Abstract
Junonia coenia densovirus (JcDNV) is an ambisense insect parvovirus highly pathogenic for lepidopteran pests at larval stages. The potential use of DNVs as biological control agents prompted us to reinvestigate the host range and cellular mechanisms of infection. In order to understand the early events of infection, we set up a functional infection assay in a cell line of the pest Lymantria dispar to determine the intracellular pathway undertaken by JcDNV to infect a permissive lepidopteran cell line. Our results show that JcDNV particles are rapidly internalized into clathrin-coated vesicles and slowly traffic within early and late endocytic compartments. Blocking late-endocytic trafficking or neutralizing the pH with drugs inhibited infection. During internalization, disruption of the cytoskeleton, and inhibition of phosphatidylinositol 3-kinase blocked the movement of vesicles containing the virus to the nucleus and impaired infection. In summary, our results define for the first time the early endocytic steps required for a productive DNV infection.
Densoviruses (DNVs) are parvoviruses highly pathogenic for arthropods, mostly insects, at larval stages. Their pathology, first described in the greater wax moth, has been associated with hypertrophied Feulgen-positive nuclei and “dense nucleosis,” which gives the virus its name (2, 49). Because DNVs are highly virulent for their insect hosts and do not replicate in vertebrates and mammals, they have been considered as potential biological tools against insect pests and vectors (7, 14, 15, 20, 23). Recently, Ren et al. (39) suggested another use for a new DNV as a tool for viral paratransgenesis in Anopheles gambiae. Discovered in an A. gambiae cell line, this virus has lost its pathogenicity for mosquito larvae but still retains dissemination and transmission capacities that have been investigated for paratransgenic malaria control.
To date, at least 30 DNVs have been discovered; they display a high degree of biochemical and structural diversity discriminating four genera in the Densovirinae subfamily (4). The DNVs have in common that they are autonomously replicating viruses with small (20 to 25 nm) icosahedral, nonenveloped particles that must deliver their enclosed single-stranded DNA genome into the nucleus of the target cells where these viruses replicate. Palindromic sequences end the 4- to 6-kb linear genomes that display a monosense or ambisense organization (for a review, see reference 4).
First described by Rivers and Longworth (40), the prototype of the genus Densovirus is the Junonia coenia DNV (JcDNV);the ambisense genome of this virus carries on one strand two open reading frames encoding three early nonstructural proteins (NS1 to NS3), primarily implicated in viral replication (11, 22, 1), and on the complementary strand, one single gene encoding the four late structural proteins (VP1 to VP4) implicated in capsid assembly. Each VP protein is able to auto-assemble and form pseudo-capsids although the role of these proteins in the infection process is largely unknown (6, 10). The sequences of insect and vertebrate parvoviruses show little overall sequence identity; however, the following domains display strong sequence homologies: the NS1 NTP-binding, nickase, and helicase domains and the N terminus of VP1 that encodes a phospholipase A2 (PLA2) motif and a basic nuclear localization signal (NLS), both of which are required for infection (27, 51, 53).
Recent studies have depicted the main events of parvovirus entry into host cells, all mediated by a wide diversity of cell surface receptors, including glycoproteins, glycans, and glycolipids, that trigger rapid clathrin-mediated endocytosis (reviewed in references 9, 18, and 37). The rapid endocytic uptake is followed by slower traffic along the endocytic compartments toward the nucleus. The endosomal pathway undertaken by parvoviruses is complex and depends on the virus, its concentration, and probably the cell type (12, 34, 46, 48). Conformational changes of the capsid probably occur in acidic vesicles, but the place where viruses escape from the endosomes is still unclear. The traffic may also require a cytoskeletal transport via interactions with microtubules, actin, or dynein motor (42, 48, 52) even though recombinant adeno-associated virus 2 (AAV2) traffic has been shown to be independent of the microtubule network (19). As the virus finally reaches the nucleus, some evidence indicates that parvovirus nuclear entry might occur independently of the nuclear pore complex (8). Once inside the nucleus and uncoated, the virus hijacks the host S-phase replicative machinery to initiate viral replication (9).
The early steps in the virus cycle are crucial for replication and therefore for the success of infection. Although the presence of the receptor is a prerequisite for the entry, several intracellular factors during particle routing also determine cell specificity and host range. Insect parvovirus entry into a host cell is poorly documented in part because very few and only nonconventional insect cell lines are permissive to JcDNV (26), and, consequently, molecular markers are missing. Curiously, cell culture infections do not form plaque lysis, which contrasts with the high pathogenicity for their larval host. In order to understand the early steps of JcDNV infection, we have characterized the crucial endocytic events leading to the productive infection of a permissive lepidopteran cell line, Lymantria dispar 652. Here, we show that the JcDNV infection pathway mainly involves rapid clathrin-mediated uptake followed by slower traffic through low-pH and late-endosomal compartments. Virus traffic along this pathway requires an intact cytoskeleton network, probably facilitating the movement of organelles containing viruses to the nucleus, where viral replication occurs.
MATERIALS AND METHODS
Cells.
L. dispar ovarian cells, IPLB-Ld 652 (17), and Spodoptera frugiperda Sf9 cells (ATCC CRL 1711) were used in this study. Cells were grown at 28°C in TC100 medium (Gibco Invitrogen), pH 6.2, supplemented with 10% heat-inactivated fetal calf serum (HyClone, Perbio).
JcDNV and VP4 virus-like particle (VLP) production and purification.
JcDNV was prepared from 20 Spodoptera littoralis injected larvae as previously described (10). Briefly, at 6 days p.i., larvae were collected and disrupted in phosphate-buffered saline (PBS) containing 2% ascorbic acid buffer. The extract was clarified at 8,000 × g for 30 min (min), and viral particles were next concentrated at 100,000 × g for 2 h. Viruses were then loaded onto a 20 to 76% Radioselectan (Sherring Laboratories) density gradient and centrifuged overnight at 100,000 × g. A band containing the concentrated viral fraction was recovered and dialysed against 1×TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA) for 48 h. Viral particles were layered on carbon-coated grids, negatively stained with 2% phosphotungstic acid, pH 7.0 (21), and examined with a Zeiss EM 10C/R transmission electron microscope (TEM) at 80 kV.
For production of VLPs containing VP4 (VP4-VLPs), Sf9 cells were infected by recombinant Autographa californica multiple nucleopolyhedrovirus virus, and VLPs were purified as described previously (10).
Virus titers were determined on L. dispar 652 cells by 50% tissue culture infective dose (TCID50) assay in 96-well plates as previously described (26). VP4-VLP concentration was assessed spectrophotometrically using a bicinchoninic protein assay kit (Pierce), and particle morphology was controlled by TEM observation.
Infection and inhibitor assays.
Infections with JcDNV (0.1 TCID50 unit/cell) were performed by first incubating cells for 1 h at 4°C to allow virus binding. Then, infected cells were switched or not to 28°C for 1 h. For the inhibitor assays, L. dispar cells were pulse treated at 28°C with the drugs (all purchased from Sigma) dynasore (DYN; 20, 40, and 80 μM), chlorpromazine (CPZ; 20 μM), methyl-β-cyclodextrin (MβCD; 5 mM), cytochalasin D (CD; 10 μM), nocodazole (ND; 30 μM), wortmannin (WTN, 1 μM), brefeldin A (BFA; 5 μg/ml), and nystatin (Nys; 50 μM) 1 h before virus adsorption (30 min for DYN), at infection (following virus adsorption), and postinfection (4 h after virus adsorption). For the infection assays, at the end of drug treatments, cells were washed and further incubated with fresh medium for 48 h postinfection to allow VP protein production; the medium contained ammonium chloride (50 μM NH4Cl), chloroquine (CQ; 10 μM) and 40 μM DYN. For all the inhibitors, a dose-response assay was done to determine the working dose, and cell viability was checked by trypan blue exclusion assay. Control cells were similarly treated with a drug solvent (dimethyl sulfoxide [DMSO] or ethanol) and infected with JcDNV.
Immunocytochemistry.
Cells were fixed with 4% paraformaldehyde for 20 min. After washes in PBS, cells were permeabilized with PBS containing 0.1% Triton (PBT). Blocking was performed with PBT containing 1% bovine serum albumin (BSA) for 30 min, and cells were then incubated for 1 h with PBT containing 0.1% BSA and a mouse polyclonal anti-JcDNV antibody (1:1,000 dilution) to reveal JcDNV or with a rabbit antitubulin antibody (Sigma) to label the microtubules. For fluorescent visualization, the secondary antibodies anti-mouse or anti-rabbit Alexa Fluor 594 and Alexa Fluor 488 (Molecular Probes-Invitrogen) were used at 1:500 and were next incubated for 1 h. Cellular cortical actin and nuclei were labeled for 30 min with fluorescein isothiocyanate (FITC)-phalloidin (Sigma) and 4′,6′-diamidino-2-phenylindole (DAPI; Sigma), respectively. Coverslips were then mounted in Prolong Gold antifade reagent (Invitrogen).
For colocalization assays, L. dispar cells were infected and concomitantly incubated with tetramethyl rhodamine isothiocyanate-conjugated transferrin (TRITC-Tfn) for 30 min, albumin conjugated with FITC (Sigma), or LysoTracker Red DND-99 (Invitrogen) for 4 h in serum-free medium to discriminate the subcellular compartments.
Images were obtained by confocal laser scanning microscopy with a Zeiss Axioplan2 microscope equipped with an LSM510 META confocal module, using a 63× (numerical aperture, 1.4) objective and filter settings optimized for the dyes. The pinhole was adjusted to 1 Airy unit. For each sample, a series of 0.3-μm-thick horizontal sections was performed. Colocalization analysis was performed from 10 cells and three z-section images per cell. Images were acquired with the same parameters. For each stack and each cell, the fluorescence areas were calculated for each fluorochrome using Metamorph software to define the percentage of colocalized JcDNV area relative to marker. We estimated the intensity correlation (Pearson's coefficient) using ImageJ software. This coefficient reflects the degree of linear relationship between the two sets of fluorochrome images (5). For the DYN assay, the integrated intensity per cell was measured for JcDNV and TRITC-Tfn on 10 cells and three z-sections per cell by using the Metamorph software. Three-dimensional (3D) reconstructions were obtained with Imaris, version 5.7.0 (Bitplane), software.
Viral replication assay.
L. dispar cells were infected as described above. To avoid any loss of cells after infection due to their modified attachment to the flasks, cells were pelleted at different times postinfection (p.i.) for 5 min at 800 × g and fixed with 4% paraformaldehyde and 2.5% glutaraldehyde for 1 h on ice. After several washes with PBS, cells were dehydrated with 30 to 100% ethanol and then embedded in 100% Unicryl resin (BB International) after progressive 30-min incubations in 1:2, 1:1, and 2:1 (vol/vol) resin-ethanol. Resin polymerization was performed for 48 h under UV light at 4°C. Freshly cut 1-μm semithin sections were first incubated for 10 min with 50 mM NH4Cl and then blocked for 10 min in freshly prepared 1% BSA before immunolabeling.
To quantify the percentage of JcDNV-positive fluorescent cells, 150 cells were viewed with a Zeiss Axiovert 200 M/Apotome instrument equipped with Zeiss-specific filters with a 40× objective (numerical aperture, 1.3). Images were obtained with an Axiocam MRm charge-coupled-device camera and analyzed with Axiovision software. Images were acquired with the same time of exposure, and images were processed with ImageJ software.
Electron microscopy.
Infected cells were fixed in 2% glutaraldehyde-0.1 M cacodylate (pH 7.4) for 1 h at 4°C and then postfixed for 1 h in 2% osmium tetroxide at room temperature. After dehydration in a graded series of ethanol solutions, cells were embedded in Epon resin. Ultrathin sections (0.1 μm) were mounted on copper grids and stained with uranyl acetate and lead citrate. Observations were performed with a Zeiss EM10CR electron microscope at 80 kV. The size of particles associated with coated pits or vesicles was measured using a Lheritier camera and Digiphot software.
Real-time qPCR.
Total DNA was extracted from 1 × 106 infected cells at different times p.i. Viral forward (5′GGAGGAGGCAACACTTCAGG 3′) and reverse (5′ TCTGCCATGGAATTTCAGCC 3′) primers were designed using Primer Express software (Applied Biosystem) optimized for the ABI prism 7000 Sequence Detection System. A 200-bp fragment was amplified from position 1710 to 1911 of the viral genome located within open reading frame 1, the major capsid protein. The β-actin gene was used as a control gene to normalize DNA quantity between samples and experiments. For β-actin, the forward (5′-TCGCGACTTGACCGACTA-3′) and reverse (5′-GAGTAACCCCTCTCGGTGAGG-3′) primers were designed from the L. dispar β-actin cDNA sequence (GI 166012628). Reactions were carried out using a Platinum SYBR Green qPCR Super Mix kit (Invitrogen). Fluorescent amplicons were detected using the ABI Prism 7000 apparatus (Applied Biosystems) using 96-well microtiter plates in a final volume of 25 μl under the following conditions: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 30 s. For each quantitative PCR (qPCR) analysis, 5 μl of diluted DNA was deposited in triplicates. A standard curve was generated after 40 cycles of PCR using seven serial 10-fold dilutions of a plasmid containing the viral genome. The linear range was obtained over 7 orders of magnitude and allowed to determine the viral DNA copy number. Data from two biological replicates, three technical replicates, and two plate permutations were analyzed by the ΔΔCT method (where CT is threshold cycle) (30).
The linear range was obtained from 1.25 × 103 to 1.25 × 109 copies of the viral genome.
RESULTS
JcDNV entry is mediated by a rapid clathrin-mediated endocytosis.
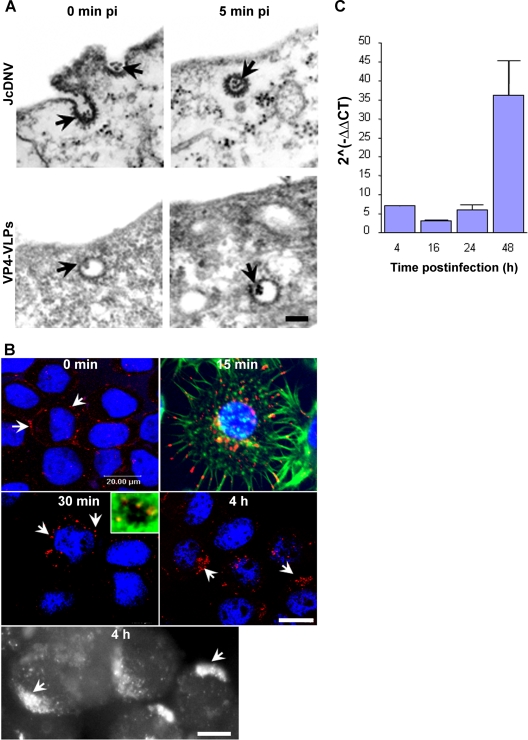
Many viruses infect their host through endocytosis, and several pathways may be exploited to gain access to the appropriate replication compartment. JcDNV uptake was first analyzed by electron microscopy. The incubation of the infected cells at 4°C (0 min p.i.) blocked virus internalization, and the viral particles were concentrated into phenotypically characteristic clathrin-coated pits at the plasma membrane (Fig. 1A). An average of 100 particles was counted on a 0.1-μm cell section, and we evaluated that roughly 10,000 viral particles might be bound/internalized to the dorsal surface of a cell, simplifying a cell to a smooth 25-μm diameter sphere. Since 0.1 TCID50 unit per cell was used to infect cells, the particle-to-infectivity ratio would be estimated at 1:100,000, meaning that most virus-containing clathrin vesicles were not infectious and were probably sent to degradation during the intracellular trafficking. Switching cells to 28°C allowed virus internalization and revealed that within 5 min p.i. most viral particles were located in characteristic clathrin-coated vesicles inside the cells. Similar to full-virus capsid, VPLs containing only the major structural protein VP4 (10) were localized within apparently similar structures, revealing no obvious role in the uptake process for the three other viral structural proteins.
FIG. 1.
Uptake and kinetics of JcDNV internalization and DNA replication within L. dispar cells. (A) Electron microscopy analysis of JcDNV (upper) and VP4-VLPs (lower) entry into L. dispar 652 cells. L. dispar cells were incubated with JcDNV virus particles or VP4-VLPs at 4°C for 1 h and then shifted to 28°C for 0 or 5 min before fixation and preparation for TEM examination. Scale bar, 100 nm. The arrows indicate either JcDNV or VLP4-VLPs. (B) Kinetics of JcDNV internalization within L. dispar cells was analyzed by immunofluorescence at various times p.i. L. dispar cells were infected with JcDNV as specified in Materials and Methods and immunolabeled with anti-JcDNV antibody (red). Confocal optical sections and epifluorescence images (black and white) of infected cells were examined at 0 min, 15 min, 30 min, and 4 h p.i. Arrows indicate JcDNV immunostaining. Nuclei were labeled with DAPI (blue). Scale bar, 20 μm. The inset is a confocal optical section of JcDNV-infected cells at 30 min p.i. Actin was labeled with phalloidin (green), and the nucleus was labeled with DAPI (blue). (C) JcDNV DNA replication kinetics. L. dispar cells were infected with JcDNV (0.1 TCID50 unit per cell) for 1h at 4°C, followed by washing to remove unbound virus, and were further incubated at 28°C. At various times p.i., total DNA was extracted and quantified with real-time PCR. Viral DNA was quantified using virus-specific primers, and the actin gene was used to quantify cellular DNA. Values represent the mean ratio of viral/cellular DNA of two different experiments.
To examine the mechanism of JcDNV uptake, we first studied the time course of the virus trafficking from the cell surface to the nucleus by confocal microscopy. Following their binding to the L. dispar cells, viral particles were observed at 15 min p.i. displaying scattered localizations within the cell, from the tip of cellular projections to perinuclear localization (Fig. 1B). At 30 min, most viral particles displayed perinuclear localization (Fig. 1B). A few particles were observed next to DAPI-stained chromatin; whether these particles were inside the nucleus or associated with nuclear membrane invaginations as observed for AAV (31) is unclear. By 4 h p.i., most of the viral staining was clustered in a crescent-shaped structure far from the nucleus (Fig. 1B).
Viruses that eventually enter into the nucleus must wait for the S phase to replicate (9, 37). We studied the kinetics of viral DNA synthesis using real-time PCR. Total DNA was extracted at various times p.i., and real-time PCR was performed with virus- and cell-specific primers to evaluate the kinetics of viral genome buildup per cell. The relative viral DNA amplification per cell (calculated by the ΔΔCT method) showed that viral DNA was detected within the cells at 4 h p.i. and remained stable until 48 h p.i. (Fig. 1C). We quantified the amount of virus detected at 4 h p.i. with a standard curve (See Materials and Methods) and estimated that 4 × 104 genome copies were detected per cell. Interestingly, the estimated number of virus copies per cell at 4 h p.i. was in a similar range as the internalized viral particles estimated by electron microscopy, thus confirming the estimation of a high particle-to-infection ratio. These results confirmed that most DNA-containing incoming particles were nonproductive and probably sent to degradation. At 16 h p.i. we could detect 2 × 104 genome copies per cell, and by 24 h, the number of genome copies had started to increase, suggesting that viral replication had begun. This was confirmed at 48 h p.i. by the exponential increase of the viral copy number.
These results define the timing of JcDNV infection; however, early viral genome replication is difficult to quantify by this method due to the overlap between viral DNA input and newly synthesized genome amplification.
Kinetics of the traffic to the nucleus: from early to late endocytic compartments.
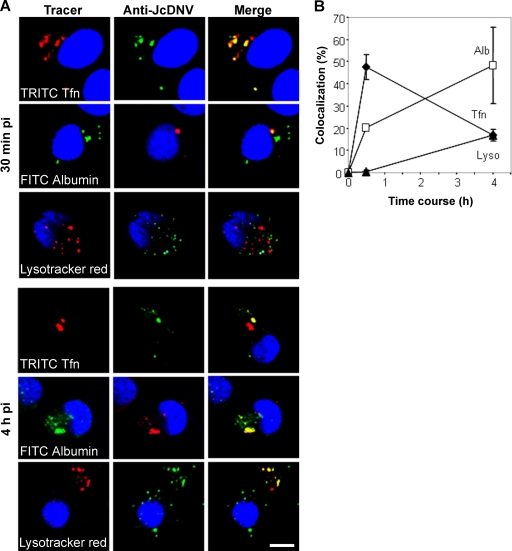
To identify which endocytic organelles were used by JcDNV for its intracellular trafficking, cells were incubated with JcDNV and with either TRITC-Tfn (an early/recycling endosome tracer), FITC-albumin (late endosome tracer), or LysoTracker (lysosome tracer). To estimate time-dependent colocalization events with different specific endocytic tracers, confocal laser images were obtained and processed with ImageJ and Metamorph analysis software. At 30 min p.i., overlay images showed that TRITC-Tfn was internalized with JcDNV, probably within early endosomes according to the perinuclear localization of large punctate structures (Fig. 2A). Fewer labeled TRITC-Tfn structures were observed per cell than in work done with vertebrate cells, which might be attributed to low expression of a Tfn receptor not characterized in insect cells. We thus analyzed JcDNV traffic through late endosomes and lysosomes by colocalization with albumin and LysoTracker, respectively (Fig. 2A). Overlay images showed that colocalization within late endosomes was partial while JcDNV was apparently excluded from lysosomes. Colocalizations were then analyzed in 3D space, quantifying for each marker the fluorescence intensity area from cell z-sectioning images. Results showed that 50% of JcDNV colocalized with Tfn at 30 min (Fig. 2B) and 20% with albumin in late endosomes. JcDNV was never found within lysosomes at this stage of infection (Fig. 2B). The same analysis was performed at 4 h p.i. Overlay images and quantification assays showed that at this time point, 20% of JcDNV was colocalized with TRITC-Tfn, probably in recycling endosomes, while 50% was colocalized with albumin within perinuclear late endosomes (Fig. 2). At that time, 20% of JcDNV colocalized with LysoTracker within lysosomes. This localization was observed far from the nucleus (Fig. 2A).
FIG. 2.
Time course of JcDNV delivery to early and late endocytic compartments: colocalization with endocytic markers. (A) Cells were concomitantly infected with JcDNV and either TRITC-Tfn, FITC-albumin, or LysoTracker red for 30 min or 4 h at 28°C. After fixation cells were labeled with anti-JcDNV antibody (green or red) and endocytic markers (red or green), and nuclei were labeled with DAPI (blue). Scale bar, 20 μm. (B). Time-dependent colocalization of JcDNV with Tfn, albumin, or LysoTracker. The percentage of colocalized JcDNV with Tfn (diamond), albumin (square), or LysoTracker (triangle) over three consecutive stacks (0.3 μm) is shown. Colocalization was significant, with a Pearson coefficient of >0.9.
At 16 h p.i. no fluorescence was detected in the cells (data not shown), suggesting that degradation of the capsids had occurred by this time. It is of note that previous qPCR analysis showed that 2 × 104 genome copies were detected per cell; as no fluorescent viral particles (representative of the input virus) were observed at 16 h p.i., the presence of viral genomes probably reflects that viral replication had already started.
JcDNV infection is mainly mediated by dynamin and clathrin-dependent endocytosis.
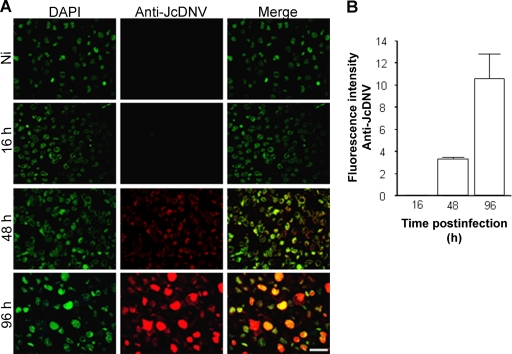
To address which functional pathway(s) leads to productive infection, we first set up an assay to analyze JcDNV endocytosis beyond entry, i.e., to productive infection. Since fluorescent viral particles had disappeared at 16 h p.i., de novo VP proteins expression at late time points was taken as a reporter of viral replication. The kinetics of VP production was first established, and viral immunostaining was detected from 48 h p.i. within the cytoplasm of infected cells, while at 96 h p.i. the typical densonucleosis was observed (Fig. 3A). We then quantified viral infectivity by determining the immunofluorescence intensity as the reporter of JcDNV infection. These quantitative results confirmed our microscopic observations (Fig. 3B). Using this assay, we tested the effects late in infection of blocking endocytic pathways with specific inhibitors. Analyzing the effects of these molecules 2 days p.i. required a low-dose exposure, thus minimizing toxic effects of inhibitors, since only live and replicating cells are taken into account.
FIG. 3.
Infection assay to quantify JcDNV infectivity. (A) Microscopic observation of the time course of viral proteins synthesis. Kinetics was assessed at different times p.i. Semithin sections of infected cells were immunolabeled with anti JcDNV antibody (red). Nuclei were labeled with DAPI (green). Scale bar, 20 μm. (B) Fluorescence quantification of JcDNV-immunopositive cells. Results are an average of three different sections. Ni, noninfected.
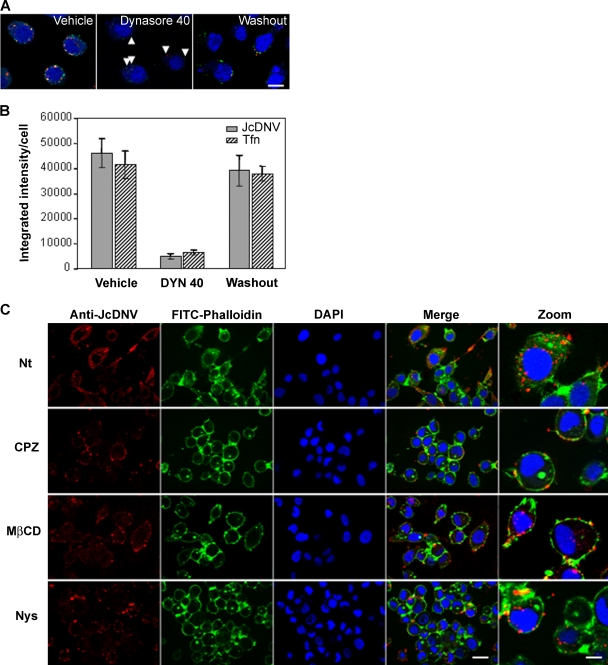
We analyzed biochemically the JcDNV early entry process and first tested the role of the large GTPase dynamin, essential for clathrin-coated-vesicle formation (reviewed in reference 24). Recently, Macia and colleagues discovered DYN, a highly specific inhibitor of dynamin's GTPase activity (32). They established a dual role for dynamin in clathrin-coated vesicle formation at an early invagination step and at a late constriction step (32). One dynamin (Shibire) is present in flies, and homologous sequences were found in expressed sequence tag collections from Bombyx mori (http://morus.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi), but no functional data are available in Lepidoptera. We thus used DYN and first established whether this molecule was active in L. dispar cells. We quantified the uptake of TRITC-Tfn and JcDNV at different DYN concentrations. As described for mammalian cells, a powerful block was obtained with 40 μM DYN (Fig. 4A and B). This effect was dose dependent, but at late times of infection (2 days p.i.) toxicity was observed with a higher concentration (80 μM) (data not shown). We thus defined 40 μM as the working dose. The inhibitory effect was also reversible, as traffic to the nucleus was partially restored 20 min after the removal of DYN from the medium (Fig. 4A and B). We also verified that this inhibition was not due to a decrease in the number of putative respective receptors at the cell surface since Tfn and JcDNV binding was the same in cells kept at 4°C with and without DYN (data not shown).
FIG. 4.
JcDNV early entry process. (A) Dynamin-dependent internalization of transferrin and JcDNV was inhibited by DYN. L. dispar cells were incubated for 30 min at 28°C with 40 μM DYN or 0.8% DMSO (vehicle). TRITC-Tfn (red) and JcDNV (green) were then added concomitantly for 30 min at 28°C in the continuous presence of DMSO or DYN (pulse) or 20 min after washout of 40 μM DYN (chase). Cells were then washed before fixation and immunolabeling of JcDNV with an anticapsid antibody (red). Nuclei were stained with Hoechst dye (blue). Scale bar, 20 μm. Cells were examined under a Zeiss confocal microscope. (B) Histogram shows the blocking and reversible effects of 40 μM DYN on Tfn and JcDNV uptake. The integrated intensity/cell of internalized JcDNV (closed bars) or Tfn (hatched bars) was quantified using Metamorph (3 stacks; n = 10 cells). (C) Effects of different inhibitors on JcDNV early entry process. L. dispar cells were pretreated or not with CPZ, MβCD, or Nys and subsequently infected with JcDNV as described in Materials and Methods. The analysis was made by immunofluorescence using anticapsid antibody (red) on 0.5-μm sections with an Apotome Axiovision microscope at various times p.i. Cortical actin was labeled with phalloidin (green) and nuclei were labeled with DAPI (blue). Scale bars, 20 μm and 5 μm (zoom). Nt, nontreated.
These data were confirmed by treating cells with a second inhibitor of clathrin-dependent endocytosis, CPZ (30, 48) which inhibits the AP2/clathrin uptake pathway by removing AP2 from the plasma membrane (Fig. 4C). (33, 50). In contrast, treatment of cells with the sterol-sequestering drugs MβCD and Nys, which specifically inhibits the caveola- and lipid raft-dependent endocytosis (36), had no effect on JcDNV entry (Fig. 4A). These results are consistent with the lack of immune colocalization between JcDNV and caveolin-1 (data not shown). All together these results confirmed that different blocks in the clathrin-mediated endocytosis strongly affect JcDNV uptake and traffic.
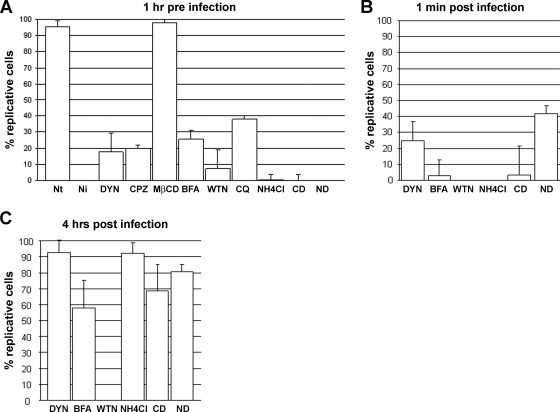
To strengthen these data, we then analyzed the effects of these inhibitors on the infection process. Infection assays showed that blocking the clathrin-dependent pathway 1 h before virus adsorption strongly affected infection since 80% of either DYN- or CPZ-treated cells were not infected while no difference in infection was observed in MβCD-treated cells compared to the controls (Fig. 5). To better define the role of dynamin on infection, DYN was then applied at infection (just after virus adsorption) and 4 h after infection, as specified in the legend of Fig. 5. When applied at infection, DYN partially affected infection (Fig. 5B). These results confirmed the role of dynamin in the virus uptake. When added 4 h after infection, DYN lost its effect and no longer affected virus replication, validating that this drug targeted the early entry process (Fig. 5C).
FIG. 5.
Effects of different inhibitors on JcDNV infection. L. dispar cells were pretreated with different inhibitors before, at, and after (4 h) JcDNV infection. Virus infectivity was quantified at 48 h p.i. as described in the text. Results represent the average of three different sections. Nt, nontreated; Ni, noninfected cells.
We were not able to perform this test with Nys due to the cytotoxic effect of this drug at 48 h p.i..
These results established that dynamin-dependent clathrin-mediated endocytosis was the main JcDNV functional entry pathway of JcDNV. The residual 20% of the DYN- and CPZ-treated cells which were infected could reflect a partial or reversible effect of the drug, allowing the recovery of the infection from cell surface-bound particles. Alternatively a clathrin- (and caveolin-) independent infectivity pathway(s) may play a minor role in the infection of L. dispar cells.
Trafficking of the virus through early and late endocytic organelles is essential for JcDNV infection.
To determine the infectious pathway, L. dispar cells were treated with drugs blocking endocytic traffic between early to late compartments. Phosphoinositide-3-OH kinase (PI3K) pathways have been reported to play a critical role in the endocytosis of adenovirus, AAV2, and clathrin-mediated endocytosis of other viruses (3, 25, 42). We hypothesized that PI3K might also play a role during JcDNV entry. L. dispar cells were treated with the specific PI3K inhibitor, the fungal toxin WTN (44). The fungal antibiotic BFA was also used to block the early to late traffic as well as retrograde traffic between the trans Golgi network and the endoplasmic reticulum (ER) (28, 41). In order to define the time course of infectious traffic and to assess for the specificity of the drugs, treatments were added before, at, and after infection. Cells were treated 1 h before infection, and we first tested the effects of drugs on virus entry by immunolabeling the cells at 30 min p.i. BFA did not impair JcDNV entry even though the presence of small punctate structures might reflect that fewer particles were internalized. WTN-treated cells displayed viral staining localized within punctate structures larger than with control cells (Fig. 4C and 6A). The resulting infection showed a strong inhibition of viral replication with both treatments (Fig. 5A). Cells were then treated at infection, and results showed that viral replication was still strongly impaired with both drugs (Fig. 5B). When added after infection (4 h p.i.), BFA lost its activity, and cells were efficiently infected while WTN treatment still impaired viral replication (Fig. 5C). Since at 4 h p.i. virus was mainly localized within late endosomes (Fig. 2), these results showed (i) that unlike WTN, BFA treatment did not affect viral replication and (ii) that impairing virus routing through late endosomes or the ER impaired infection. With respect to WTN, the effects were not restricted to traffic since blocking the phosphatidylinositol pathway affected DNA replication. Further experiments are required to analyze the function of these pathways on viral replication.
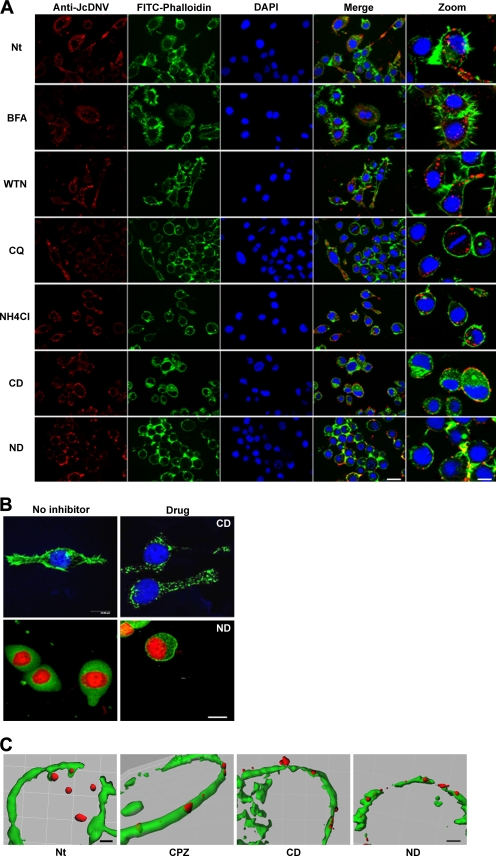
FIG. 6.
Effects of different inhibitors on JcDNV trafficking. (A) L. dispar cells were untreated or pretreated with BFA, WTN, NH4Cl, CQ, CD, or ND and subsequently infected with JcDNV as described in Materials and Methods. The analysis was made as described in the legend of Fig. 4. Scale bars, 20 μm and 5 μm (zoom). (B) Effects of drugs disrupting the cytoskeleton on L. dispar cells. L. dispar cells were incubated with 10 μM CD and 30 μM ND for 1 h and then fixed to observe microtubule (anti-β-tubulin; green) and microfilament (FITC-phalloidin; green) morphology by confocal analysis. Nuclei are in blue or red (DAPI). Scale bar, 20 μm. (C) Colocalization analysis was performed on images over five consecutive stacks (0.5 μm) by using the 3D imaging analysis software Imaris, version 5.7.0 (Bitplane). The threshold was set manually. Nt, nontreated. Scale bars, 6 μm (Nb) and 3 μm (CP2, CD, ND).
JcDNV productive infection in L. dispar cells requires transit through late endocytic compartments.
It has been shown that endosomal acidification is essential for parvovirus infection, triggering capsid structural rearrangements and escape of the virus from endosomes into the cytoplasm (13, 34, 38, 41, 53). To determine the role of a low-pH-dependent step in JcDNV infectivity, we examined the effects on infection of two weak bases known to raise the pH in endosomes and lysosomes, CQ and ammonium chloride (NH4Cl) (35, 43). Thirty minutes after treatments, viral staining appeared diffuse in both types of treated cells compared to the control, which might be due to the swelling of acidic vesicles (Fig. 6A). Full inhibition of virus replication was observed when NH4Cl treatment was applied before infection while this effect was partial with CQ, suggesting a dose effect of this drug in L. dispar cells (Fig. 5A). Treatment with NH4Cl performed at infection strongly inhibited infection (Fig. 5B) while a loss in activity was observed 4 h p.i. (Fig. 5C). Since raising the pH at 4 h p.i. no longer affected viral replication, it is possible that viral escape had already occurred from late endosomes or, alternatively, that that virus escape might be pH independent. Further experiments with pH-independent molecular markers and virus mutants will help to solve this important question.
JcDNV infection requires an intact host cytoskeleton.
It has been reported that host cell cytoskeleton components are involved in early events during infection with viruses from several families including parvoviruses (41, 47). To determine whether actin and microtubule networks were involved in the transport of JcDNV particles to the nucleus, L. dispar cells were treated 1 h prior to infection either with the actin-polymerization inhibitor CD or the microtubule-disrupting agent ND. Both drugs strongly affected L. dispar microfilaments and microtubule organization; actin labeling was punctiform, while a tubulin network became visible, thus contrasting with the diffuse labeling in nontreated cells (Fig. 6B). By 30 min p.i. in both ND- and CD-treated cells, JcDNV immunostaining was maintained within cells at the plasma membrane, contrasting with control cells displaying a predominant perinuclear staining (Fig. 6A). As exemplified by overlay images and 3D analysis using Imaris software (Fig. 6C), virus colocalized at the plasma membrane with cortical actin, suggesting that host cytoskeleton was required for internalization. We further explored the effects of both drugs on the infection, i.e., the ability of the virus to traffic to the nucleus. No staining was detected when these inhibitors were added 1 h before infection, showing that infection was totally impaired by both treatments (Fig. 5A). Disrupting the cytoskeleton before infection might reduce virus internalization below an infectious threshold if we consider the high particle-to-infectivity ratio. Treating the cells just after virus adsorption still affected infection even though inhibition was partial in ND-treated cells (Fig. 5B). This loss in efficiency of ND might be explained by a limited role of microtubules in virus uptake. In that case, viral particles internalized before cytoskeleton disruption might lead to a productive infection while their traffic to the nucleus might depend more on the actin network. When performed at 4 h p.i., both treatments displayed only a little effect on virus replication (Fig. 5C), showing that virus traffic was not dependent at this time point on cytoskeleton and/or that cytoplasmic JcDNV escape had already occurred.
DISCUSSION
DNVs are considered as potential microbiological agents against insect pests. We chose JcDNV as our model to study its potentiality. This potential use raised several underlying questions about the cellular interactions that also control insect susceptibility. JcDNV has a potentially broad host spectrum; understanding the first steps of infection, i.e., the elucidation of host factors involved in binding and entry processes, might help to understand the infection and the pathogenesis of new hosts. The aim of this work was to understand the early cellular events that lead to JcDNV infection of a permissive insect cell line.
We first observed a clear clathrin-mediated uptake of viral particles by electron microscopy. To link viral entry and trafficking with infection, we designed a functional assay to block important cellular functions with drugs and analyzed the resulting effect on virus replication. We have applied a variety of different molecules to block the viral nuclear translocation at different intracellular steps. This assay validates and strengthens an analysis made at an early time of entry, when the sole microscopic observation of fixed cells could have been misleading.
Here, we describe for the first time the entry of an insect parvovirus within a host cell, and we demonstrate that viral entry is a rate-limiting step in the infection process. Alternative strategies remain to be set up with these lepidopteran cells; few sequences are available, thus excluding an RNA interference targeting approach, and low transfection efficiencies are at the moment the main obstacle to the expression of dominant negative proteins.
JcDNV infection proceeds mainly through clathrin-coated vesicles.
Many studies have shown that parvovirus uptake requires receptor-mediated endocytosis allowing virus internalization via the clathrin-dependent endocytic pathway (18). Our results demonstrate that JcDNV internalization is rapid and that the infection pathway also proceeds mainly via clathrin-dependent endocytosis probably through a cell surface receptor. Neither the cellular receptor nor the receptor binding sites on the capsid for any insect parvovirus have been characterized so far. Based on their capsid structures and sequences, DNV potential interacting residues and host range determinants could be predicted (6, 45). No prediction, however, can be made for a cellular receptor, as illustrated by the diversity of characterized parvoviral receptors (9) and the structure of the capsids. Furthermore, insect parvovirus capsids have relatively smooth surfaces compared to the surfaces of vertebrate parvoviruses (6, 45). Interestingly, our results show that only the major viral structural protein VP4 could mediate VP4-VLP binding and internalization via a similar mechanism even though we cannot exclude a role for other structural protein(s) in the cell membrane attachment and/or in the infectious routing of viral particles.
Infectious pathway requires late endosomal trafficking.
We have shown that viral particles enter cells rapidly within vesicles and then accumulate around the nucleus within 30 min p.i. Most of the particles remain within endosomal compartments at least for 4 h. Several points are unsolved concerning the sorting of viral particles. We could assume that the time for the virus to reach the nucleus is 4 h p.i. because it mainly depends on the traffic through late endocytic compartments. Impairing JcDNV routing through late endosomes or the ER strongly inhibits infection and shows that the “default” infectious pathway requires this step.
Acidification is known to be essential for the parvoviruses entry and may induce capsid conformational changes essential for infection (13, 38, 41). In vertebrate parvoviruses, the VP1-associated PLA2 activity seems to be directly or indirectly responsible for late endosomal escape (53). Like most parvoviruses, JcDNV presents the conserved PLA2 and the basic NLS-like motifs in the VP1 region (16, 27, 53). We showed that neutralizing the endosomal pH at 4 h p.i. had no effect on infection, thus restricting the role of acidification and/or the time of viral escape. Combining the observations that blocking early to late endosomal/ER traffic and acidification impaired infection might suggest that acidification is important for infection; whether this effect is direct remains unclear. JcDNV capsid mutations within PLA2 or NLS-like domains did not abolish infection albeit the in vivo infectivity was decreased (A. Abd-Alla, personal communication), suggesting an important role for these domains in DNV pathogenesis. Defining the role of these motifs will help elucidate the role of the pH in the infection process.
A low-pH environment probably initiates JcDNV release from vesicles to the cytoplasm. Whether this release preferentially occurs within late endosomes or lysosomes or both is unclear. Localization of the lysosomes far from the nucleus and the fact that no viral labeling is detected at 16 h p.i. most likely suggest a massive degradation of viral particles in this compartment. We cannot rule out a lysosomal requirement for the infection process that could be hidden by the high number of viral particles used in these experiments. The use of more cellular markers will help to clarify the virus endosomal traffic and escape mechanisms.
Trafficking toward the nucleus requires an intact cytoskeleton.
Whether JcDNV particles require an intact cytoskeleton network to enter the cells or whether this network facilitates the movement of virus-containing vesicles within the cytoplasm is unclear and remains to be studied more carefully. Previous studies have shown that cytoskeleton elements and PI3K play an important role in controlling the AAV intracellular movement or the internalization of adenovirus (25, 42). Our results are consistent with these observations since disrupting cytoskeleton strongly inhibits viral entry and infection. However, the role of PI3K on traffic remains unclear since blocking this pathway affects more than virus trafficking. As already observed, blocking the phosphatidylinositol pathway might affect multiple functions, including DNA replication (54). Recently, WTN was shown to target polo-like kinase and block cell cycle progression (29). Targeting one or both pathways might explain the inhibition of viral DNA replication.
Concluding remarks.
As already observed for other parvoviruses, even though this part is poorly documented, JcDNV infection is very inefficient if we consider the high particle-to-infectivity ratio. It has been suggested that escape from endosomes is the major limiting step in infection (31, 34). In the JcDNV infection process, most particles entering the cells do not replicate and are eliminated. We were not able to clearly detect incoming viral proteins within the nucleus even though a few of them were detected close to chromatin as early as 30 min p.i. This observation might reflect either that DNA entering the nucleus is naked or that it is associated with rearranged viral proteins.
It has been suspected that parvoviruses escape from all the endocytic organelles, which probably reflects the complexity of a trafficking process that depends on the cellular context encountered by the virus in vivo. In the natural course of infection, lepidopteran larvae are infected through ingestion of contaminated food. JcDNV must first traffic through polarized intestinal cells although these cells do not replicate the virus (unpublished data). Alternative trafficking pathways might aid the virus to cross epithelial barriers and reach target tissues to replicate.
Acknowledgments
This work was supported by the Ecole Pratique des Hautes Etudes (A.V. M.D., and T.D.) and INRA.
We thank A. Abd-Alla for sharing unpublished data. We are grateful to Michel Vidal (DIMNP lab, UMII) for his help and for providing TRITC-Tfn and to Fabrice Raynaud (CRBM, Montpellier) for providing LysoTracker. We also thank Vicky Diakou, Nicole Lautredou-Audouy, and Volker Baker and all the members of the MRI facility (Montpellier) for their help during image acquisition and analysis. Special thanks go to M. Bergoin and A. S. Gosselin for their support and all the very helpful discussions. We are extremely grateful to V. Ollendorff and P. Lemaire for their critical comments about the manuscript.
Footnotes
Published ahead of print on 18 February 2009.
REFERENCES
- 1.Abd-Alla, A., F. X. Jousset, Y. Li, G. Fediere, F. Cousserans, and M. Bergoin. 2004. NS-3 protein of the Junonia coenia densovirus is essential for viral DNA replication in an Ld 652 cell line and Spodoptera littoralis larvae. J. Virol. 78790-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amargier, A., C. Vago, and G. Meynadier. 1965. Histopathological study of a new type of viral disease demonstrated in the lepidoptera Galleria mellonella. Arch. Gesamte Virusforsch. 15659-667. [In French.] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 742777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergoin, M., and P. Tijssen. 2000. Molecular biology of Densovirinae. Contrib. Microbiol. 412-32. [DOI] [PubMed] [Google Scholar]
- 5.Bolte, S., and F. P. Cordelieres. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224213-232. [DOI] [PubMed] [Google Scholar]
- 6.Bruemmer, A., F. Scholari, M. Lopez-Ferber, J. F. Conway, and E. A. Hewat. 2005. Structure of an insect parvovirus (Junonia coenia Densovirus) determined by cryo-electron microscopy. J. Mol. Biol. 347791-801. [DOI] [PubMed] [Google Scholar]
- 7.Carlson, J., E. Suchman, and L. Buchatsky. 2006. Densoviruses for control and genetic manipulation of mosquitoes. Adv. Virus Res. 68361-392. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, S., A. R. Behzad, J. B. Carroll, and N. Pante. 2006. Parvoviral nuclear import: bypassing the host nuclear-transport machinery. J. Gen. Virol. 873209-3213. [DOI] [PubMed] [Google Scholar]
- 9.Cotmore, S. F., and P. Tattersall. 2007. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 70183-232. [DOI] [PubMed] [Google Scholar]
- 10.Croizier, L., F. X. Jousset, J. C. Veyrunes, M. Lopez-Ferber, M. Bergoin, and G. Croizier. 2000. Protein requirements for assembly of virus-like particles of Junonia coenia densovirus in insect cells. J. Gen. Virol. 811605-1613. [DOI] [PubMed] [Google Scholar]
- 11.Ding, C., M. Urabe, M. Bergoin, and R. M. Kotin. 2002. Biochemical characterization of Junonia coenia densovirus nonstructural protein NS-1. J. Virol. 76338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 762014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 751824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Far, M., Y. Li, G. Fediere, S. Abol-Ela, and P. Tijssen. 2004. Lack of infection of vertebrate cells by the densovirus from the maize worm Mythimna loreyi (MlDNV). Virus Res. 9917-24. [DOI] [PubMed] [Google Scholar]
- 15.Fediere, G. 2000. Epidemiology and pathology of Densovirinae. Contrib. Microbiol. 41-11. [PubMed] [Google Scholar]
- 16.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83973-978. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin, R. H., G. J. Tompkins, and P. McCawley. 1978. Gypsy moth cell lines divergent in viral susceptibility. I. Culture and identification. In Vitro 14485-494. [DOI] [PubMed] [Google Scholar]
- 18.Harbison, C. E., J. A. Chiorini, and C. R. Parrish. 2008. The parvovirus capsid odyssey: from the cell surface to the nucleus. Trends Microbiol. 16208-214. [DOI] [PubMed] [Google Scholar]
- 19.Hirosue, S., K. Senn, N. Clement, M. Nonnenmacher, L. Gigout, R. M. Linden, and T. Weber. 2007. Effect of inhibition of dynein function and microtubule-altering drugs on AAV2 transduction. Virology 36710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirunkanokpun, S., J. O. Carlson, and P. Kittayapong. 2008. Evaluation of mosquito densoviruses for controlling Aedes aegypti (Diptera: Culicidae): variation in efficiency due to virus strain and geographic origin of mosquitoes. Am. J. Trop. Med. Hyg. 78784-790. [PubMed] [Google Scholar]
- 21.Home, R. W., S. Brenner, A. P. Waterson, and P. Wildy. 1959. The icosahedral form of an adenovirus. J. Mol. Biol. 184-86. [Google Scholar]
- 22.Jourdan, M., F. X. Jousset, M. Gervais, S. Skory, M. Bergoin, and B. Dumas. 1990. Cloning of the genome of a densovirus and rescue of infectious virions from recombinant plasmid in the insect host Spodoptera littoralis. Virology 179403-409. [DOI] [PubMed] [Google Scholar]
- 23.Jousset, F. X., C. Barreau, Y. Boublik, and M. Cornet. 1993. A parvo-like virus persistently infecting a C6/36 clone of Aedes albopictus mosquito cell line and pathogenic for Aedes aegypti larvae. Virus Res. 2999-114. [DOI] [PubMed] [Google Scholar]
- 24.Kirchhausen, T., E. Macia, and H. E. Pelish. 2008. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 43877-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, E., D. Stupack, R. Klemke, D. A. Cheresh, and G. R. Nemerow. 1998. Adenovirus endocytosis via αv integrins requires phosphoinositide-3-OH kinase. J. Virol. 722055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., F. X. Jousset, C. Giraud, F. Rolling, J. M. Quiot, and M. Bergoin. 1996. A titration procedure of the Junonia coenia densovirus and quantitation of transfection by its cloned genomic DNA in four lepidopteran cell lines. J. Virol. Methods 5747-60. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., Z. Zadori, H. Bando, R. Dubuc, G. Fediere, J. Szelei, and P. Tijssen. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 822821-2825. [DOI] [PubMed] [Google Scholar]
- 28.Lippincott-Schwartz, J., L. Yuan, C. Tipper, M. Amherdt, L. Orci, and R. D. Klausner. 1991. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67601-616. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., N. Jiang, J. Wu, W. Dai, and J. S. Rosenblum. 2007. Polo-like kinases inhibited by wortmannin. Labeling site and downstream effects. J. Biol. Chem. 2822505-2511. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 31.Lux, K., N. Goerlitz, S. Schlemminger, L. Perabo, D. Goldnau, J. Endell, K. Leike, D. M. Kofler, S. Finke, M. Hallek, and H. Buning. 2005. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J. Virol. 7911776-11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macia, E., M. Ehrlich, R. Massol, E. Boucrot, C. Brunner, and T. Kirchhausen. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10839-850. [DOI] [PubMed] [Google Scholar]
- 33.Maginnis, M. S., B. A. Mainou, A. Derdowski, E. M. Johnson, R. Zent, and T. S. Dermody. 2008. NPXY motifs in the beta1 integrin cytoplasmic tail are required for functional reovirus entry. J. Virol. 823181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani, B., C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, and C. Ros. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 801015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misinzo, G., P. Meerts, M. Bublot, J. Mast, H. M. Weingartl, and H. J. Nauwynck. 2005. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J. Gen. Virol. 862057-2068. [DOI] [PubMed] [Google Scholar]
- 36.Moriyama, T., J. P. Marquez, T. Wakatsuki, and A. Sorokin. 2007. Caveolar endocytosis is critical for BK virus infection of human renal proximal tubular epithelial cells. J. Virol. 818552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Op De Beeck, A., and P. Caillet-Fauquet. 1997. Viruses and the cell cycle. Prog. Cell Cycle Res. 31-19. [DOI] [PubMed] [Google Scholar]
- 38.Parker, J. S., and C. R. Parrish. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 741919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren, X., E. Hoiczyk, and J. L. Rasgon. 2008. Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog 4e1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivers, C. F., and J. F. Longworth. 1972. A non-occluded virus of Junonia coenia (Nymphalidae: Lepidoptera). J. Invertebr. Pathol. 20369-370. [Google Scholar]
- 41.Ros, C., C. J. Burckhardt, and C. Kempf. 2002. Cytoplasmic trafficking of minute virus of mice: low-pH requirement, routing to late endosomes, and proteasome interaction. J. Virol. 7612634-12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by Rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 749184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seglen, P. O., and P. B. Gordon. 1979. Inhibition of cell spreading by lysosomotropic amines. FEBS Lett. 105345-348. [DOI] [PubMed] [Google Scholar]
- 44.Shpetner, H., M. Joly, D. Hartley, and S. Corvera. 1996. Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J. Cell Biol. 132595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 61355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 8011040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suikkanen, S., T. Aaltonen, M. Nevalainen, O. Valilehto, L. Lindholm, M. Vuento, and M. Vihinen-Ranta. 2003. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 7710270-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316267-280. [DOI] [PubMed] [Google Scholar]
- 49.Vago, C., J. M. Quiot, and J. Luciani. 1966. Infection of lepidopteran tissue culture with the purified virus of the densonucleus. C R Acad. Sci. Hebd. Seances Acad. Sci. D 263799-800. [In French.] [PubMed] [Google Scholar]
- 50.Veyrat-Durebex, C., L. Pomerleau, D. Langlois, and P. Gaudreau. 2005. Internalization and trafficking of the human and rat growth hormone-releasing hormone receptor. J. Cell Physiol. 203335-344. [DOI] [PubMed] [Google Scholar]
- 51.Vihinen-Ranta, M., L. Kakkola, A. Kalela, P. Vilja, and M. Vuento. 1997. Characterization of a nuclear localization signal of canine parvovirus capsid proteins. Eur. J. Biochem. 250389-394. [DOI] [PubMed] [Google Scholar]
- 52.Vihinen-Ranta, M., W. Yuan, and C. R. Parrish. 2000. Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport. J. Virol. 744853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1291-302. [DOI] [PubMed] [Google Scholar]
- 54.Zewail, A., M. W. Xie, Y. Xing, L. Lin, P. F. Zhang, W. Zou, J. P. Saxe, and J. Huang. 2003. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. USA 1003345-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]