattachment.
Fed larvae were collected within 24 h of repletion.
Values for fed larvae versus fed nymphs labeled for the same antigen are not significantly different.
P = 0.035 for flat versus fed nymphs.
RpoS-mediated repression begins during nymphal tick feeding.
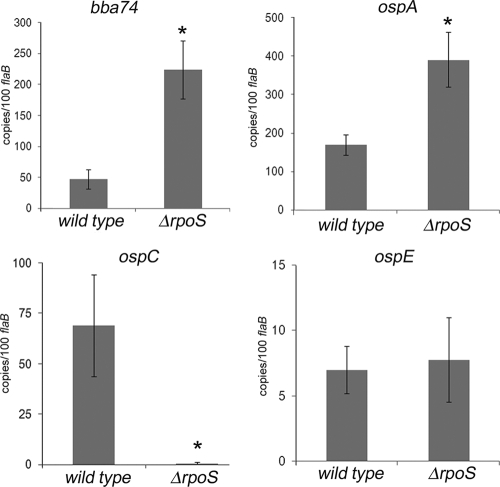
Because RpoS-dependent genes also are induced during nymphal feeding (19, 36, 69, 76, 77), we could not state unequivocally that expression of bba74 during this stage is dependent solely on σ70. Moreover, the data presented thus far left open the question of whether the RpoS-mediated repression of bba74 observed in DMC-cultivated spirochetes is induced by signals conveyed by the blood meal or occurs only after spirochetes have been transmitted to their mammalian host. To address these issues, we infected naïve I. scapularis larvae with wild-type or rpoS mutant B. burgdorferi using the immersion feeding method developed by Policastro and Schwan (72); once fed on a naïve mouse, rpoS mutant spirochetes survived the larval molt and persisted in flat nymphs at levels comparable to the wild-type parent (data not shown). Flat nymphs infected by immersion with either wild-type or rpoS mutant spirochetes were fed on naïve mice and examined by qRT-PCR and IFA. As shown in Fig. 4, the normalized transcript copy numbers for both bba74 and ospA increased dramatically (4.75-fold and 2.3-fold, respectively) in the rpoS mutant compared to its wild-type counterpart while ospE was expressed at similar levels by both isolates. Transcripts for ospC were detected only in ticks infected with the wild-type isolate, thereby confirming that the absolute RpoS dependence of this lipoprotein observed in other systems (19, 26, 42, 105) also pertains in ticks. IFAs revealed that a significant proportion of wild-type strain 297 spirochetes in fed nymphs were negative for BBA74 (26%) and OspA (23%), whereas virtually all of the rpoS mutant spirochetes expressed these two antigens (Table 2). OspE, on the other hand, was detected at comparable levels in both isolates during feeding. Consistent with the qRT-PCR data, OspC was not detected in the RpoS-deficient organisms.
FIG. 4.
qRT-PCR analysis of bba74, ospA, ospC, and ospE in fed nymphal ticks infected with either wild-type or rpoS mutant B. burgdorferi strain 297. RNAs used to generate cDNA for each isolate were obtained from pools of 30 fed nymphs ∼72 h postattachment. Values represent the average copy number for each gene (± standard deviation) normalized per 100 copies of flaB. Statistical significance was determined using an unpaired t test; average values are considered significantly different when P is ≤0.05 (indicated by asterisks).
TABLE 2.
Immunofluorescent labeling of wild-type and rpoS mutant B. burgdorferi organisms within fed Ixodes nymphs
| Antigen | Percentage of antigen-labeled spirochetes in the indicated population of FlaB-labeled spirochetesa
|
Pb | |
|---|---|---|---|
| Wild type | rpoS mutant | ||
| BBA74 | 74.13 ± 6.43 | 91.18 ± 5.48 | 0.002 |
| OspA | 77.33 ± 2.91 | 94.22 ± 3.11 | 0.004 |
| OspE | 53.90 ± 4.94 | 51.27 ± 72.12 | NS |
| OspC | 49.43 ± 2.25 | 0.6 ± 1.07 | 0.005 |
A minimum of 1,000 FlaB-labeled organisms were counted for each double-labeling experiment. Values presented are the average of three biologically independent experiments ± standard deviations.
NS, not significant.
Loss of bba74 does not appear to affect spirochetal infectivity or transit between tick and mouse.
In an attempt to functionally characterize BBA74, we took advantage of the serendipitous observation that B31-A3 contains a point mutation at position 118 of the coding sequence, resulting in a mature polypeptide truncated at amino acid 40 (data not shown). Immunoblot analysis confirmed that B31-A3 does not express full-length BBA74, whereas a polypeptide of the appropriate size (apparent molecular mass of 25 kDa) was readily detected in its isogenic parent, B31-MI (data not shown). The virulence of this natural mutant was compared to that of B31-MI following syringe inoculation. As shown in Table 3, the ID50 values for B31-A3 and B31-MI were highly similar (890 and 1,450, respectively). Because bba74 is induced in fed ticks, it was, of course, possible that the gene is involved in transmission and/or acquisition of spirochetes. However, B31-A3 isolates containing this point mutation (K. Tilly, personal communication) have been taken through the tick-mouse infectious cycle without any obvious defect (8, 91, 95).
TABLE 3.
Infectivity of isolates B31-A3 and B31-MI
| Isolate and dose (no. of spirochetes/ mouse [ID50])a | No. of culture-positive samples/ total no. of samplesb
|
|||
|---|---|---|---|---|
| Ear | Joint | Bladder | All sites | |
| B31-A3 (890 ± 155) | ||||
| 104 | 3/3 | 2/3 | 3/3 | 8/9 |
| 103 | 2/3 | 2/3 | 2/3 | 6/9 |
| 102 | 0/3 | 0/3 | 0/3 | 0/9 |
| B31-MI (1,450 ± 150) | ||||
| 104 | 3/3 | 3/3 | 2/3 | 8/9 |
| 103 | 1/3 | 1/3 | 1/3 | 3/9 |
| 102 | 0/3 | 0/3 | 0/3 | 0/9 |
C3H/HeJ mice (three per group) were needle inoculated with each isolate at the dosage indicated. B31-A3 harbors a truncated BBA74 protein while isolate B31-MI expresses the full-length protein. ID50 values were calculated using an algorithm provided by the National Center for Biotechnology Information.
Based on the presence of spirochetes 3 to 5 weeks following culture in BSK medium.
DISCUSSION
The complex changes that B. burgdorferi undergoes in response to either arthropod- or mammalian host-specific signals are mediated by only two transcriptional pathways. The first utilizes the constitutively expressed sigma factor, σ70, while the second is controlled by the Rrp2-RpoN/RpoS signaling network (17-19, 32, 33, 83, 103, 105). The majority of the genes controlled by σ70 encode proteins that are related to cell maintenance and general metabolism and, therefore, are likely to be required throughout the enzootic cycle. In addition to these housekeeping genes, σ70 also controls the differential expression of genes involved in host adaptation. Some of the earliest and best-studied examples of σ70-dependent upregulation are the ospE alleles (also referred to as erpA, erpC, and erpP in B31-MI). Differential expression of these genes is regulated in a temperature-dependent manner in vitro that is mirrored by their expression profiles during the enzootic cycle (1, 3, 35, 39, 60, 89, 93). σ70 also transcribes a subset of genes whose expression is downregulated in response to either mammalian host-specific (i.e., ospA) (1, 35, 41, 71, 77) or arthropod-specific (i.e., ospD and p66) (24, 91) signals. As is well recognized in other bacteria, changes in promoter recognition by σ70 in B. burgdorferi likely involve the binding of trans-acting factors. The borrelial genome encodes at least four putative DNA binding proteins, Hbb (48, 94), Gac (47), BosR/Fur (10, 46), and the putative YbaB ortholog, EbfC (5, 55), which could contribute either positively or negatively to transcription. The constitutive nature of σ70 enables the spirochete to modulate the expression of genes transcribed by this sigma factor at any point during the enzootic cycle.
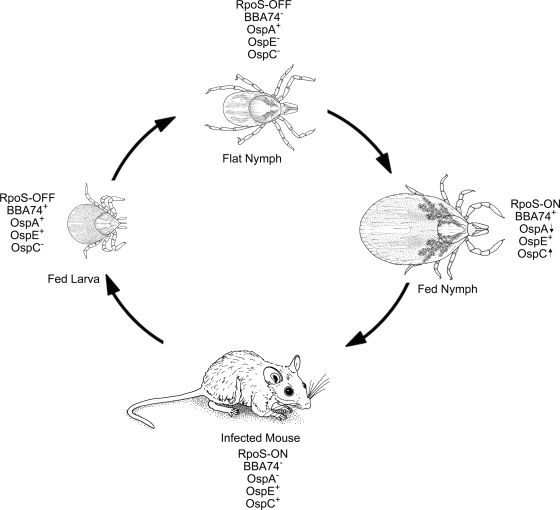
The second transcriptional pathway in B. burgdorferi involves the spirochete's two alternate sigma factors, RpoN and RpoS. Unlike σ70, these sigma factors must be either activated (RpoN) or transcribed de novo (RpoS) in order to modulate gene expression. Although Fisher et al. (32) contended that RpoN controls the expression of a large number of genes independently of RpoS, more recent data demonstrating a substantial overlap between the RpoN and RpoS regulons (9, 66) argues that the contribution of RpoN to borrelial gene regulation is largely confined to the induction of rpoS, in line with the model originally proposed by Hubner et al. (42). Microarray analyses by us (19) and others (9, 32, 66) have identified more than 100 genes whose expression is absolutely dependent on RpoS (19). The strict requirement for RpoS ensures that these genes are transcribed only under conditions when this alternate sigma factor is present. RpoS-dependent gene expression begins during the nymphal blood meal (19) and continues within the mammalian host (34, 40, 43, 52, 54, 104); we have designated this period within the enzootic cycle as the RpoS-ON state (Fig. 5). RpoS also is required for the downregulation of 33 σ70-dependent genes in response to mammalian host signals, among which are the tick-phase genes ospA, bba62, and lp6.6 (17, 19). Relief from RpoS-mediated repression occurs almost immediately following acquisition of spirochetes by feeding naïve larvae (16, 19, 35, 76), thereby giving rise to a period within the enzootic cycle that we have designated as the RpoS-OFF state (Fig. 5). By synchronizing the reciprocal regulation of tick- and mammalian host-phase-specific genes with these alternating RpoS states, B. burgdorferi has evolved a facile means of precisely coordinating the transcriptome changes required for transitioning between its mammalian host and arthropod vector. The demonstration here that RpoS-deficient spirochetes survive and replicate at levels comparable to their wild-type counterparts within flat and fed nymphs infected as larvae by immersion provides further support for our contention that RpoS is not required for physiological adaptation during the enzootic cycle but, rather, controls the expression of key virulence determinants involved in cycling between the arthropod and mammalian hosts (18, 19).
FIG. 5.
Expression profiles of BBA74, OspA, OspE, and OspC in relation to the on and off states of RpoS during the enzootic cycle. Expression of each antigen is based on qRT-PCR and IFA data. Up and down arrows are used to indicate the decreasing and increasing expression of OspA and OspC, respectively, by spirochetes during the nymphal blood meal.
The expression profile of bba74 illustrates the complexity of gene regulation that the spirochete can achieve by integrating elements of both the σ70- and RpoN/RpoS-dependent regulatory networks. bba74 is transcribed when rpoS is either not expressed (i.e., at 23°C in vitro and within fed larvae) or not present (i.e., rpoS mutant). Based on these findings, we conclude that transcription of bba74 is strictly σ70 dependent, and, therefore, differential expression of this gene likely involves modulating promoter recognition by the housekeeping sigma factor. Like ospA, bba74 is not expressed during murine infection, i.e., during the RpoS-ON state. The avirulence of RpoS-deficient spirochetes (18) precludes a direct examination of mechanism(s) controlling downregulation of bba74 and ospA during infection; using DMCs as a surrogate for infection, however, we have demonstrated, here and elsewhere (17, 19), that repression of both of these genes within the mammalian host is mediated by RpoS. Expression of bba74 and ospA resumes during the larval blood meal, when newly acquired spirochetes assume the RpoS-OFF state. In contrast to ospA, expression of bba74 declines to virtually undetectable levels following the larval molt when rpoS is no longer expressed (19). Based on the observation that bba74 is well expressed at 23°C in vitro (Fig. 1A), we hypothesize that the lack of expression of this gene within flat nymphs is due to an RpoS-independent form of suppression induced by arthropod-specific signals. Release from this suppression would then be triggered by the influx of the next blood meal as demonstrated by the high levels of BBA74-positive spirochetes in fed nymphs. As feeding progresses and spirochetes prepare for transmission to the mammalian host, expression of bba74 once again becomes subject to RpoS-mediated repression, as is evident by the marked increase in BBA74 positivity in fed nymphs infected with rpoS mutant spirochetes. By bracketing the transcription of bba74 between two forms of repression that are synchronized with the spirochete's RpoS-ON and RpoS-OFF states, B. burgdorferi is able to restrict its expression to the larval and nymphal blood meals. The interrelationship between the σ70 and RpoS transcriptional networks discerned from our analyses of bba74 also provides a fresh perspective on ospE, the other σ70-dependent gene examined herein. Like bba74, ospE is not expressed within flat nymphs and is induced during nymphal feeding. Because it is not subject to RpoS-mediated repression, ospE continues to be transcribed during murine infection and larval acquisition (Fig. 5). This expression pattern ensures that OspE is present during all stages of the enzootic cycle when the spirochete requires its complement-inactivating (11, 49) and plasminogen-binding (12) activities.
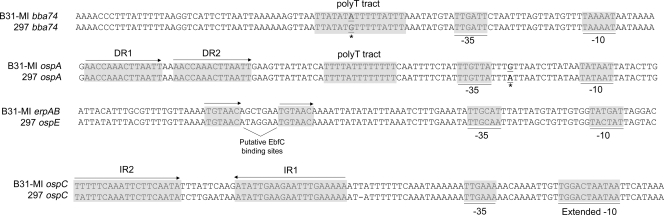
A comparison of the upstream sequences for bba74 and the three paradigmatic genes examined herein helps to explain how spirochetes achieved the versatile transcriptional programs involved their expression (Fig. 6). Mutagenesis studies in B. burgdorferi (26, 27), as well as in Escherichia coli (98), have demonstrated that the −10 motif is a critical determinant of sigma factor recognition and/or promoter selectivity. Consistent with their σ70 dependence, the promoters for bba74, ospA, and ospE all contain similar −10 consensus motifs that are distinct from the extended −10 sequence [TG(G/A)(G/A) ATA(T/A)ATT] required for promoter recognition of ospC and other RpoS-dependent genes (19). The high degree of similarity between the bba74 and ospA −10 motifs compared to the same region of ospE could account for the markedly higher transcript levels for these two genes in vitro and within fed ticks. The ospA upstream sequence contains two motifs, a poly(T) tract and the direct repeats DR1 and DR2, which could function as cis-acting elements for the downregulation of this gene (Fig. 6). If one assumes that the RpoS-dependent downregulation of bba74 and ospA occurs via a common trans-acting factor, then DR1 and DR2 cannot be involved because they are not present upstream of bba74. We (17, 19) along with others (13, 84) have proposed that the poly(T) tracts shared by both bba74 and ospA represent a candidate repressor binding site. ospE is thought to be regulated, in part, by EbfC, a chromosomally encoded YbaB DNA binding protein ortholog (55) induced during tick feeding and murine infection (5, 59). The absence of EbfC recognition sites (TGTAACA) within the bba74 upstream region leads us to predict that bba74 and ospE are regulated differently despite the fact that both are downregulated within flat nymphs and induced during the nymphal blood meal. The observation that ospE displays a classic temperature-inducible expression pattern in vitro (1, 2, 28, 39, 89) while bba74 does not (Fig. 1A) supports this supposition.
FIG. 6.
Alignment of the bba74, ospA, ospE, and ospC upstream sequences from strains B31-MI and 297. The σ70 consensus −35 and −10 sites for ospA, bba74, and ospE are based on those of E. coli (50, 99) while the RpoS-dependent extended −10 for ospC is from Eggers et al. (26). The ospA direct repeats (DR1 and DR2) and ospC operator region (IR1 and IR2) are based on Sohaskey et al. (84) and Xu et al. (101), respectively. Poly(T) tracts upstream of ospA and bba74 are based on Caimano et al. (19). Putative EbfC binding sites are based on Babb et al. (5). Transcriptional start sites (+1) for ospA (84), ospC (57), and ospE (39) are based on previously published reports while the start site for bba74 is based on primer extension analyses presented here. Asterisks are used to indicate nucleotide polymorphisms between the ospA and bba74 genes of strains B31-MI and 297, confirmed by sequencing.
The reciprocal expression of OspA and OspC as spirochetes alternate between the arthropod vector and mammal host is a central paradigm of differential gene regulation by B. burgdorferi (75, 90). Numerous studies demonstrating that spirochetes within flat nymphs express abundant amounts of OspA and no OspC, while the converse pattern holds during mammalian infection, are often interpreted to mean that expression of these two lipoproteins is mutually exclusive. On the other hand, qRT-PCR and IFA studies, including those presented here, have shown that spirochetal populations within fed nymphs express both antigens simultaneously (31, 64, 76, 77), while Ohnishi et al. (64) demonstrated by double labeling that this is true for individual organisms as well. How can one explain the ostensible dichotomy created by the findings that strict reciprocal expression of OspA and OspC does not occur during all stages of the enzootic cycle? The heterogeneity in OspA and OspC expression levels observed during nymphal feeding could also be viewed as problematic for our concept of RpoS as the coordinator of the ospA ↔ ospC transcriptional switch. One potential explanation is that upregulation of OspC occurs during the nymphal blood meal while RpoS-dependent downregulation of OspA occurs within the mammalian host. Two lines of evidence, however, argue strongly that these two transcriptional changes occur concurrently: (i) IFAs revealing that a substantial proportion of spirochetes within fed nymphs are OspA−/OspC+ (30, 64, 76) (Tables 1 and 2) and (ii) our demonstration herein that expression of ospA is markedly enhanced in rpoS mutant spirochetes during the nymphal blood meal (Fig. 4 and Table 2).
We propose that the “deviation”' from the OspA/OspC paradigm results instead from the concatenation of transcriptional and translational events that ensue when spirochetes transition from the RpoS-OFF to -ON state (Fig. 5). The substantial proportion of spirochetes within engorged nymphs that remain OspC− (∼50 to 80%) (64, 76) (Tables 1 and 2) indicates that, for unknown reasons, many organisms either do not initiate the RpoS program or do so very slowly. For those organisms that do demonstrate an RpoS-ON state, one would predict that the appearance of OspC, which is dependent solely on the transcription and translation of ospC mRNA, would occur much more rapidly than the downregulation of OspA, which requires the synthesis and/or activation of an unidentified repressor, the turnover of residual ospA mRNA, and the dilution of the large amounts of OspA lipoprotein present prior to the burst of replication that accompanies the blood meal. Srivastava and de Silva (86) proposed a similar scenario based on the use of flow cytometry to study the expression of OspA and OspC by wild-type and rpoN mutant spirochetes following a temperature shift in vitro. This sequence of events serves equally well to explain the heterogeneity of BBA74 expression within fed nymphs. A key question that remains unresolved is how spirochetes achieve the high degree of OspA−/OspC+ uniformity that is characteristic of acute infection. Ohnishi et al. (64) concluded that the selectivity for OspA−/OspC+ spirochetes does not occur within the tick but within the dermis of the infected murine host. Previous reports (32, 36) demonstrating that a lack of OspC does not impair the ability of spirochetes to traverse the midgut and penetrate the salivary glands are in accord with this conclusion. A recent study by Battisti et al. (8) suggests that, in addition to its role as a TROSPA ligand, OspA serves a critical antibody-shielding role within nature, where B. burgdorferi-infected nymphal ticks may feed on reservoir hosts that have been previously infected; in this case, the continued expression of OspA during the nymphal blood meal would provide a survival advantage to spirochetes that would not be reflected within laboratory-infected nymphs fed on naïve hosts. Given the minute fraction of organisms that reach the salivary glands (22, 32, 36, 56, 69, 70; also our unpublished findings), it is reasonable to presume that the transcriptional events initiated by feeding do generate an infection-competent phenotype that is denoted by an as yet undetermined antigen expression pattern, including downregulation of BBA74.
We took advantage of the serendipitous discovery that B31-A3 is a natural BBA74 mutant to investigate the biological function of the corresponding gene product. The virulence of B31-A3 by needle inoculation is in line with our findings that bba74 is not expressed during murine infection. In light of the high levels of expression of BBA74 within both fed larvae and nymphs, we were surprised to learn, however, that B31-A3 is able to successfully complete the tick-mouse cycle (8, 36, 91). We cannot dismiss the possibility that bba74 encodes either a nonessential gene product or that its loss is compensated for by another borrelial gene. Alternatively, it is possible that insertional inactivation of bba74 within a clonal wild-type background may yield results that differ from those comparing B31-A3 and B31-MI. Moreover, naturally infected ticks collected from sites of endemicity often have substantially lower spirochete burdens (15) than those generated experimentally. Therefore, a comparison of wild-type- and Δbba74 mutant-infected ticks containing spirochete burdens closer to those found in nature may reveal a difference in survival rates between the two isolates. Lastly, BBA74 may be required for adaptation within a tick microenvironment that cannot be readily detected using qRT-PCR and IFA. Stewart et al. (91) recently reported that ospD exhibits a blood meal-restricted expression profile similar, though not identical, to that of bba74. As with BBA74, OspD-deficient spirochetes did not exhibit an obvious phenotype and were able to complete the tick-mouse infectious cycle, leading the authors to propose that OspD may have a nonessential, but nevertheless important, role in intercellular cell signaling and/or nutrient scavenging. Unlike the OspD lipoprotein, BBA74 is located entirely within the periplasm and, therefore, would not function at the host-pathogen interface. bba74 and ospD point to the existence of a class of gene products required by the spirochete for adaptation to the common physiological demands and/or environmental stresses imposed by the blood meal during both stages of the tick life cycle.
Supplementary Material
Acknowledgments
We thank Cynthia Gonzalez for her superb technical assistance, Juan Salazar for helpful suggestions and advice on the statistical analyses, and Kit Tilly for kindly providing strain B31-A3 and data regarding the original isolation and tick transmissibility of this isolate.
This work was supported in part by NIH grants AI-29735 (J.R. and M.J.C.) and AI45801 (I.S.).
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 1012240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 671526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol. Microbiol. 18507-520. [DOI] [PubMed] [Google Scholar]
- 4.Alverson, J., S. F. Bundle, C. D. Sohaskey, M. C. Lybecker, and D. S. Samuels. 2003. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 481665-1677. [DOI] [PubMed] [Google Scholar]
- 5.Babb, K., T. Bykowski, S. P. Riley, M. C. Miller, E. DeMoll, and B. Stevenson. 2006. Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 1884331-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babb, K., N. El Hage, J. C. Miller, J. A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control expression of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 694146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57521-525. [PMC free article] [PubMed] [Google Scholar]
- 8.Battisti, J. M., J. L. Bono, P. A. Rosa, M. E. Schrumpf, T. G. Schwan, and P. F. Policastro. 2008. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect. Immun. 765228-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boardman, B. K., M. He, Z. Ouyang, H. Xu, X. Pang, and X. F. Yang. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 763844-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boylan, J. A., J. E. Posey, and F. C. Gherardini. 2003. Borrelia oxidative stress response regulator, BosR: A distinctive Zn-dependent transcriptional activator. Proc. Natl. Acad. Sci. USA 10011684-11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brissette, C. A., A. E. Cooley, L. H. Burns, S. P. Riley, A. Verma, M. E. Woodman, T. Bykowski, and B. Stevenson. 2008. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 298(Suppl. 1)257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brissette, C. A., K. Haupt, D. Barthel, A. E. Cooley, A. Bowman, C. Skerka, R. Wallich, P. F. Zipfel, P. Kraiczy, and B. Stevenson. 2009. The Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77300-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 713371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 257-65. [DOI] [PubMed] [Google Scholar]
- 15.Brunet, L. R., A. Spielman, and S. R. Telford. 1995. Short report: density of Lyme disease spirochetes within deer ticks collected from zoonotic sites. Am. J. Trop. Med. Hyg. 53300-302. [DOI] [PubMed] [Google Scholar]
- 16.Bykowski, T., M. E. Woodman, A. E. Cooley, C. A. Brissette, V. Brade, R. Wallich, P. Kraiczy, and B. Stevenson. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 754227-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caimano, M. J., C. H. Eggers, C. A. Gonzalez, and J. D. Radolf. 2005. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J. Bacteriol. 1877845-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caimano, M. J., C. H. Eggers, K. R. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 726433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 686677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 673181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 891111-1119. [DOI] [PubMed] [Google Scholar]
- 23.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 937973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cugini, C., M. Medrano, T. G. Schwan, and J. Coburn. 2003. Regulation of expression of the Borrelia burgdorferi β3-chain integrin ligand, P66, in ticks and in culture. Infect. Immun. 711001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Silva, A. M., S. R. Telford, III, L. R. Brunet, S. W. Barthold, and E. Fikrig. 1996. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J. Exp. Med. 183271-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2004. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 1867390-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2006. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 591859-1875. [DOI] [PubMed] [Google Scholar]
- 28.El-Hage, N., and B. Stevenson. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 1844536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fikrig, E., and S. Narasimhan. 2006. Borrelia burgdorferi—traveling incognito? Microbes Infect. 81390-1399. [DOI] [PubMed] [Google Scholar]
- 30.Fingerle, V., U. Hauser, G. Liegl, B. Petko, V. Preac-Mursic, and B. Wilske. 1995. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus. J. Clin. Microbiol. 331867-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fingerle, V., G. Liegl, U. Munderloh, and B. Wilske. 1998. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus ticks removed from humans. Med. Microbiol. Immunol. 187121-126. [DOI] [PubMed] [Google Scholar]
- 32.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 1025162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 34.Gilmore, R. D., Jr., R. R. Howison, V. L. Schmit, A. J. Nowalk, D. R. Clifton, C. Nolder, J. L. Hughes, and J. A. Carroll. 2007. Temporal expression analysis of the Borrelia burgdorferi paralogous gene family 54 genes BBA64, BBA65, and BBA66 during persistent infection in mice. Infect. Immun. 752753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3799-808. [DOI] [PubMed] [Google Scholar]
- 36.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: A protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 1013142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haugen, S. P., W. Ross, and R. L. Gourse. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 112237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 693618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodzic, E., S. Feng, K. J. Freet, and S. W. Barthold. 2003. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 715042-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodzic, E., S. Feng, K. J. Freet, D. L. Borjesson, and S. W. Barthold. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 703382-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 9812724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes, J. L., C. L. Nolder, A. J. Nowalk, D. R. Clifton, R. R. Howison, V. L. Schmit, R. D. Gilmore, Jr., and J. A. Carroll. 2008. Borrelia burgdorferi surface-localized proteins expressed during persistent murine infection are conserved among diverse Borrelia spp. Infect. Immun. 762498-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyde, J. A., J. P. Trzeciakowski, and J. T. Skare. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J. Bacteriol. 189437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Indest, K. J., R. Ramamoorthy, M. Sole, R. D. Gilmore, B. J. Johnson, and M. T. Philipp. 1997. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect. Immun. 651165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katona, L. I., R. Tokarz, C. J. Kuhlow, J. Benach, and J. L. Benach. 2004. The fur homologue in Borrelia burgdorferi. J. Bacteriol. 1866443-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knight, S. W., and D. S. Samuels. 1999. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J. 184875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobryn, K., D. Z. Naigamwalla, and G. Chaconas. 2000. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 37145-155. [DOI] [PubMed] [Google Scholar]
- 49.Kraiczy, P., C. Skerka, P. F. Zipfel, and V. Brade. 2002. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: a new protein family involved in complement resistance. Wien. Klin. Wochenschr. 114568-573. [PubMed] [Google Scholar]
- 50.Lacour, S., A. Kolb, and P. Landini. 2003. Nucleotides from −16 to −12 determine specific promoter recognition by bacterial σS-RNA polymerase. J. Biol. Chem. 27837160-37168. [DOI] [PubMed] [Google Scholar]
- 51.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36587-609. [DOI] [PubMed] [Google Scholar]
- 52.Li, X., X. Liu, D. S. Beck, F. S. Kantor, and E. Fikrig. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 743305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, X., U. Pal, N. Ramamoorthi, X. Liu, D. C. Desrosiers, C. H. Eggers, J. F. Anderson, J. D. Radolf, and E. Fikrig. 2007. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol. Microbiol. 63694-710. [DOI] [PubMed] [Google Scholar]
- 54.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim, K., A. Tempczyk, J. F. Parsons, N. Bonander, J. Toedt, Z. Kelman, A. Howard, E. Eisenstein, and O. Herzberg. 2003. Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR. Proteins 50375-379. [DOI] [PubMed] [Google Scholar]
- 56.Lima, C. M., N. S. Zeidner, C. B. Beard, C. A. Soares, M. C. Dolan, G. Dietrich, and J. Piesman. 2005. Differential infectivity of the Lyme disease spirochete Borrelia burgdorferi derived from Ixodes scapularis salivary glands and midgut. J. Med. Entomol. 42506-510. [DOI] [PubMed] [Google Scholar]
- 57.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marconi, R. T., S. Y. Sung, C. A. Norton Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 1785615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medrano, M. S., Y. Ding, X. G. Wang, P. Lu, J. Coburn, and L. T. Hu. 2007. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 1892653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller, J. C., and B. Stevenson. 2004. Increased expression of Borrelia burgdorferi factor H-binding surface proteins during transmission from ticks to mice. Int. J. Med. Microbiol. 293(Suppl. 37)120-125. [DOI] [PubMed] [Google Scholar]
- 61.Montgomery, R. R., S. E. Malawista, K. J. M. Feen, and L. K. Bockenstedt. 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117307-310. [DOI] [PubMed] [Google Scholar]
- 63.Mulay, V., M. J. Caimano, D. Liveris, D. C. Desrosiers, J. D. Radolf, and I. Schwartz. 2007. Borrelia burgdorferi BBA74, a periplasmic protein associated with the outer membrane, lacks porin-like properties. J. Bacteriol. 1892063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohnishi, J., J. Piesman, and A. M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA 98670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 711689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ouyang, Z., J. S. Blevins, and M. V. Norgard. 2008. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 1542641-2658. [DOI] [PubMed] [Google Scholar]
- 67.Pal, U., and E. Fikrig. 2003. Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes Infect. 5659-666. [DOI] [PubMed] [Google Scholar]
- 68.Pal, U., X. Li, T. Wang, R. R. Montgomery, N. Ramamoorthi, A. M. Desilva, F. Bao, X. Yang, M. Pypaert, D. Pradhan, F. S. Kantor, S. Telford, J. F. Anderson, and E. Fikrig. 2004. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119457-468. [DOI] [PubMed] [Google Scholar]
- 69.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piesman, J., B. S. Schneider, and N. S. Zeidner. 2001. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 394145-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piesman, J., N. S. Zeidner, and B. S. Schneider. 2003. Dynamic changes in Borrelia burgdorferi populations in Ixodes scapularis (Acari: Ixodidae) during transmission: studies at the mRNA level. Vector Borne Zoonotic Dis. 3125-132. [DOI] [PubMed] [Google Scholar]
- 72.Policastro, P. F., and T. G. Schwan. 2003. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J. Med. Entomol. 40364-370. [DOI] [PubMed] [Google Scholar]
- 73.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, F. J. De La, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 1026972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwan, T. G. 1996. Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect. Agents Dis. 5167-181. [PubMed] [Google Scholar]
- 76.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 922909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seshu, J., J. A. Boylan, F. C. Gherardini, and J. T. Skare. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 721580-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seshu, J., M. D. Esteve-Gassent, M. Labandeira-Rey, J. H. Kim, J. P. Trzeciakowski, M. Höök, and J. T. Skare. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 591591-1601. [DOI] [PubMed] [Google Scholar]
- 80.Shi, Y., Q. Xu, K. McShan, and F. T. Liang. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 761239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 1784909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skare, J. T., E. S. Shang, D. M. Foley, D. R. Blanco, C. I. Champion, T. Mirzabekov, Y. Sokolov, B. L. Kagan, J. M. Miller, and M. A. Lovett. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Investig. 962380-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 1892139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sohaskey, C. D., W. R. Zuckert, and A. G. Barbour. 1999. The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol. Microbiol. 3341-51. [DOI] [PubMed] [Google Scholar]
- 85.Spouge, J. I., R. I. Shrager, and D. S. Dimitrov. 1996. HIV-1 infection kinetics in tissue cultures. Math. Biosci. 1381-22. [DOI] [PubMed] [Google Scholar]
- 86.Srivastava, S. Y., and A. M. de Silva. 2008. Reciprocal expression of ospA and ospC in single cells of Borrelia burgdorferi. J. Bacteriol. 1903429-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 1131093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stevenson, B., J. L. Bono, T. G. Schwan, and P. A. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 662648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 634535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stevenson, B., L. K. von, S. P. Riley, A. E. Cooley, M. E. Woodman, and T. Bykowski. 2006. Evolving models of Lyme disease spirochete gene regulation. Wien. Klin. Wochenschr. 118643-652. [DOI] [PubMed] [Google Scholar]
- 91.Stewart, P. E., A. Bestor, J. N. Cullen, and P. A. Rosa. 2008. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect. Immun. 761970-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Strother, K. O., A. Broadwater, and A. de Silva. 2005. Plasmid requirements for infection of ticks by Borrelia burgdorferi. Vector Borne Zoonotic Dis. 5237-245. [DOI] [PubMed] [Google Scholar]
- 93.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 924269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tilly, K., J. Fuhrman, J. Campbell, and D. S. Samuels. 1996. Isolation of Borrelia burgdorferi genes encoding homologues of DNA-binding protein HU and ribosomal protein S20. Microbiology 1422471-2479. [DOI] [PubMed] [Google Scholar]
- 95.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, M. J. Vanraden, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 743554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tilly, K., P. A. Rosa, and P. E. Stewart. 2008. Biology of infection with Borrelia burgdorferi. Infect. Dis. Clin. N. Am. 22217-234, v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 725419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Typas, A., G. Becker, and R. Hengge. 2007. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 631296-1306. [DOI] [PubMed] [Google Scholar]
- 99.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weening, E. H., N. Parveen, J. P. Trzeciakowski, J. M. Leong, M. Hook, and J. T. Skare. 2008. Borrelia burgdorferi lacking dbpBA exhibits an early survival defect during experimental infection. Infect. Immun. 765694-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu, Q., K. McShan, and F. T. Liang. 2007. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol. Microbiol. 64220-231. [DOI] [PubMed] [Google Scholar]
- 102.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 371470-1479. [DOI] [PubMed] [Google Scholar]
- 103.Yang, X. F., S. M. Alani, and M. V. Norgard. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 1001100-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang, X. F., A. Hubner, T. G. Popova, K. E. Hagman, and M. V. Norgard. 2003. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect. Immun. 715012-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang, X. F., M. C. Lybecker, U. Pal, S. M. Alani, J. Blevins, A. T. Revel, D. S. Samuels, and M. V. Norgard. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 1874822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.