Abstract
The RND-type efflux pumps are responsible for the multidrug resistance phenotype observed in many clinically relevant species. Also, RND pumps have been implicated in physiological processes, with roles in the virulence mechanisms of several pathogenic bacteria. We have previously shown that the BepC outer membrane factor of Brucella suis is involved in the efflux of diverse drugs, probably as part of a tripartite complex with an inner membrane translocase. In the present work, we characterize two membrane fusion protein-RND translocases of B. suis encoded by the bepDE and bepFG loci. MIC assays showed that the B. suis ΔbepE mutant was more sensitive to deoxycholate (DOC), ethidium bromide, and crystal violet. Furthermore, multicopy bepDE increased resistance to DOC and crystal violet and also to other drugs, including ampicillin, norfloxacin, ciprofloxacin, tetracycline, and doxycycline. In contrast to the ΔbepE mutant, the resistance profile of B. suis remained unaltered when the other RND gene (bepG) was deleted. However, the ΔbepE ΔbepG double mutant showed a more severe phenotype than the ΔbepE mutant, indicating that BepFG also contributes to drug resistance. An open reading frame (bepR) coding for a putative regulatory protein of the TetR family was found upstream of the bepDE locus. BepR strongly repressed the activity of the bepDE promoter, but DOC released the repression mediated by BepR. A clear induction of the bepFG promoter activity was observed only in the BepDE-defective mutant, indicating a regulatory interplay between the two RND efflux pumps. Although only the BepFG-defective mutant showed a moderate attenuation in model cells, the activities of both bepDE and bepFG promoters were induced in the intracellular environment of HeLa cells. Our results show that B. suis harbors two functional RND efflux pumps that may contribute to virulence.
Brucella is a facultative intracellular pathogen taxonomically classified within the Alphaproteobacteria, along with other intracellular pathogens, such as Rickettsia, Bartonella, and several plant symbionts and pathogens (7). Brucella spp. are the etiological agents of brucellosis, a major zoonotic disease distributed worldwide and transmitted from domestic, farm, and wild animals to humans. Brucella enters the host through the nasal, oral, and pharyngeal cavities and from there is transported to the proximal lymph nodes. Early during infection, host innate immunity mechanisms contribute to reduce the initial number of infecting brucellae (26). Once in contact with the organism, Brucella is able to invade professional and nonprofessional phagocytes (10). Within the cells, Brucella is found in a membrane-associated vacuole called a Brucella-containing vacuole. The Brucella-containing vacuole is able to subvert the normal phagocytic pathway to form a vacuole with endoplasmic reticulum markers suitable for Brucella sp. replication (13, 50). This strategy helps the bacteria to escape from the bactericidal mechanisms used by the host (7).
Host barriers range from antimicrobial products of the innate immune system to toxic compounds, such as bile salts, in the gastrointestinal tract. These barriers constitute effective defense mechanisms that a pathogen must overcome to survive, colonize, and replicate. In several bacterial species, low outer membrane permeability to hydrophobic (and toxic) compounds accompanies active efflux of the noxious agent (37), allowing pathogens to eliminate toxic compounds by pumping them from the cytoplasm back to the external environment. Efflux pumps have been classified into five families according to amino acid sequence homology and their mechanisms of action. Three-component pumps of gram-negative bacteria traverse both inner and outer membranes and form a continuous channel through which the substrate is transported without a periplasmic intermediate. These systems operate with an inner membrane transporter (IM), a protein from the membrane fusion protein (MFP) family that is mostly periplasmically anchored to the inner membrane, and an outer membrane factor (OMF), which is recruited by the IM-MFP complex to form the channel (1, 4, 63). Within the IM components, those belonging to the resistance-nodulation-cell division (RND) superfamily have been shown to exhibit unusually broad substrate spectra, resulting in a multiple drug resistance phenotype (42, 55). The uncontrolled expression of RND-MFP-OMF tripartite efflux pumps has been associated with the multiple drug resistance phenotype in many clinically relevant strains (48, 69). In addition, recent works have reported a role of the RND-MFP-OMF in bacterial pathogenesis (49).
Compared with other gram-negative bacteria and with the closely related Ochrobactrum, Brucella shows an outer membrane with higher permeability to hydrophobic compounds (38, 65). This characteristic has been associated with the properties of the lipopolysaccharide, which allows exposure of hydrophobic patches (41). Conversely, this feature was associated with an increase in resistance to polycations and EDTA (40). The elevated permeability to hydrophobic molecules of the Brucella outer membrane makes efflux pumps particularly relevant to survival within the host. In a previous study, we showed that the unique OMF identified in Brucella suis, named BepC (for Brucella efflux protein C) is involved in drug efflux. A mutational approach has shown that a BepC-deficient mutant was more sensitive to hydrophobic compounds, including antimicrobials, several dyes, and intercalators. Furthermore, survival of the bepC mutant was affected in the mouse model, probably due to its inability to expel toxic compounds during the course of infection (54). Since BepC must interact with an inner membrane translocase (for example, an RND-MFP complex), a question arises as to which partners of BepC are involved in the efflux of toxic compounds.
Analysis of the of B. suis genome sequence revealed the presence of at least six putative RND-MFP translocases that may interact with BepC. In this study, we show that two RND-MFP translocases of B. suis are involved in the efflux of several toxic compounds that partially account for the defects observed in the bepC mutant. In addition, we present evidence of notable regulatory interplay between the two translocases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All Brucella strains used in this study were derived from B. suis 1330 (ATCC 23444T) and are listed in Table 1. The Brucella strains were grown in tryptic soy broth (TSB) (Bacto) medium or in modified minimal medium E (MME) (29) in combination with the appropriate antibiotics (chloramphenicol, 6 μg ml−1; kanamycin, 25 μg ml−1). Escherichia coli strain DH5α was used as the recipient strain for all cloning experiments and was routinely grown in Luria-Bertani (Bacto) medium with the appropriate antibiotics (ampicillin, 50 μg ml−1; chloramphenicol, 50 μg ml−1; and kanamycin, 50 μg ml−1). All bacterial strains were grown at 37°C and 200 rpm when needed. DNA manipulation was performed according to standard techniques (58). All experiments with viable brucellae were performed in a biosafety level 3 containment facility. Since the plasmid pFC138 with cloned bepDE conferred a reduction in sensitivity to some antibiotics, including tetracycline, the stocks and cultures of Brucella harboring cloned bepDE were destroyed immediately after these observations were made.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| B. suis | ||
| 1330 | Biotype 1; ATCC 23444T; wild-type strain | ATCC |
| ΔbepE | Deletion of bepE gene in 1330 | This study |
| ΔbepG | Deletion of bepG gene in 1330 | This study |
| ΔbepE ΔbepG | Deletion of both bepE and bepG in 1330 | This study |
| ΔbepE ΔbepC | Deletion of both bepE and bepC in 1330 | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector; Ampr. | Promega Inc. |
| pBBAD22k | Broad-host-range vector | 61 |
| pSDM3005 | pBGS19 derivative containing sacB gene from B. subtilis | 66 |
| pKGFP | Broad-host-range vector containing a promoterless gfpmut3 | 28 |
| pK18mobsacB | Allelic-exchange vector | 59 |
| pFC26 | pGEM-T Easy with a 2,553-bp fragment containing the bepC gene | This study |
| pFC70 | pGEM-T Easy with a 4.6-kb fragment containing the bepD and bepE genes | This study |
| pFC138 | pBBAD22k with a 4.6-kb fragment containing the bepD and bepE genes | This study |
| pSDΔbepE | pSDM3005 with a 488-bp fragment containing 5′ and 3′ regions of bepE gene | This study |
| pSDΔbepG | pSDM3005 with a 548-bp fragment containing 5′ and 3′ regions of bepG gene | This study |
| pFC165 | pGEM-T Easy containing a deletion of the bepC gene with 367 bp upstream and 778 bp downstream of the bepC gene | This study |
| pFC166 | pK18mobsacB containing a SphI-PstI fragment of pFC165 | This study |
| pAV45 | Broad-host-range vector containing gfpmut3 under the control of tacp; Cmr | A. Vergunst |
| pbepD-GFP | pKGFP with a 194-bp fragment containing the intergenic region between bepD and bepR genes (bepDE promoter activity) | This study |
| pbepR-GFP | pKGFP with a 194-bp fragment containing the intergenic region between bepD and bepR genes (bepR promoter activity) | This study |
| pbepRD-GFP | pKGFP with a 824-bp fragment containing the bepDE promoter and bepR gene | This study |
| pbepF-GFP | pKGFP with a 462-bp fragment containing the bepFG promoter | This study |
Gene cloning and generation of unmarked deletion mutants.
The bepD-bepE locus, corresponding to BR0291-BR0292 according to The Institute for Genomic Research annotation (http://www.tigr.org) (GenBank accession no. AAN29240.1 and AAN29241.1), was amplified from a B. suis 1330 (ATCC 23444T) genomic DNA sample using bepDfw (CGATTTCCGTCAGTGTGGA) and bepErv (AAATACCGCCGCCCGTA). MFP-RND loci were amplified using Pfx DNA Polymerase (Invitrogen Life Technologies) for accurate amplification and cloned into the pGEM-T Easy vector (Promega Inc.) according to the manufacturer's procedure to obtain pFC70 containing bepDE. The fidelity of the amplification reaction was confirmed by sequencing both inserts. Thereafter, a 4,667-bp fragment was excised from the pFC70 plasmid with SpeI and SphI and subcloned into the pBBAD22K plasmid (pFC138). Isogenic mutants in bepE or bepG were generated by unmarked deletions using a similar deletion strategy. A PCR fragment of approximately 250 bp was obtained in the 5′ region of bepE using the bepE5′fw (CGGGATCCGCCCAATATCCCGAGCTGAC) and bepE5′linkrv (TCCAAGACTGCTACGTATCGCAGTGCGAAGGTCACGGTCA) primers for the bepE deletion and bepG5′fw (CGGGATCCTGTCCGCCACGAGTTTCTGA) and bepG5′linkrv (TCCAAGACTGCTACGTATCGCCGATGCTCTACGTGGTGCTA) for the bepG deletion. A second amplicon of approximately 250 bp was generated in the 3′ regions of both bepE and bepG using bepE3′rv (GGACTAGTGCGCGATCAGGCTGTAGAA) and bepE3′linkfw (GCGATACGTAGCAGTCTGGACTGGTGACGCTCATCGGTCT) or bepG3′rv (GGACTAGTTACGTTCTGGCCGTCGGTGA) and bepG3′linkfw (GCGATACGTAGCAGTCTGGAGGATCAATTCCCCACCGACA) for each gene. Both fragments including the 5′ and the 3′ region of each gene contained complementary regions (underlined in the bepE5′linkrv, bepG5′linkrv, bepE3′linkfw, and bepG3′linkfw sequences) and were ligated by overlapping PCR and amplified using the external oligonucleotides for each sequence. The resulting fragment was cut with BamHI (sites underlined in bepE5′fw and bepG5′fw) and SpeI (bepE3′rv and bepG3′rv) and cloned into pSDM3005 (66), which carries the suicide sacB gene encoding the levansucrase from Bacillus subtilis. The plasmid was introduced into B. suis 1330 by electroporation. The integration of the suicide vector into the chromosome was selected for resistance to kanamycin (25 μg/ml) and sucrose sensitivity (10%) in tryptic soy agar (TSA) plates. A single colony was grown overnight in TSB without antibiotics and plated on TSA-sucrose. The sucrose-resistant and kanamycin-sensitive colonies were selected. The excision of the plasmid and the generation of the mutant strain by allelic exchange were confirmed by colony PCR and Southern blotting.
A bepC mutant was generated by allelic exchange of an unmarked deletion. The pFC26 plasmid harboring a 2,512-bp DNA region from B. suis 1330 containing bepC and surrounding regions was used as a template. Briefly, the oligonucleotides GCGGGATCCTGATCGGCTCTGCAAACAA and GCGGGATCCCATCTCAGAACAAACGAATCCA were used in an inverse PCR outward from the start and stop codons of the bepC gene (boldface in the oligonucleotide sequences). The amplicon was cut with BamHI (the sites are underlined in the oligonucleotide sequences) and religated to obtain the pFC165 plasmid. The fidelity of the reaction was confirmed by sequencing. A 1,215-bp SphI and PstI fragment was cut from pFC165 and ligated into the pK18mobsacB vector (59), which carries the sacB counterselecter marker, and introduced into B. suis 1330 by electroporation. The integration of the pFC166 plasmid into the chromosome was selected for resistance to kanamycin (25 μg/ml) and sucrose sensitivity in TSA plates. A single colony was grown overnight in TSB without antibiotics and plated on TSA-sucrose. Sucrose-resistant (and kanamycin-sensitive) colonies were selected. The bepC unmarked deletion mutant was confirmed by colony PCR. To generate a bepE bepC double mutant, the pSDΔbepE plasmid (Table 1) was introduced into the bepC unmarked deletion mutant, and the bepE deletion allelic exchange was generated as described above.
Construction of GFP reporter plasmids.
The promoter regions of the bepDE and bepFG genes were amplified using Pfu DNA polymerase (Promega Inc.). A region containing exactly the intergenic region between bepD and divergently transcribed bepR was amplified with the oligonucleotides pr_bepDf (CGGGATCCCGCATGGTGGGAATTCGC) and pr_bepDr (CGGGATCCCGGCTTTTGTTCTGCGCAT). The same region, but including the complete open reading frame coding for BepR, was amplified with the oligonucleotides pr_bepDf and pr_bepRDr (CGGGATCCGGTGGGAATTCGCCCTTACT). Both amplicons were digested with BamHI (the recognition sequence [underlined] was included in the oligonucleotides) and ligated into the pKGFP (Kanr) vector (28) upstream of the promoterless green fluorescent protein (GFP) gene to generate the pbepD-GFP, pbepR-GFP, and pbepRD-GFP plasmids. The 462 bp upstream of bepF was amplified using the oligonucleotides pr_bepFf (CGGGATCCCGCAACCAGCTTGTCAATTCGA) and pr_bepFr (CGGGATCCCGATGAAAATGCCGGAACCA). The amplicon was cloned into the BamHI site (the recognition site is underlined) of the pKGFP vector to generate pbepF-GFP. In all cases, the sequence and the correct orientation of each amplicon were confirmed by sequencing.
Antimicrobial sensitivity assays.
The MICs for B. suis 1330 and for the corresponding isogenic mutants were evaluated following the recommendations established by the CLSI (Clinical Laboratory Standards Institute) standard M7-A7. The MICs were determined by the agar dilution test method using TSA (Bacto) containing graded concentrations of the drug to be tested. An inoculum of 104 CFU per spot was applied in triplicate in each plate. All plates were incubated for 48 h at 37°C. The MIC was considered to be the concentration of the first plate in which no visible growth was observed.
Cell infection assay.
J774 or HeLa cells were seeded in 24-well plates and inoculated (at multiplicities of infection of 20:1 and 50:1, respectively) with the wild-type B. suis 1330 or the respective isogenic mutants. A standard gentamicin protection assay was performed as previously described (45).
Promoter activity assay.
In vitro assay of GFP reporter activity was performed as follows. B. suis harboring the indicated plasmid containing each GFP transcriptional fusion was grown for 16 h in TSB, and a 1:100 dilution of this culture was made in 2 ml of TSB. For MME, cells were grown for 48 h in TSB, and a 1:100 dilution was made in 2 ml fresh MME. In all cases, the cells were harvested at early exponential phase for fluorescence activity determination. The cultures were washed twice with phosphate-buffered saline and concentrated five times in phosphate-buffered saline. Fluorescence was determined using a Mithras LB940 multiplate reader (Berthold Technologies). The results are expressed as relative fluorescence units divided by the optical density at 600 nm (RFU/OD600). Analysis of expression in the presence of deoxycholate (DOC) was performed in MME. All determinations were made at least three times in triplicate. For in vivo observation, HeLa cells were infected with B. suis harboring the indicated construction as indicated above. After either 5 or 48 h, the cells were fixed with 3% paraformaldehyde for 30 min. The cells were observed with a fluorescence microscope (Leica DM IRB). Statistical significance was analyzed by one-way analysis of variance. P values of <0.05 were considered statistically significant.
Bioinformatics analysis.
Computer-assisted analysis of nucleotide sequences was performed with VectorNTI (Invitrogen Life Technologies), and the Basic Local Alignment Search Tool (BLAST) algorithm (3) was used to compare the sequences with the nucleotide and amino acid sequences currently deposited in GenBank. A domain search was performed using the KEGG database (http://www.kegg.com). Protein sequence alignment was performed using ClustalX (30) and T_coffee (44).
RESULTS
Sequence analysis.
The B. suis 1330 genome sequence annotation suggests the presence of six putative RND-MFP translocases. BLAST analysis (3) against the Swiss-Prot protein database (20) with the six putative proteins from the RND family showed that AAN29241.1 (BepE) and AAN33534.1 (BepG) had the strongest homology to AcrB (36) from E. coli (47 and 45% identity, respectively) and MexB (51) from Pseudomonas aeruginosa (45 and 43%, respectively). Both AcrB and MexB, together with their cognate MFP components, have been shown to confer a multidrug-resistant phenotype. BepE and BepG also showed considerable identity to the aromatic hydrocarbon TtgB transporter (56) from Pseudomonas putida (43 and 37%, respectively) and the broad-spectrum CmeB RND efflux pump (34) from Campylobacter jejuni (51 and 38%). Sequence analysis using the DAS server (14) revealed that BepE and BepG possess the classical pattern of an RND transporter (15) with 12 hydrophobic transmembrane segments (TMS) and two large loops (∼300 amino acids) between TMS 1 and 2 and TMS 7 and 8. Multiple alignments of B. suis BepE and BepG with the MexB and AcrB RND proteins also showed the characteristic pattern of the RND family described by Putman et al. (55), with high levels of amino acid conservation between the Bep proteins and AcrB and MexB (Fig. 1). An open reading frame encoding an MFP protein was identified upstream of and in the same orientation as both the bepE and bepG genes (Fig. 2). Accordingly, the products of these genes were named BepD and BepF. Both BepD and BepF contain most of the traits conserved among the MFP proteins (2, 6, 17): an amino-terminal region with a hydrophilic amino acid stretch, followed by a nonconserved hydrophobic region at the amino-terminal end. However, BepD, but not BepF, showed a predicted lipoprotein signal peptidase cleavage site motif in its amino-terminal region. Similar to what was observed for the RND partners, the first homologues of BepD and BepF were the MFP proteins AcrA (36) from E. coli (34 and 35%, respectively), AcrE from E. coli (34 and 32%), TtgA (56) from P. putida (36 and 32%), and CmeA (34) from C. jejuni (27 and 28%).
FIG. 1.
Multiple-sequence alignment of the BepE and BepG proteins with AcrB from E. coli and MexB from P. aeruginosa. The regions displayed were selected according to the motifs described by Putman et al. (55) for the RND superfamily. x indicates any amino acid; capital letters denote amino acids that occur in 70% of the examined sequences; lowercase letters represent amino acids that occur in 40% of the sequences used to generate the motifs. The sequences were aligned using ClustalX and T-coffee software and manually edited with GeneDoc (43).
FIG. 2.
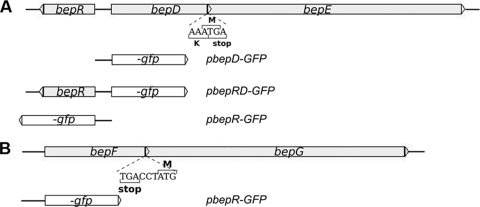
Schematic representation of the bepDE (A) and bepFG (B) genomic sequences from B. suis. The open arrowheads represent coding sequences and indicate the sense of transcription. The overlapping region between bepD and bepE and the proximity of bepF and bepG are shown. The bepF and bepG genes are separated by 2 bases. In both cases, the organization suggests the presence of an operon. Transcriptional fusions to the GFP gene used to analyze in vitro and in vivo bepDE and bepFG expression are shown.
A divergent open reading frame (named bepR) with similarity to those of many known bacterial transcriptional repressors from the TetR family was located upstream of the bepD gene (Fig. 2). BepR has the characteristic DNA binding helix-turn-helix motif (23) near its amino-terminal region and showed significant similarity to AcrR from E. coli (35) (33%), TtgR from P. putida (18) (33%), MtrR (46) from Neisseria gonorrhoeae (27%), and QacR (22) from Staphylococcus aureus (26%), which are involved in the regulation of inner membrane efflux translocases. Genome analysis of the adjacent regions of bepFG showed no evidence of a putative regulator from the TetR family or any other type.
The genes of RND-MFP translocases are mostly organized in operons. Based on the genetic arrangements of the bepDE and bepFG loci and sequence similarities with other efflux pumps of the RND family, it is very likely that both bepDE and bepFG are transcribed as operons (Fig. 2).
BepDE and BepFG are implicated in the resistance to structurally unrelated compounds.
Our sequence analysis suggests that both the putative translocases BepDE and BepFG of B. suis might be implicated in antimicrobial resistance. To gain insight into the possible functions of these translocases, deletion mutants in the RND genes (bepE and bepG) were generated as described in Materials and Methods. To investigate the effects of the bepE and bepG deletions on the sensitivity profile of B. suis to different dyes, detergents, and antimicrobials, the MICs were assayed by the agar dilution test. Within the antimicrobial group, we tested different classes of antibiotics, including those used to treat human brucellosis. The ΔbepE mutant showed increased sensitivity to DOC (MIC, 250 μg/ml), ethidium bromide (MIC, 3.125 μg/ml), and crystal violet (MIC, 3.125 μg/ml) in comparison with the wild-type strain of B. suis (MICs, 1,000, 25, and 6.25 μg/ml, respectively) (Table 2). We did not observe differences between the MICs of the rest of the evaluated drugs for the wild type and the ΔbepE mutant (Table 2). The ΔbepG mutant showed no differences in the resistance profile compared with that of the wild-type strain (data not shown). However, the mutant in both RND transporters (ΔbepE ΔbepG) showed a marked increase in sensitivity to nalidixic acid (MIC < 0.48 μg/ml) and sodium dodecyl sulfate (SDS) (MIC, 62.5 μg/ml) compared with B. suis 1330 (MICs, 15.625 and 1,000 μg/ml, respectively), two compounds for which no differences were observed between the single mutants and the wild type. Interestingly, the ΔbepE ΔbepG double mutant showed higher sensitivity to DOC (MIC, 62.5 μg/ml) than the single ΔbepE isogenic mutant (MIC, 250 μg/ml) (Table 2).
TABLE 2.
MIC determinations of antimicrobials, dyes, and detergents for B. suis 1330 and derivative strains
| Drug | MICa (μg/ml) for B. suis
|
|||
|---|---|---|---|---|
| 1330 | ΔbepE | ΔbepE (pFC138) | ΔbepE ΔbepG | |
| Antimicrobials | ||||
| Ampicillin | 0.78 | 0.78 | 6.25 | 0.78 |
| Chloramphenicol | >25 | >25 | >25 | >25 |
| Ciprofloxacin | 0.313 | 0.313 | 0.626 | ND |
| Doxycycline | 0.325 | 0.325 | 0.650 | ND |
| Erythromycin | 0.078 | 0.078 | 0.078 | ND |
| Nalidixic acid | 15.625 | 15.625 | 15.625 | 0.48 |
| Norfloxacin | 0,625 | 0,625 | 1.25 | ND |
| Novobiocin | 12.5 | 12.5 | 50 | 12.5 |
| Thiamphenicol | 3.125 | 3.125 | 50 | 3.125 |
| Tetracycline | <0.19 | <0.19 | 1.56 | <0.19 |
| Trimethoprim | 50 | 50 | 50 | 50 |
| Polymyxin B | 17.5 | 17.5 | 140 | 17.5 |
| Rifampin | 0.15 | 0.15 | 0.15 | ND |
| Streptomycin | 0.52 | 0.52 | 0.52 | ND |
| Dyes | ||||
| Acriflavine | 12.5 | 12.5 | 50 | 12.5 |
| Crystal violet | 6.25 | 3.125 | 100 | 3.125 |
| Ethidium bromide | 25 | 3.125 | 12.5 | 3.125 |
| Detergents | ||||
| DOC | 1,000 | 250 | 4,000 | 62.5 |
| SDS | 250 | 250 | 1,000 | 62.5 |
The MICs of the different antimicrobials were determined by the agar dilution test in triplicate in TSA following CLSI standards. Three different independent experiments gave similar results. The numbers in boldface indicate higher or lower MICs than those for the wild-type B. suis 1330. ND, not determined.
The resistance of the B. suis ΔbepE mutant to ethidium bromide was restored with pFC138 containing the bepDE locus. Moreover, resistance to DOC and crystal violet increased severalfold in the presence of multicopy bepDE compared with the wild-type strain. In addition, the pFC138 plasmid conferred increased resistance to other compounds to which the B. suis ΔbepE (and the ΔbepG) single mutant showed no phenotype. Among these compounds, we found an increase in the resistance of B. suis to ampicillin, norfloxacin, ciprofloxacin, novobiocin, polymyxin B, tetracycline, doxycycline, thiamphenicol, acriflavine, and SDS. Taken together, these observations indicate that the RND-MFP BepDE translocase is involved in resistance to several unrelated toxic compounds. In addition, the effect of the bepG deletion in the bepE mutant background on the sensitivity of B. suis to nalidixic acid, SDS, and DOC suggests that the BepFG translocase also contributes to resistance to some drugs.
Resistance conferred by BepDE is dependent on BepC.
RND transporters require an OMF to expel the substrate. As was noted previously, the downstream regions of bepDE and bepFG do not contain a gene that might encode an OMF. In fact, our previous report indicated that BepC, which is encoded by an open reading frame separated from those of inner membrane translocases, is the unique OMF present in B. suis (54). In order to investigate whether the resistance conferred by BepDE requires BepC to exert its function, we made use of the increased resistance to DOC conferred by bepDE cloned into pFC138 and tested whether inactivation of the bepC gene prevents this effect. We generated a ΔbepE ΔbepC double mutant, which was transformed with the pFC138 plasmid. As expected, the sensitivity to DOC of the B. suis ΔbepE ΔbepC double mutant (MIC, 31.25 μg/ml) was significantly higher than that of the B. suis ΔbepE single mutant (MIC, 250 μg/ml). The pFC138 plasmid did not increase the resistance to DOC of the B. suis ΔbepE ΔbepC double mutant (MIC, 31.25 μg/ml), strongly suggesting that BepC is necessary for BepDE to confer resistance. The increased sensitivity to DOC of the ΔbepE ΔbepC double mutant may account for the sensitivity phenotype of the mutant defective in both BepDE and BepFG RND translocases (ΔbepE ΔbepG) (Table 2), suggesting that both BepDE and BepFG interact with BepC to form tripartite pumps.
Survival of RND mutants in macrophages J774 and HeLa cells.
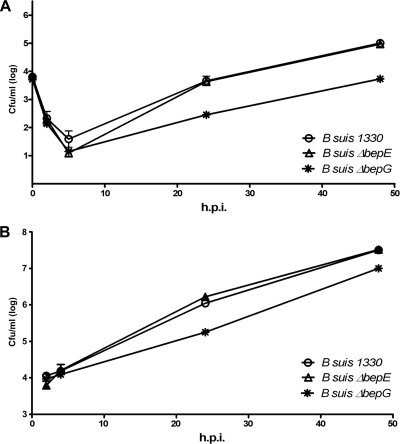
To determine the roles of BepDE and BepFG translocases in the survival of B. suis in model cells, the abilities of bepE and bepG isogenic mutants to enter and replicate in J774 macrophages and HeLa cells were analyzed. The wild-type strain showed the typical biphasic curve of viable brucellae recovered after infection of cultured cells. A slightly lower number of CFU of the BepE-deficient strain than of the wild type were recovered from HeLa cells after 5 h of infection, but the CFU level was able to recover after 24 h, displaying a level similar to that of the wild-type strain (Fig. 3A). Similarly, no significant differences in CFU numbers between the wild type and the ΔbepE mutant were observed in J774 macrophages (Fig. 3B). The B. suis ΔbepG mutant showed a lower number of recovered brucellae than the wild-type strain recovered from HeLa cells after 5, 24, and 48 h of infection (Fig. 3A). This suggests that the ΔbepG mutant is more sensitive to the initial intracellular killing than the wild type. The intracellular survival of the B. suis ΔbepE ΔbepG double mutant did not differ from that of the ΔbepG isogenic mutant (data not shown), indicating that in this case there was not the same additive effect that we found for the sensitivity phenotype to certain drugs (Table 2). As in HeLa cells, a lower number of CFU of the B. suis ΔbepG mutant than of the wild-type strain were recovered from J774 macrophages (Fig. 3B), which reinforces the idea that BepFG is required for optimal intracellular growth.
FIG. 3.
Intracellular replication of B. suis ΔbepE and ΔbepG mutants in HeLa cells (A) and murine J774 macrophages (B). The numbers of viable intracellular bacteria were determined 2, 5, 24, and 48 h postinfection (p.i.). The experiment was repeated three times, and the data presented are the results of a representative experiment done in triplicate. The results are expressed as geometric means and standard deviations (error bars) of plate counts.
bepDE and bepFG expression in vitro and in vivo.
To further assess the biological significance of BepDE and BepFG, in vitro and in vivo expression analyses were carried out. Sequence analysis of the bepDE upstream region showed the presence of a divergent gene named bepR, encoding a putative repressor from the TetR family that may be involved in the regulation of bepDE. Our analysis using the promoter prediction software BPROM (SoftBerry Inc.) indicated the presence of two overlapping and divergent putative promoters within the 172-bp intergenic region between bepR and bepD (Fig. 2). To investigate the expression (in vitro and in vivo) of both bepDE and bepR, we constructed three different transcriptional fusions to the GFP reporter gene in the pKGFP vector containing the promoterless gfpmut3 reporter gene (28). The GFP gene was placed under the control of the putative promoters within the intergenic region between the bepR and bepD genes in both 5′-3′ (pbepD-GFP) and 3′-5′ (pbepR-GFP) orientations (Fig. 2A) to analyze the expression of bepDE and bepR, respectively. In order to prevent a stoichiometric imbalance between the promoter activity of bepDE expressed in a multicopy plasmid and a single chromosomal copy of the putative repressor gene (bepR), we also generated a construction with the bepDE transcriptional fusion, together with a complete copy of the bepR gene (pbepRD-GFP) (Fig. 2A).
To validate the reporter expression analysis, quantification of the fluorescence levels of B. suis harboring the pKGFP promoterless vector (28) and the pAV45 plasmid containing the GFP gene under the control of the tac promoter (Annette Vergunst, personal communication) was performed over a time course experiment. The normalized RFU count of B. suis containing pAV45 was fivefold higher (5 × 104 RFU/OD600) than that of bacteria containing pKGFP (1 × 104 RFU/OD600) after 10 h of incubation, indicating that pKGFP is suitable to analyze promoter activities in B. suis.
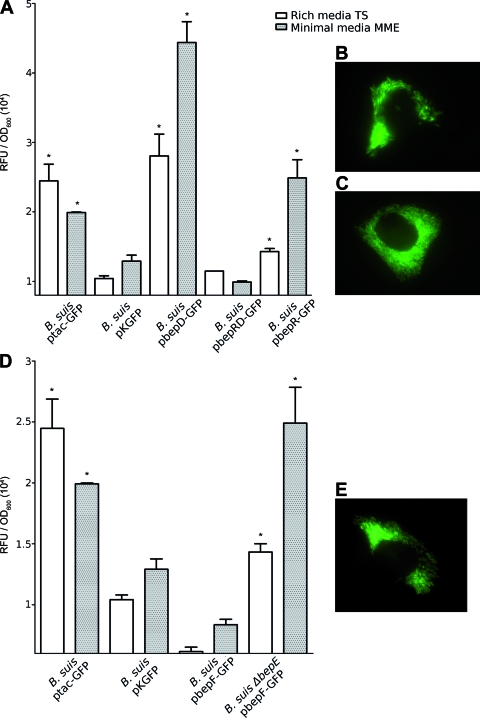
The expression of the bepDE promoter (in pbepD-GFP) was sixfold higher than that of the negative control in both rich and minimal media (Fig. 4A). However, with the pbepRD-GFP plasmid, which also carries the bepR gene, we saw only fluorescence levels similar to those of the negative control in both media tested (Fig. 4A). These results strongly suggest that BepR acts as a repressor, preventing bepDE expression. As judged by the effect of cloned bepR on bepDE expression, a considerable level of bepR expression was predicted. In fact, significant expression of the bepR promoter was observed in both media (Fig. 4A).
FIG. 4.
In vitro and in vivo expression of bepDE and bepR (A, B, and C) and bepFG (D and E). Bacteria containing the GFP promoter fusion were incubated in rich (TS) or minimal (MME) medium for 12 h, and the fluorescence was determined in a Mithras LB 940 (Berthold Technologies). The results are expressed as RFU relative to the OD600 plus the standard deviation of each bacterial culture. All samples were compared with the negative control, pKGFP (*, P < 0.05). (B, C, and E) Active expression of intracellular bacteria after 48 h of infection with B. suis pbepRD-GFP (B), B. suis pbepR-GFP (C), and B. suis pbepF-GFP (E).
To analyze the relevance of BepDE in vivo, we sought to determine whether bepDE is expressed in HeLa cells by using the pbepRD-GFP reporter plasmid. HeLa cells infected with B. suis harboring pbepRD-GFP were fixed after 5 and 48 h postinfection and examined by fluorescence microscopy. HeLa cells were also infected with brucellae containing the empty pKGFP vector as a negative control. B. suis harboring pbepRD-GFP showed consistent reporter expression, as judged by the fluorescent bacteria observed after 5 h (data not shown) and 48 h (Fig. 4B) of infection. Reporter expression was not observed in HeLa cells infected with B. suis harboring pKGFP (data not shown). This leads to the conclusion that the intracellular environment induces bepDE. The same strategy was used to analyze the expression of bepR in HeLa cells. B. suis harboring pbepR-GFP also showed a clear level of fluorescent bacteria (Fig. 4C), indicating that the repressor gene is also expressed in vivo within HeLa cells.
To evaluate the in vitro expression of bepFG, the GFP reporter gene was placed under the control of the 462-bp upstream region of bepF to generate the pbepF-GFP plasmid. Expression from the bepFG putative promoter over the level of the pKGFP-containing bacteria was not detected in rich or minimal medium, indicating that bepFG is not expressed in vitro under these conditions (Fig. 4D). However, bepFG promoter expression was clearly induced intracellularly in HeLa cells after 5 h (data not shown) or 48 h (Fig. 4E) of infection.
We were unable to identify any putative regulator in the genome sequences upstream or downstream of bepFG. We showed that a single deletion of the bepG gene had no effect on the sensitivity profile of B. suis. Nevertheless, the ΔbepE ΔbepG double mutant showed a further reduction in resistance to some drugs (Table 2). One possibility is that disruption of bepE induces bepFG expression, contributing to resistance to some drugs in the absence of BepDE. In this scenario, an additional mutation in bepG might result in a more severe phenotype. To test this hypothesis, we transformed the B. suis ΔbepE isogenic mutant with the reporter fusion cloned into pbepF-GFP to evaluate the expression of bepFG in a bepDE-defective background. Interestingly, the promoter activity of bepFG significantly increased in the ΔbepE mutant background (Fig. 4D). This observation indicates that the absence of the BepDE translocase induces bepFG expression.
Expression of bepDE is induced in the presence of the DOC bile salt component.
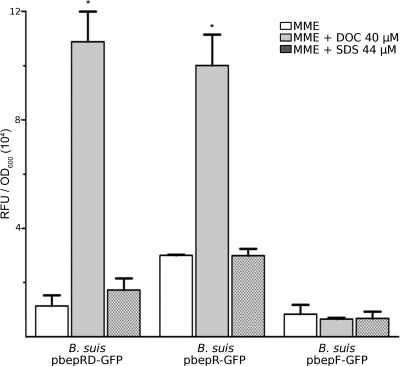
We showed that the activity of the bepDE promoter in the presence of stoichiometric amounts of bepR is not significant in either rich or minimal medium. Likewise, expression of the bepFG promoter in these media was also below the level of the negative control. Detergent activities of conjugated and unconjugated bile salts encountered in the intestine are a primary defense mechanism against pathogenic bacteria, and DOC (an unconjugated bile salt) is one of the major components of bile (8). Since DOC seems to be a substrate of both BepDE and BepFG and might be a physiological substrate of these pumps encountered by Brucella in the intestine, we aimed to evaluate whether this compound could induce the expression of the RND efflux pumps. Surprisingly, after 12 h of incubation in the presence of 40 μM DOC, reporter expression from the bepDE promoter cloned into pbepRD-GFP showed a ninefold increase compared with the expression in the absence of DOC (Fig. 5). In addition, a significant increase in the activity of the bepR promoter was observed (Fig. 5). Conversely, the bepFG promoter was not induced by DOC (Fig. 5). As expected, expression of bepFG was also induced by the bepE deletion in the presence of DOC (not shown), confirming that BepFG may compensate for the absence of BepDE (see above). The presence of 44 μM SDS did not activate the bepDE promoter (Fig. 5), implying that the activation mediated by DOC is substrate specific and is not due merely to the detergent activity of DOC.
FIG. 5.
Induction of bepDE expression by DOC. Samples were incubated for 12 h in the presence or absence of 40 μM DOC or 44 μM SDS. The results are expressed as RFU relative to the OD600 plus the standard deviation of each bacterial culture. *, P < 0.05 compared with the same samples incubated in the absence of substrate.
DISCUSSION
BepDE and BepFG: two RND-MFP pumps.
Detailed analysis of the BepE and BepG peptide sequences suggests that both proteins are complete and functional RND transporters. Comparison of these proteins with the AcrB (36) and MexB (31) proteins showed high conservation of key residues involved in protein structure (19, 39, 70) and proton relay transfer (24) that sustain a possible role of BepE and BepG in multidrug efflux. A similar conclusion was drawn for the BepD and BepF MFP components. The deletion approach confirmed that the BepDE and probably the BepFG translocases are involved in resistance to several compounds. Deletion of bepE makes B. suis more sensitive to DOC, crystal violet, and ethidium bromide. In contrast, deletion of bepG did not confer any particular sensitivity phenotype. However, the ΔbepE ΔbepG double mutant showed a more severe sensitivity phenotype to DOC than the single ΔbepE mutant. Furthermore, the ΔbepE ΔbepG double mutant was clearly more sensitive to SDS and nalidixic acid than the wild-type B. suis, two drugs for which no differences were observed between the single mutants and the wild-type strain. Additional evidence of multidrug efflux mediated by BepDE was the effect of multicopy bepDE in the ΔbepE mutant, conferring an increase in the resistance of B. suis to ampicillin, norfloxacin, ciprofloxacin, novobiocin, polymyxin B, tetracycline, doxycycline, thiamphenicol, acriflavine, and SDS compared with the B. suis wild-type strain. Taken together, these results show that BepDE is able to confer resistance to several toxic compounds, such as dyes, detergents, and antibiotics. Moreover, the sensitivity phenotypes of the double mutant are consistent with the idea that efflux of some drugs in the bepE single mutant of B. suis might be partially or totally compensated for by BepFG. Therefore, BepFG may contribute to the resistance to some drugs, like DOC, SDS, and nalidixic acid, in the absence of BepDE. The interplay between the two translocases was further supported by the in vitro expression analysis (see below).
BepC is the unique OMF harbored in the B. suis genome. This is a situation similar to that seen in E. coli, where RND-MFP translocases rely on the presence of TolC to exert their functions (33, 67). On the other hand, four out of the six RND-MFP translocases described in P. aeruginosa are encoded by an MFP-RND-OMF operon (52). We have previously shown that BepC is responsible for resistance to several drugs, dyes, and detergents (54). Our genetic analysis showed that BepDE requires BepC to confer DOC resistance. Since the bepC mutant showed an augmented sensitivity to DOC compared with that of the single bepE mutant, another translocase may be a partner of BepC contributing to DOC efflux. The further sensitivity to DOC of the ΔbepE ΔbepG double mutant supports the idea of BepFG as a second partner of BepC to exclude DOC and probably other compounds.
Regulation: in vitro and in vivo expression of bepDE and bepFG.
To give more insight into the roles of the RND-MFP translocases, we investigated the regulation of these pumps by using transcriptional fusions of GFP to the bepDE and bepFG putative promoters. At first glance, the context of bepDE showed the presence of a gene encoding a putative repressor from the TetR family named BepR, while the regions around bepFG did not suggest the presence of any putative regulator. No bepFG expression was observed in vitro in either rich or minimal medium. We found that bepDE promoter activity was completely repressed by BepR in both culture media. However, the expression of both promoter fusions showed early induction in the intracellular environment of HeLa cells, suggesting that both the BepDE and BepFG pumps are functional in vivo. Surprisingly, only the ΔbepG mutant showed a moderate but reproducible attenuation in HeLa cells. The observed interplay between these systems would explain these data in part, i.e., that both BepDE and BepFG can transport (or efflux) a set of substrates but the transport of other, more specific physiological substrates inside the cell can be performed only by BepFG. Hence, although there are some interchangeable functions between BepDE and BepFG, the transport of a specific substrate by BepFG would explain the moderate attenuation of the ΔbepG mutant in model cells. Our data are consistent with the idea that both BepDE and BepFG might protect brucellae from some toxic intracellular compound. Alternatively, the possibility exists that these RND systems are involved in the transport of other metabolites within the host cell. It has been proposed that RND pumps participate in the transport of autoinducers, such as acyl-homoserine lactones (11, 47). In fact, Brucella produces a C12 acyl-homoserine lactone that may need a transport mechanism to exert its function as a signaling molecule (62).
The mechanism by which the expression of bepDE and bepFG is induced within the host cell is unknown. In some cases, the expression of RND efflux pumps has been shown to be responsive to general stress conditions or induced by a specific substrate recognized and transported by the pump (23). During cell infection, early phagosome acidification (9, 27, 53) and the oxidative burst (21) generated by the macrophage are necessary for virulence gene activation in Brucella spp. Our preliminary data indicate that the expression from the bepDE or bepFG promoter is not induced at low pH, and only a 10% increase in the expression of bepDE was observed in the presence of hydrogen peroxide. Further studies are required to better define the effects of these or other stress conditions on bepDE and bepFG expression.
bepDE induction by DOC.
Although a considerable level of bepDE promoter activity was observed with the reporter plasmid that lacks a copy of bepR, expression of bepDE was strongly repressed in the presence of equivalent amounts of bepR, indicating that BepR is a local repressor of bepDE expression. Interestingly, 40 μM DOC released the repression mediated by BepR. The simplest interpretation, based on evidence accumulated for other TetR regulators (23), is that DOC binds directly to BepR, promoting BepR release from the bepDE promoter. In other species, DOC and other components of bile salts were shown to be efficient inductors of RND systems (32, 57). SDS, which is also a detergent and a substrate of BepDE, was not able to activate bepDE expression, indicating that induction by DOC is substrate specific.
One puzzling observation is the increased expression of bepR in the presence of DOC. The concomitant increase of bepR expression, along with bepDE, was somehow unexpected. However, a similar observation was described in the regulatory circuit of other RND systems. Induction conditions increased the transcription of the acrR repressor gene, along with the acrAB efflux genes (35). It was proposed that AcrR functions as a secondary modulator to fine-tune the level of acrAB transcription to prevent an excess of acrAB expression and, as a consequence, deleterious effects on cell viability. In the TetR/TetA system, the increased expression of the tetR repressor gene in the presence of the inductor ensures strict control of tetA transcription at all times (25). It has also been reported that the expression of bpeR of Burkholderia pseudomallei was induced in late exponential phase, probably due to the accumulation of a metabolite that is a substrate of the BpeAB-OprB efflux system (12).
It is becoming clear that DOC and other bile salts may represent physiological substrates of BepDE and BepFG. Brucellosis can be acquired by ingestion of contaminated products. Therefore, during the initial stages of infection, Brucella is exposed to antimicrobials, including bile salts and fatty acids. The RND pumps may contribute to resistance to DOC and other bile components, along with the bile salt hydrolase that has recently been described in Brucella abortus (16). Although our results strongly suggest a role of BepDE and BepFG in protecting bacteria from some intracellular antimicrobial compound, a function of the RND pumps in survival during the extracellular stage of Brucella infection cannot be ruled out.
bepFG expression is induced in the absence of BepDE.
It was interesting to find that disruption of bepE induces bepFG expression. A possible explanation for this activation is that bepDE and bepFG are under the control of a common DOC-independent level of regulation, resulting in a compensatory mechanism in the absence of BepDE. We hypothesize that an unknown global regulator may mediate this compensatory mechanism. The affinity of this hypothetical regulator for the bepDE promoter would be significantly higher than for the bepFG promoter. Thus, upon activation of the global regulator, an initial increase of bepDE expression is induced. In the absence of BepDE, the inductor is accumulated in the cytoplasm, favoring the ability of the regulator to bind and activate the bepFG promoter.
Resistance to antibiotics mediated by RND systems in B. suis.
The resistance profile of B. suis in the presence of multicopy bepDE indicates that BepDE is able to exclude antibiotics, including tetracycline, doxycycline, and fluoroquinolones.
Antibiotic resistance associated with mutations in repressors from the TetR family has been previously described in E. coli (68) and Klebsiella pneumoniae (60). This implies that a “resistance” phenotype to tetracyclines, fluoroquinolones, or other antibiotics due to the selection of a strain with bepR mutations can be predicted. Therefore, special attention should be paid to a possible effect of bepDE overexpression on resistance to antibiotics, such as tetracyclines and fluoroquinolones, two broad types of antibiotics used in clinical treatment for brucellosis. In fact, resistance to fluoroquinolone in two Brucella strains has been associated with an energy-dependent efflux mechanism (64). The agents recommended by the World Health Organization guidelines for more than a decade have included rifampin (rifampicin) or streptomycin plus doxycycline for the management of human brucellosis (5). Although tetracyclines have been proven to be efficient in Brucella treatment during randomized trials, our analyses suggest that a tetracycline resistance mechanism in Brucella is possible.
Acknowledgments
We thank Adrian A. Vojnov, Alfonso Soler Bistue, and Marcelo E. Tolmasky for critical reading of the manuscript. We also thank Annete Vergunst for kindly providing the pAV45 and pSDM3005 plasmids. We thank Daniela Marta Russo, Patricia Abdián, Nicolas Vozza, Veronica Ruiz, Lorena Haurigot, and Julia Sabio y Garcia for helpful discussions and support. We especially thank Eric Botella, Gilles Patey, and Gisèle Bourg for support and help in the cell infection experiments. We thank Maria Soledad Ramirez and Alejandro Petroni for providing ciprofloxacin and doxycycline. We thank Marta Bravo and Jimena Ortega for DNA sequencing.
This work was supported by the University of Buenos Aires (UBACyT X-245) and Agencia de Promoción Científica y Tecnológica (PICT8266 and PICT20334). INSERM Espri26 is supported by INSERM, La Region Languedoc Roussillon, La Ville de Nimes, and the University of Montpellier 1. A.Z. is a member of CONICET and a professor of the University of Buenos Aires; F.A.M. was supported by a University of Buenos Aires fellowship and the Programme Alβan, the European Union Programme of High Level Scholarships for Latin America, scholarship no. E05D053527AR. D.M.P. was supported by a CONICET fellowship.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Aires, J. R., and H. Nikaido. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 1871923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S.-I. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 27925939-25942. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, C. 2003. Channel-tunnels: outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria. Rev. Physiol. Biochem. Pharmacol. 147122-165. [DOI] [PubMed] [Google Scholar]
- 5.Ariza, J., M. Bosilkovski, A. Cascio, J. D. Colmenero, M. J. Corbel, M. E. Falagas, Z. A. Memish, M. R. H. Roushan, E. Rubinstein, N. V. Sipsas, J. Solera, E. J. Young, and G. Pappas. 2007. Perspectives for the treatment of brucellosis in the 21st century: the Ioannina recommendations. PLoS Med. 4e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avila-Sakar, A. J., S. Misaghi, E. M. Wilson-Kubalek, K. H. Downing, H. Zgurskaya, H. Nikaido, and E. Nogales. 2001. Lipid-layer crystallization and preliminary three-dimensional structural analysis of AcrA, the periplasmic component of a bacterial multidrug efflux pump. J. Struct. Biol. 13681-88. [DOI] [PubMed] [Google Scholar]
- 7.Batut, J., S. G. E. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2933-945. [DOI] [PubMed] [Google Scholar]
- 8.Begley, M., C. G. M. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29625-651. [DOI] [PubMed] [Google Scholar]
- 9.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 991544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celli, J., and J.-P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 793-97. [DOI] [PubMed] [Google Scholar]
- 11.Chan, Y. Y., and K. L. Chua. 2005. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J. Bacteriol. 1874707-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, Y. Y., T. M. C. Tan, Y. M. Ong, and K. L. Chua. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 481128-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comerci, D. J., M. J. Martínez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 3159-168. [DOI] [PubMed] [Google Scholar]
- 14.Cserzo, M., F. Eisenhaber, B. Eisenhaber, and I. Simon. 2004. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics 20136-137. [DOI] [PubMed] [Google Scholar]
- 15.Das, D., Q. S. Xu, J. Y. Lee, I. Ankoudinova, C. Huang, Y. Lou, A. DeGiovanni, R. Kim, and S.-H. Kim. 2007. Crystal structure of the multidrug efflux transporter AcrB at 3.1A resolution reveals the N-terminal region with conserved amino acids. J. Struct. Biol. 158494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delpino, M. V., M. I. Marchesini, S. M. Estein, D. J. Comerci, J. Cassataro, C. A. Fossati, and P. C. Baldi. 2007. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 75299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1763825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 391100-1106. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, C. A., and H. Nikaido. 2003. 3D structure of AcrB: the archetypal multidrug efflux transporter of Escherichia coli likely captures substrates from periplasm. Drug Resist. Updat. 69-13. [DOI] [PubMed] [Google Scholar]
- 20.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 313784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gee, J. M., M. W. Valderas, M. E. Kovach, V. K. Grippe, G. T. Robertson, W.-L. Ng, J. M. Richardson, M. E. Winkler, and R. M. Roop II. 2005. The Brucella abortus Cu, Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect. Immun. 732873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grkovic, S., M. H. Brown, K. M. Hardie, N. Firth, and R. A. Skurray. 2003. Stable low-copy-number Staphylococcus aureus shuttle vectors. Microbiology 149785-794. [DOI] [PubMed] [Google Scholar]
- 23.Grkovic, S., M. H. Brown, N. J. Roberts, J. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 1218665-18673. [DOI] [PubMed] [Google Scholar]
- 24.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 1831734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48345-369. [DOI] [PubMed] [Google Scholar]
- 26.Ko, J., and G. A. Splitter. 2003. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin. Microbiol. Rev. 1665-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J.-P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 9915711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, S., S. Ouahrani-Bettache, M. Layssac, J. Teyssier, and J. P. Liautard. 1999. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 676695-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulakov, I. K., D. O'Callaghan, M. Ramuz, J. P. Liautard, and A. G. Skavronskaia. 1996. A genetic and molecular biological study of the pathogenicity factors in Brucella. Zh. Mikrobiol. Epidemiol. Immunobiol. 334-39. [PubMed] [Google Scholar]
- 30.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 31.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 391948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., M. Akiba, O. Sahin, and Q. Zhang. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 491067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, J., S. Huang, and Q. Zhang. 2002. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect. 4325-331. [DOI] [PubMed] [Google Scholar]
- 34.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 462124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19101-112. [DOI] [PubMed] [Google Scholar]
- 36.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1645-55. [DOI] [PubMed] [Google Scholar]
- 37.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2489-493. [DOI] [PubMed] [Google Scholar]
- 38.Martínez de Tejada, G., and I. Moriyón. 1993. The outer membranes of Brucella spp. are not barriers to hydrophobic permeants. J. Bacteriol. 1755273-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 1861258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriyon, I., and D. T. Berman. 1982. Effects of nonionic, ionic, and dipolar ionic detergents and EDTA on the Brucella cell envelope. J. Bacteriol. 152822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriyón, I., C. Gamazo, and R. Díaz. 1987. Properties of the outer membrane of Brucella. Ann. Inst. Pasteur Microbiol. 13889-91. [DOI] [PubMed] [Google Scholar]
- 42.Murakami, S., and A. Yamaguchi. 2003. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13443-452. [DOI] [PubMed] [Google Scholar]
- 43.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNET News 41-4. [Google Scholar]
- 44.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 45.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 331210-1220. [DOI] [PubMed] [Google Scholar]
- 46.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11769-775. [DOI] [PubMed] [Google Scholar]
- 47.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1811203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piddock, L. J. V. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piddock, L. J. V. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 50.Pizarro-Cerdá, J., E. Moreno, V. Sanguedolce, J. L. Mege, and J. P. Gorvel. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 662387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 1757363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 159-71. [DOI] [PubMed] [Google Scholar]
- 53.Porte, F., J. P. Liautard, and S. Köhler. 1999. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun. 674041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posadas, D. M., F. A. Martín, J. V. Sabio y García, J. M. Spera, M. V. Delpino, P. Baldi, E. Campos, S. L. Cravero, and A. Zorreguieta. 2007. The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect. Immun. 75379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 1803323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 481609-1619. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 60.Schneiders, T., S. G. Amyes, and S. B. Levy. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 472831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sukchawalit, R., P. Vattanaviboon, R. Sallabhan, and S. Mongkolsuk. 1999. Construction and characterization of regulated l-arabinose-inducible broad host range expression vectors in Xanthomonas. FEMS Microbiol. Lett. 181217-223. [DOI] [PubMed] [Google Scholar]
- 62.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 703004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Touzé, T., J. Eswaran, E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53697-706. [DOI] [PubMed] [Google Scholar]
- 64.Turkmani, A., A. Psaroulaki, A. Christidou, G. Samoilis, T. A. Mourad, D. Tabaa, and Y. Tselentis. 2007. Uptake of ciprofloxacin and ofloxacin by 2 Brucella strains and their fluoroquinolone-resistant variants under different conditions. An in vitro study. Diagn. Microbiol. Infect. Dis. 59447-451. [DOI] [PubMed] [Google Scholar]
- 65.Velasco, J., J. A. Bengoechea, K. Brandenburg, B. Lindner, U. Seydel, D. González, U. Zähringer, E. Moreno, and I. Moriyón. 2000. Brucella abortus and its closest phylogenetic relative, Ochrobactrum spp., differ in outer membrane permeability and cationic peptide resistance. Infect. Immun. 683210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290979-982. [DOI] [PubMed] [Google Scholar]
- 67.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 874776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 451515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Webber, M. A., and L. J. Piddock. 2003. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 519-11. [DOI] [PubMed] [Google Scholar]
- 70.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 1876804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]