Abstract
The nucleoprotein (NP), which has multiple functions during the virus life cycle, possesses regions that are highly conserved among influenza A, B, and C viruses. To better understand the roles of highly conserved NP amino acids in viral replication, we conducted a comprehensive mutational analysis. Using reverse genetics, we attempted to generate 74 viruses possessing mutations at conserved amino acids of NP. Of these, 48 mutant viruses were successfully rescued; 26 mutants were not viable, suggesting a critical role of the respective NP amino acids in viral replication. To identify the step(s) in the viral life cycle that is impaired by these NP mutations, we examined viral-genome replication/transcription, NP localization, and incorporation of viral-RNA segments into progeny virions. We identified 15 amino acid substitutions in NP that inhibited viral-genome replication and/or transcription, resulting in significant growth defects of viruses possessing these substitutions. We also found several NP mutations that affected the efficient incorporation of multiple viral-RNA (vRNA) segments into progeny virions even though a single vRNA segment was incorporated efficiently. The respective conserved amino acids in NP may thus be critical for the assembly and/or incorporation of sets of eight vRNA segments.
Influenza A virus consists of eight negative-sense single-stranded viral genomic-RNA segments and encodes at least 11 proteins (reviewed in reference 39). These genomic RNAs are incorporated into virions as ribonucleoprotein (RNP) complexes, which consist of the viral RNA (vRNA) associated with three viral polymerase subunit proteins (PA, PB1, and PB2) and nucleoprotein (NP). Upon binding to cell surface receptors, virions are internalized by receptor-mediated endocytosis. After fusion of the viral and endosomal membranes, the viral RNPs (vRNPs) are released into the cytoplasm and transported to the nucleus, where viral-genome replication and transcription take place (34). Newly synthesized vRNAs are associated with the NP and form vRNPs in the nucleus (4). Subsequently, the vRNPs are transported to the cytoplasm and packaged into the progeny virus particles, which then bud from the cells.
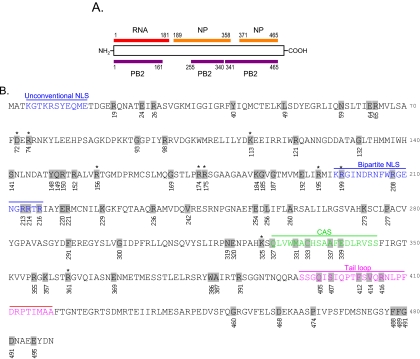
NP, a basic protein composed of 498 amino acids, is a major component of vRNPs (reviewed in reference 33). It contains an RNA-binding region at its N terminus (residues 1 to 181) (1, 19) and two domains, responsible for NP-NP self-interaction, at residues 189 to 358 and 371 to 465 (8) (Fig. 1A). Both of these NP functions are important to maintain the organization of vRNPs. Besides its structural role, NP is involved in many other functions throughout the virus replication cycle. In the early stages of the viral life cycle, NP facilitates vRNP import into the nucleus via its two nuclear localization signals (NLSs), an unconventional NLS (residues 3 to 13) and a bipartite NLS (residues 198 to 216) (35). NP also plays a role in RNA synthesis in the nucleus (15). It is required for the synthesis of longer RNAs, although three polymerase proteins are sufficient to synthesize short RNAs (14). NP also interacts with the viral polymerase proteins PB1 and PB2 (3), suggesting a potential role in the regulation of polymerase activity. Export of vRNPs from the nucleus to the cytoplasm is promoted via an interaction between NP and M1/NS2 (2, 22, 27, 29, 38, 40, 42). NP has an important role in RNP export; besides binding to the M1 protein, NP contains a cytoplasmic accumulation signal (residues 327 to 345), which interacts with F actin and causes cytoplasmic retention of NP late in infection (2, 5). In addition, NP contains a nuclear export signal that is recognized by the nuclear export receptor CRM1 (9). Overexpression of CRM1 biases transfected NP toward cytoplasmic accumulation, and the two proteins interact in in vitro binding assays (9).
FIG. 1.
Individual residues targeted by mutagenesis. (A) The NP regions responsible for binding to RNA (red), NP itself (orange), and PB2 (purple) are shown. (B) Amino acid sequence of WSN NP. Amino acids tested in this study are shaded. The unconventional NLS, the bipartite NLS, the cytoplasmic accumulation signal (CAS), and the NP tail loop, which mediates NP oligomerization, are shown in different colors. NP mutants flagged with an asterisk were further tested for intracellular localization and incorporation efficiency of a vRNA segment(s) into VLPs, in addition to replicative ability and polymerase activity.
The NP possesses regions that are highly conserved among influenza A, B, and C viruses (23). Mena et al. (23) used mutational analysis to identify several amino acid residues that are important for vRNA replication in the conserved regions of NP. For most of the conserved amino acids, however, the biological significance and the role in the viral life cycle remain unknown. To close this gap in knowledge, we attempted to generate 74 mutant viruses possessing mutations at conserved residues of NP, and for the mutations that did not support viral growth, we studied the steps at which the mutations negatively affected NP functions.
MATERIALS AND METHODS
Cells.
293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. Madin-Darby canine kidney (MDCK) cells were maintained in minimum essential medium containing 5% newborn calf serum. NP-expressing MDCK (MDCK-NP) cells were maintained in minimum essential medium containing 5% newborn calf serum and 5 μg/ml puromycin. All cells were maintained at 37°C in 5% CO2.
MDCK-NP cells stably expressing NP from A/WSN/33 (H1N1; WSN) were established by infecting MDCK cells with retroviruses that were generated by cotransfecting 293T cells with the murine leukemia virus retroviral vector pMX (30) containing the NP gene, the internal ribosome entry site (IRES) sequence, and the puromycin resistance gene (designated pMX-WSNNP-IRES-puro [see below]) and plasmids expressing Gal/Pol, NF-κB, and vesicular stomatitis virus G protein (17), which were kind gifts from Bill Sugden (University of Wisconsin—Madison). A stable MDCK cell clone expressing NP was selected in medium containing 0.15 mg/ml puromycin (Roche, Mannheim, Germany) by indirect immunostaining with a monoclonal antibody to NP.
Plasmid construction.
The NP segment of WSN virus was reverse transcribed with an oligonucleotide complementary to the conserved 3′ end of the vRNA (16) and a pair of NP gene-specific oligonucleotide primers containing BsmBI sites. The PCR product was cloned into the pT7Blueblunt vector (Novagen, Madison, WI), and the resultant plasmid was referred to as pT7-WSN-NP. After digestion with BsmBI, the fragment containing the NP gene was cloned into the BsmBI sites of the pHH21 vector, which contains the human RNA polymerase I promoter and the mouse RNA polymerase I terminator separated by BsmBI sites (24), resulting in the generation of pPolI-NP (all plasmids derived from pHH21 for the expression of vRNA are referred to as PolI constructs in this report).
pCAGGs/MCS-NP, for the expression of the NP protein, was generated by inserting the BsmBI-digested NP fragment into the BsmBI sites of a modified pCAGGS/MCS (18, 25) vector. The substitution mutations listed in Table 1 were first introduced into the pT7-WSN-NP plasmid by in vitro site-directed mutagenesis (Stratagene, La Jolla, CA) and then subcloned into the pHH21 and pCAGGS/MCS vectors by using the unique BsmBI restriction enzyme sites. All mutations were verified by sequencing.
TABLE 1.
Phenotypic analyses of mutant NP proteins
| NPa | Virus rescueb | Mean virus titer at 48 h postinfection (log10 PFU/ml) ±SDc
|
Groupd | Relative luciferase activity at 48 h posttransfection (%)e
|
||
|---|---|---|---|---|---|---|
| 33°C | 37°C | 33°C | 37°C | |||
| Wild type | + | 7.99 ± 0.01 | 8.85 ± 0.07 | I | 100.00 ± 6.88 | 100.00 ± 13.51 |
| R19A | + | 7.28 ± 0.02 | 9.00 ± 0.00 | I | 94.68 ± 4.14 | 88.26 ± 1.30 |
| E24A | + | 6.65 ± 0.05 | 8.67 ± 0.05 | I | 131.38 ± 14.44 | 95.84 ± 4.20 |
| Y40A | + | 7.98 ± 0.05 | 8.41 ± 0.01 | I | 91.83 ± 4.06 | 81.73 ± 5.45 |
| L49A | + | 8.12 ± 0.01 | 8.54 ± 0.07 | I | 151.00 ± 7.27 | 142.78 ± 8.76 |
| R65A | + | 6.29 ± 0.07 | 8.49 ± 0.02 | I | 138.46 ± 3.43 | 108.60 ± 17.82 |
| R98A | + | 8.30 ± 0.01 | 8.55 ± 0.06 | I | 108.01 ± 1.14 | 89.65 ± 3.67 |
| R121A | + | 6.37 ± 0.20 | 7.94 ± 0.02 | I | 144.83 ± 4.15 | 104.00 ± 2.76 |
| G132A | + | 7.62 ± 0.05 | 9.11 ± 0.02 | I | 145.46 ± 10.02 | 106.67 ± 4.86 |
| S141A | + | 8.09 ± 0.01 | 8.87 ± 0.05 | I | 133.72 ± 2.09 | 100.26 ± 3.02 |
| Q149A | + | 3.65 ± 0.01 | 7.51 ± 0.16 | I | 135.63 ± 6.60 | 102.19 ± 2.52 |
| G169A | + | 7.89 ± 0.03 | 8.43 ± 0.21 | I | 126.76 ± 2.21 | 94.92 ± 3.96 |
| K184A | + | 6.40 ± 0.08 | 7.68 ± 0.02 | I | 145.34 ± 11.35 | 96.27 ± 5.72 |
| G185A | + | 6.58 ± 0.16 | 7.46 ± 0.06 | I | 153.55 ± 6.32 | 95.09 ± 4.86 |
| G187A | + | 6.31 ± 0.23 | 8.43 ± 0.01 | I | 114.82 ± 5.33 | 99.31 ± 1.40 |
| R214A | + | 7.21 ± 0.01 | 8.01 ± 0.07 | I | 122.85 ± 4.95 | 87.02 ± 14.18 |
| R216A | + | 6.32 ± 0.06 | 6.92 ± 0.04 | I | 111.38 ± 4.64 | 85.36 ± 6.68 |
| E220A | + | 5.22 ± 0.06 | 8.57 ± 0.19 | I | 62.84 ± 3.39 | 103.46 ± 8.18 |
| K229A | + | 6.92 ± 0.01 | 8.03 ± 0.09 | I | 113.40 ± 0.68 | 97.47 ± 5.12 |
| R236A | + | 5.07 ± 0.05 | 7.61 ± 0.00 | I | 107.59 ± 2.69 | 90.13 ± 14.06 |
| V242A | + | 7.67 ± 0.13 | 8.15 ± 0.11 | I | 136.01 ± 4.81 | 101.33 ± 2.46 |
| L256A | + | 6.33 ± 0.10 | 6.97 ± 0.07 | I | 143.86 ± 3.42 | 87.44 ± 1.70 |
| P277A | + | 8.31 ± 0.01 | 8.79 ± 0.03 | I | 131.35 ± 3.02 | 98.03 ± 2.96 |
| F291A | + | 7.15 ± 0.05 | 8.41 ± 0.01 | I | 126.04 ± 3.53 | 89.44 ± 5.27 |
| G300A | + | 5.02 ± 0.06 | 7.75 ± 0.01 | I | 100.10 ± 3.52 | 86.88 ± 3.82 |
| N319A | + | 7.75 ± 0.11 | 8.36 ± 0.05 | I | 107.45 ± 2.23 | 81.28 ± 4.23 |
| Q327A | + | 6.74 ± 0.09 | 7.19 ± 0.27 | I | 125.66 ± 0.88 | 95.25 ± 7.13 |
| C333A | + | 8.06 ± 0.05 | 8.9 ± 0.01 | I | 105.19 ± 4.78 | 99.49 ± 6.63 |
| K357A | + | 7.78 ± 0.01 | 8.7 ± 0.08 | I | 105.22 ± 0.25 | 81.12 ± 1.17 |
| E369A | + | 7.27 ± 0.21 | 7.98 ± 0.02 | I | 114.80 ± 4.74 | 142.64 ± 1.72 |
| W386A | + | 5.44 ± 0.11 | 7.53 ± 0.05 | I | 88.39 ± 4.83 | 125.05 ± 8.59 |
| G460A | + | 6.68 ± 0.06 | 8.88 ± 0.04 | I | 118.59 ± 17.56 | 138.93 ± 10.31 |
| P474A | + | 6.04 ± 0.03 | 8.79 ± 0.08 | I | 115.52 ± 3.98 | 116.35 ± 3.24 |
| G490A | + | 7.63 ± 0.04 | 8.86 ± 0.13 | I | 113.78 ± 11.54 | 104.90 ± 6.71 |
| E495A | + | 7.21 ± 0.09 | 8.3 ± 0.02 | I | 73.02 ± 1.39 | 83.08 ± 7.90 |
| R26A | + | 1.54 ± 0.09 | 3.81 ± 0.03 | II | 79.44 ± 0.71 | 85.62 ± 1.65 |
| *R74A | + | 3.11 ± 0.09 | 4.42 ± 0.08 | II | 139.73 ± 9.61 | 109.25 ± 2.07 |
| *R175A | + | 2.91 ± 0.01 | 4.11 ± 0.05 | II | 170.32 ± 5.96 | 108.56 ± 6.54 |
| E192A | + | 4.16 ± 0.02 | 6.03 ± 0.01 | II | 78.03 ± 3.16 | 82.87 ± 1.66 |
| R221A | + | 4.56 ± 0.03 | 4.75 ± 0.13 | II | 111.94 ± 4.57 | 93.17 ± 4.51 |
| M331A | + | 1.00 ± 0.00 | 4.77 ± 0.1 | II | 99.45 ± 8.38 | 83.98 ± 7.56 |
| R391A | + | 2.89 ± 0.03 | 6.57 ± 0.02 | II | 70.66 ± 2.04 | 63.56 ± 0.46 |
| S407A | + | 1.00 ± 0.00 | 6.19 ± 0.03 | II | 5.40 ± 0.39 | 86.15 ± 0.28 |
| V414A | + | 1.69 ± 0.12 | 6.85 ± 0.06 | II | 47.29 ± 6.11 | 92.27 ± 1.14 |
| D468A | + | 3.54 ± 0.34 | 6.67 ± 0.10 | II | 97.46 ± 2.33 | 81.45 ± 5.87 |
| D491A | + | 5.09 ± 0.07 | 5.82 ± 0.09 | II | 52.09 ± 0.67 | 73.18 ± 4.46 |
| N59A | + | ND | ND | III | 159.52 ± 4.80 | 54.32 ± 1.72 |
| E64A | + | ND | ND | III | 193.40 ± 14.30 | 0.03 ± 0.00 |
| E320A | + | ND | ND | III | 145.78 ± 6.37 | 51.05 ± 1.47 |
| *D72A | − | ND | ND | IV | 121.17 ± 1.30 | 135.07 ± 1.05 |
| G93A | − | ND | ND | IV | 131.30 ± 10.53 | 55.13 ± 2.74 |
| *K113A | − | ND | ND | IV | 183.46 ± 12.21 | 106.16 ± 5.07 |
| Y148A | − | ND | ND | IV | 100.87 ± 3.15 | 82.89 ± 10.15 |
| R150A | − | ND | ND | IV | 12.46 ± 0.79 | 32.17 ± 1.44 |
| R152A | − | ND | ND | IV | 100.52 ± 1.78 | 77.61 ± 14.19 |
| *R156A | − | ND | ND | IV | 123.85 ± 6.03 | 95.24 ± 3.16 |
| *R174A | − | ND | ND | IV | 214.89 ± 17.15 | 100.56 ± 6.91 |
| *R195A | − | ND | ND | IV | 160.59 ± 14.23 | 105.41 ± 5.63 |
| *R199A | − | ND | ND | IV | 152.91 ± 10.97 | 102.41 ± 6.38 |
| R208A | − | ND | ND | IV | 0.19 ± 0.01 | 0.13 ± 0.01 |
| R213A | − | ND | ND | IV | 32.42 ± 0.36 | 38.29 ± 7.36 |
| E254A | − | ND | ND | IV | 0.26 ± 0.03 | 0.37 ± 0.13 |
| A260R | − | ND | ND | IV | 0.04 ± 0.00 | 0.02 ± 0.01 |
| K273A | − | ND | ND | IV | 0.19 ± 0.02 | 0.04 ± 0.01 |
| *K325A | − | ND | ND | IV | 157.38 ± 7.37 | 134.67 ± 12.73 |
| A337R | − | ND | ND | IV | 0.16 ± 0.00 | 0.03 ± 0.00 |
| E339A | − | ND | ND | IV | 0.16 ± 0.01 | 0.03 ± 0.01 |
| R355A | − | ND | ND | IV | 52.91 ± 0.47 | 45.07 ± 1.06 |
| *R361A | − | ND | ND | IV | 105.57 ± 0.49 | 104.60 ± 20.58 |
| A387R | − | ND | ND | IV | 0.15 ± 0.00 | 0.04 ± 0.00 |
| Q405A | − | ND | ND | IV | 0.16 ± 0.01 | 0.05 ± 0.01 |
| F412A | − | ND | ND | IV | 0.40 ± 0.32 | 0.03 ± 0.00 |
| R416A | − | ND | ND | IV | 0.21 ± 0.10 | 0.03 ± 0.00 |
| F488A | − | ND | ND | IV | 0.20 ± 0.01 | 8.05 ± 1.02 |
| F489A | − | ND | ND | IV | 0.16 ± 0.01 | 0.23 ± 0.22 |
We selected 74 amino acids that are conserved among influenza A, B, and C virus NPs for mutagenesis. NP mutants flagged with an asterisk were further tested for intracellular localization, and incorporation efficiency of a vRNA segment(s) into VLPs, in addition to replicative ability and polymerase activity.
Viruses were generated using an established plasmid-based reverse-genetics system. +, NP mutant virus recovery was verified by plaque assay, cytopathic effect, and NP gene sequencing; −, no replicating virus was recovered.
MDCK cells were infected with wild-type WSN or NP mutant virus at an MOI of 0.0001. At 48 h postinfection, the culture supernatants were harvested and subjected to plaque assays in MDCK cells. ND, plaques were not detected.
Based on virus rescue and viral-replication properties, NP mutants were categorized into four groups. Group I, at either 33°C or 37°C, viruses were attenuated by less than 2 log units; group II, mutant viruses were attenuated by more than 2 log units at both temperatures tested; group III, NP mutant viruses were rescued but did not form plaques at 48 h postinfection; group IV, NP mutant virus were not rescued.
The abilities of mutant NPs to support vRNA replication/transcription in an in vitro assay were examined in 293T cells as described in Materials and Methods. Since luciferase levels reflect the overall transcription and replication activity of the polymerase complex, we defined the polymerase activity levels as high, normal, and low for mutants that exhibited >120%, 80% to 120%, and <80%, respectively, of the wild-type activity.
pPolI-WSN-NA-firefly-luciferase produces a virus-like RNA in which the coding region for firefly luciferase is flanked by the packaging signals of the NA segment (12). This plasmid was generated by inserting the firefly luciferase open reading frame between the BamHI and XhoI sites of pPolI-WSN-NA-MCS, which was constructed by modifying the start codon of pPolINA(183)GFP(157) (12) (in essence, we replaced ATATG at nucleotide positions 113 to 117 and 161 to 165 with GCGCG) and by replacing the sequence from the initiation of the enhanced green fluorescent protein (GFP) gene to the StuI site on pPolI-WSN-NA(183)GFP(157). pPolI-HA-GFP, for the generation of a virus-like RNA encoding GFP, was generated as described previously (36).
pCAGGS-Renilla, for the expression of Renilla luciferase, was generated by inserting the open reading frame of the Renilla luciferase gene into the BsmBI sites of pCAGGs/MCS.
The pPolI-NP(Met−) plasmid, used to generate vRNA that did not encode NP due to the lack of a start codon, was generated by changing the 1st through the 7th ATG codons to TAG and by changing the 14th and 15th ATG codons to TAG and TGA, respectively, using in vitro site-directed mutagenesis (Stratagene, La Jolla, CA).
The plasmid pMX-WSNNP-IRES-puro was constructed as follows. First, the puromycin resistance gene was inserted into XbaI and SalI sites of the plasmid pIRES (Clontech, Mountain View, CA), and then a fragment containing the IRES sequence and the puromycin resistance gene was inserted into EcoRI and SalI sites of the pMX retroviral vector (30). This plasmid was designated pMX-IRES-puro. Finally, the WSN NP gene was cloned into EcoRI and MluI sites of pMX-IRES-puro.
Plasmid-driven reverse genetics.
All viruses used in this study were generated by reverse genetics, using plasmids expressing the eight vRNA segments, the three polymerase proteins, and NP, as described by Neumann et al. (24). At 48 h posttransfection, viruses were harvested and used to inoculate MDCK cells for the production of stock viruses. The NP genes of transfectant viruses were sequenced to confirm the origins of the genes and the presence of the intended mutations and to ensure that no unwanted mutations were present.
Virus-like particles (VLPs) were generated from 293T cells transfected with eight PolI constructs producing eight vRNA segments [i.e., PB1, PB2, PA, NA, M, NS, HA-GFP, and NP(Met−), which lacks a start codon] and five protein expression constructs producing PB1, PB2, PA, hemagglutinin (HA), and mutant or wild-type NP (see Fig. 3). These plasmids were mixed with transfection reagent (2 μl of Trans IT LT-1 [Mirus, Madison, WI] per μg of DNA), incubated at room temperature for 20 min, and added to 106 293T cells. Six hours later, the DNA transfection reagent mixture was replaced by 1 ml Opti-MEM (Invitrogen, Grand Island, NY). Forty-eight hours after transfection, the supernatant was harvested, treated with trypsin, and used to determine the number of VLPs containing the test genes, as described below.
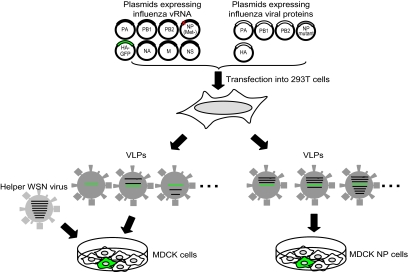
FIG. 3.
System to test vRNP virion incorporation efficiencies. VLPs were recovered from 293T cells transfected with eight PolI constructs for the eight RNA segments PB1, PB2, PA, NA, M, NS, HA-GFP, and NP(Met−), which lacks a start codon. These VLPs were used to infect MDCK cells in the presence of helper virus (left). In this experimental setting, GFP-positive MDCK cells are representative of VLPs that contain at least one vRNA segment (i.e., the HA-GFP vRNA). In a parallel experiment, VLPs derived from 293T cells were used to infect MDCK cells expressing NP (right). In this experimental setting, GFP-positive MDCK cells are indicative of VLPs that possess at least four vRNAs (i.e., the PB2, PB1, PA, and HA-GFP vRNAs).
Replicative properties of viruses.
MDCK cells were infected with wild-type WSN or NP mutant virus at a multiplicity of infection (MOI) of 0.0001 at 33°C and 37°C, respectively. At 48 h postinfection, the culture supernatants were collected and subjected to plaque assays in MDCK cells for virus titration.
Luciferase assay.
293T cells were transfected with plasmids for the expression of the viral proteins PA, PB1, PB2, and NP (wild-type or mutated NP) and pPolI-WSN-NA-firefly-luciferase. Plasmid pCAGGS-Renilla was used as an internal control for the dual-luciferase assay. As a negative control, 293T cells were transfected with the same plasmids, with the exception of the NP expression plasmid. After transfection, the cells were incubated at 33°C or 37°C for 48 h, and then luciferase activity was measured with a dual-luciferase reporter system (Promega, Madison, WI) on a Glomax microplate luminometer (Promega, Madison, WI) according to the manufacturer's instructions.
Immunostaining assay.
MDCK cells were transfected with pCAGGs/MCS plasmids expressing wild-type or mutant NP. At 9 h and 24 h posttransfection, the cells were fixed and indirect immunofluorescence assays were performed. Briefly, the cells were washed twice with phosphate-buffered saline (PBS), fixed with 3.7% paraformaldehyde (in PBS) for 20 min at room temperature, washed again, and permeabilized with 0.5% Triton X-100 for 10 min. After being blocked with 10% bovine serum albumin for 20 min at room temperature, the cells were incubated with an anti-NP monoclonal antibody (347/3) for 30 min. The cells were then incubated with fluorescein isothiocyanate-labeled goat anti-mouse antibody immunoglobulin G (1:200 dilution; Roche) for 30 min, washed, and mounted with 10 mM p-phenylenediamine in glycerol-PBS (9:1), pH 8.5. To avoid the detection of newly synthesized NP, protein synthesis was inhibited by adding cycloheximide (50 mg/ml; Sigma) to the growth medium at 9 h posttransfection. Twenty-four hours later, indirect immunofluorescence assays were performed. Samples were observed under a fluorescence microscope or a confocal laser microscope (LSM510META; Carl Zeiss, Jena, Germany).
Determination of the number of VLPs.
To determine the number of VLPs containing at least one vRNA (i.e., HA-GFP vRNA), culture supernatants of 293T cells transfected with plasmids for VLP production (as described above) were used to infect MDCK cells. To provide polymerase and NP that supported HA-GFP vRNA replication and transcription, cells were coinfected with WSN virus at an MOI of 0.1 (see Fig. 3). Sixteen hours postinfection, the infected cells were fixed, and the cells expressing GFP were counted under a fluorescence microscope.
To determine the number of VLPs containing a set of four vRNAs (i.e., PB1, PB2, PA, and HA-GFP vRNAs), the culture supernatants of transfected cells were used to infect MDCK-NP cells that stably expressed WSN NP (see Fig. 3). GFP expression from the HA-GFP vRNA would occur only if VLPs contained the HA-GFP vRNA together with the PB2, PB1, and PA vRNAs. GFP-positive cells, therefore, represented VLPs that possessed at least the PB2, PB1, PA, and HA-GFP vRNAs. As described above, the number of GFP-expressing cells was determined at 16 h postinfection.
RESULTS
Generation of NP mutant viruses.
For mutagenesis, we selected amino acids that are highly conserved among influenza A, B, and C viruses, as well as amino acids that may be critical for RNA binding, as suggested by the recently published crystal structure of influenza A virus NP (41). In total, 74 amino acid residues were selected for testing (Fig. 1B). Using plasmid-driven reverse genetics (24), we were able to generate 48 mutant viruses (Table 1). The remaining 26 mutant viruses were not viable (Table 1), suggesting that the respective mutations in NP are critical for the viral life cycle.
Replicative properties of NP mutant viruses.
For all viable mutant viruses, we next examined their replicative abilities in cell culture. Briefly, MDCK cells were infected with wild-type WSN or NP mutant viruses at an MOI of 0.0001 and incubated at 33°C and 37°C. Forty-eight hours later, the culture supernatants were harvested and subjected to plaque assays in MDCK cells for virus titration. Based on their abilities to support viral replication, the NP mutants could be divided into four groups. As shown in Table 1, groups I, II, and III include all 48 recovered NP mutant viruses, while group IV contains the 26 nonviable mutant viruses.
Group I contains 34 mutant viruses whose titers were less than 2 log units lower than that of wild-type WSN virus at either 33°C or 37°C. Seventeen viruses (possessing NP mutations R19A, Y40A, L49A, R98A, G132A, S141A, G169A, R214A, V242A, P277A, F291A, N319A, C333A, K357A, E369A, G490A, and E495A) were attenuated by less than 1 log unit at both 33°C and 37°C. Mutant viruses Q149A, K184A, G185A, R216A, R236A, L256A, G300A, Q327A, and W386A were clearly attenuated at both 33°C and 37°C, with virus titers reduced by more than 1 log unit compared to wild-type WSN virus, whereas E24A, R65A, R121A, G187A, E220A, K229A, G460A, and P474A were attenuated only at 33°C. Of these viruses, mutants Q149A, E220A, R236A, G300A, and W386A were attenuated by more than 2.5 log units at 33°C. The NP sequences of these mutant viruses were reconfirmed, and one unexpected mutation was found at position 140 (His to Arg) in the NP protein of mutant R65A (the significance of this substitution is currently unknown).
The NP mutant viruses in group II were significantly attenuated, i.e., their titers were more than 2 log units lower than that of wild-type WSN virus at both 33°C and 37°C. Eleven NP mutant viruses (R26A, R74A, R175A, E192A, R221A, M331A, R391A, S407A, V414A, D468A, and D491A) comprised this group. The titers of four mutants (R26A, R74A, R175A, and M331A) were more than 4 log units lower than that of wild-type WSN virus at both 33°C and 37°C. Four other viruses (R391A, S407A, V414 A, and D468A) were attenuated by more than 4 log units at 33°C but only 2 to 3 log units at 37°C.
Group III contains three mutant viruses, N59A, E64A, and E320A, which were unable to form plaques, although cytopathic effects were observed in MDCK cells infected with these mutants. The 50% tissue culture infective doses of N59A, E64A, and E320A were 5.5, 3.5, and 6.5 log10 50% tissue culture infective doses/ml at 33°C, respectively.
Group IV includes the 26 nonviable mutants (D72A, G93A, K113A, Y148A, R150A, R152A, R156A, R174A, R195A, R199A, R208A, R213A, E254A, A260R, K273R, K325A, A337R, E339R, R355A, R361A, R387A, Q405A, F412A, R416A, F488A, and F489A), attesting to a critical role of these amino acids in the viral life cycle.
Polymerase activities of replication complexes containing NP mutants.
As stated above and shown in Table 1, we identified NP amino acid substitutions that caused viral growth defects. To assess the effects of these substitutions on vRNA transcription activity, we tested the activities of NP mutants by using a minireplicon system. pPolI-WSN-NA-firefly-luciferase synthesizes a modified influenza virus NA vRNA in which the coding region for firefly luciferase replaces the native NA coding sequences. The luciferase levels thus reflect the overall transcription and replication activities of the polymerase complex upon cotransfection of cells with pPolI-WSN-NA-firefly-luciferase and plasmids for the expression of the viral PA, PB1, PB2, and NP (wild-type or mutant NP), which support both replication and transcription from a vRNA template (32). At 48 h posttransfection, luciferase activity was measured. We found that NP mutants in group I, whose mutations had no or moderate effects on virus replicative ability, had at least 80% and 60% of the luciferase activity of wild-type NP at 37°C and 33°C, respectively (Table 1). NP mutant E220A showed reduced activity to support polymerase function at 33°C but not at 37°C. Other mutants in group I (i.e., Q149A, R236A, and G300A) showed no defect in supporting polymerase function compared to wild-type NP at 33°C, even though the respective virus titers were more than 2 log units lower than that of wild-type WSN virus at 33°C. Hence, these NP mutations do not affect genome replication and transcription, but other steps in the viral life cycle.
In group II, six NP mutants, R26A, E192A, R391A, S407A, V414A, and D491A, showed lower activity to support polymerase function at 33°C than wild-type NP, suggesting that these mutations may cause virus attenuation at low temperatures. Interestingly, R74A and R175A showed normal activity to support polymerase function at 37°C, and even higher activity at 33°C, compared to wild-type NP, whereas the respective viruses were attenuated by more than 4 log units. These results suggest that these mutations may inhibit steps other than genome replication and transcription, such as transport, assembly, and/or virion incorporation.
Mutants in group III showed some temperature sensitivity, expressing more than 145% of wild-type luciferase activity at 33°C but less than 55% at 37°C. Specifically, mutant E64A lost the ability to support reporter gene transcription at 37°C but expressed 193% of wild-type luciferase activity at 33°C. Notably, these mutant viruses could not form plaques at either 33°C or 37°C, even though they expressed higher luciferase activity than wild-type WSN virus at 33°C.
In group IV, many of the variants (i.e., R150A, R208A, R213A, E254A, A260R, K273A, A337R, E339A, R355A, A387R, Q405A, F412A, R416A, F488A, and F489A) showed no or very low luciferase activity at both 33°C and 37°C. Hence, these mutations likely lost the ability to support viral polymerase activity, resulting in the growth defect of the mutant viruses (Table 1). The remaining mutants expressed reasonable levels of luciferase activity at either 33°C or 37°C. Of these mutants, D72A, K113A, R156A, R174A, R195A, R199A, K325A, and R361A showed normal or high polymerase activity at both 33°C and 37°C despite the observed failure of virus rescue (Table 1). These data suggest a role of the respective amino acids in processes such as intracellular transport, assembly, and/or packaging.
Localization of mutant NPs.
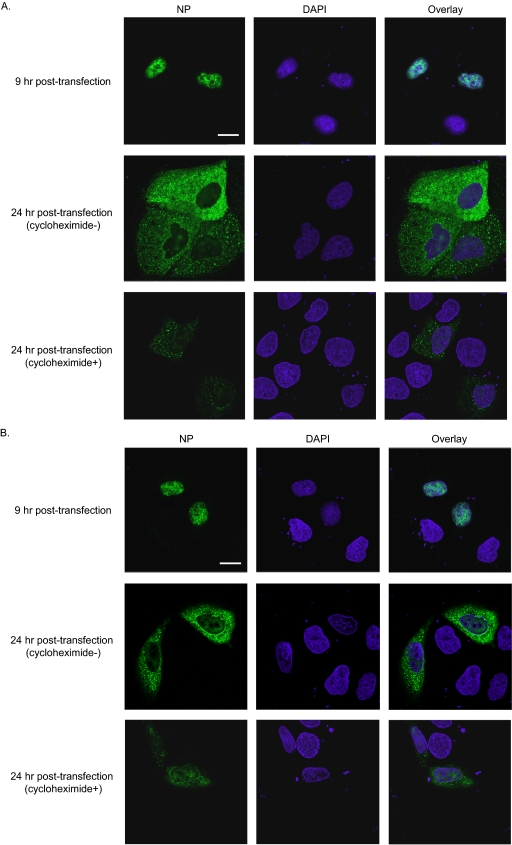
We found that 10 NP variants (D72A, R74A, K113A, R156A, R174A, R175A, R195A, R199A, K325A, and R361A) caused viral growth defects with no apparent effect on genome replication and transcription. For these variants, therefore, we assessed intracellular localization. MDCK cells were transfected with plasmids expressing mutant NPs, and indirect immunofluorescence assays were performed 9 h and 24 h after transfection. As shown in Fig. 2A and B and Fig. S1 in the supplemental material, 9 hours after transfection, wild-type NP and all NP mutants tested localized to the nuclei of transfected MDCK cells. Twenty-four hours after transfection, wild-type NP and all variants tested localized both to the nucleus and to the cytoplasm regardless of the addition of a translation inhibitor, cycloheximide. Since the localization of these mutant NPs was indistinguishable from that of the wild-type NP, their attenuated growth properties cannot be explained by defects in nuclear export.
FIG. 2.
Localization of NP mutants. MDCK cells transfected with plasmids expressing mutant NP or wild-type NP were subjected to indirect immunofluorescence assays. NP and DNA were stained with an anti-NP antibody and DAPI (4′,6′-diamidino-2-phenylindole), respectively. Samples were observed under a confocal laser microscope. Nine hours after transfection, wild-type and mutant NP exclusively localized to the nucleus. Shown are wild-type NP (A) and mutant R175A as a representative example of the mutants tested (B). Twenty-four hours after transfection, wild-type NP and all mutant NPs tested localized to both the nucleus and the cytoplasm, regardless of the addition of the translational inhibitor cycloheximide at 9 h posttransfection. The localization of the mutant NP proteins was indistinguishable from that of wild-type NP. Scale bars, 20 μm.
Incorporation efficiency of a single vRNA segment into VLPs containing mutant NPs.
For some NP mutants, the growth defects of the corresponding viruses cannot be explained by deficiencies in replication or intracellular transport. Therefore, we next examined the effects of the respective mutations on virus assembly. To determine the incorporation efficiency of vRNA segments into VLPs, we transfected 293T cells with eight PolI plasmids for the PB1, PB2, PA, NA, M, NS, HA-GFP, and NP(Met−) vRNA segments. The NP(Met−) vRNA produced an NP gene whose start codon was replaced with a stop codon to avoid expression of wild-type NP; the HA-GFP vRNA encodes the GFP reporter flanked by the packaging signals of the HA segment (36). Cells were cotransfected with five protein expression plasmids for PB1, PB2, PA, HA, and mutant or wild-type NP. The resultant VLPs were replication incompetent because they possessed mutant HA and NP vRNA segments instead of wild-type HA and NP vRNAs (Fig. 3). The supernatants of transfected cells were harvested at 48 h posttransfection and used to infect MDCK cells, together with a helper virus to provide a functional polymerase complex that supports the replication/transcription of HA-GFP vRNA. The number of VLPs containing the HA-GFP vRNA segment was determined by counting the cells expressing GFP at 16 h postinfection. As shown in Table 2, the number of VLPs possessing HA-GFP vRNA produced from cells expressing R74A, R156A, R175A, R195A, R199A, or R361A was at least as high as that from cells expressing wild-type NP. These results suggest that the mutations had no or little effect on the incorporation efficiency of a single vRNA segment into VLPs. In contrast, cells expressing D72A, K113A, R174A, or K325A produced VLPs containing HA-GFP vRNA less efficiently than did the wild-type NP, indicating that these NP mutations may impair vRNA incorporation into VLPs, resulting in the observed failure of virus rescue.
TABLE 2.
Virion incorporation efficiencies of vRNA segmentsa
| NP | No. of VLPs possessing at least one vRNA (per ml)b | No. of VLPs possessing at least four vRNAs (per ml)c |
|---|---|---|
| Wild type | 31,900 | 17,800 |
| D72A | 450 | 0 |
| R74A | 62,500 | 390 |
| K113A | 2,160 | 0 |
| R156A | 167,000 | 11 |
| R174A | 8,980 | 0 |
| R175A | 100,000 | 0 |
| R195A | 26,200 | 0 |
| R199A | 51,000 | 0 |
| K325A | 5,700 | 0 |
| R361A | 21,080 | 0 |
293T cells were transfected with eight PolI constructs for eight RNA segments [i.e., PB1, PB2, PA, NA, M, NS, HA-GFP, and NP(Met−), which lacks a start codon] and five protein expression constructs for PB1, PB2, PA, HA, and mutant or wild-type NP. Forty-eight hours later, VLPs were harvested, treated with TPCK trypsin, and used to determine the virion incorporation efficiencies of viral RNA segments as described in notes b and c. The results shown are representative data from three independent experiments.
VLPs and WSN helper virus were used to infect MDCK cells. With helper virus providing polymerase proteins and wild-type NP, GFP-positive MDCK cells represented VLPs that possessed at least one vRNA, i.e., HA-GFP vRNA (Fig. 3).
Infectious VLPs were used to infect MDCK-NP cells. In this approach, GFP expression from HA-GFP vRNA requires the expression of the PB2, PB1, and PA proteins from the respective vRNA segments. GFP-positive MDCK cells therefore represented VLPs that contained at least four vRNAs (i.e., the HA-GFP, PB1, PB2, and PA vRNAs).
Incorporation efficiency of multiple vRNA segments into VLPs containing mutant NPs.
As described above, we found that R74A, R156A, R175A, R195A, R199A, and R361A support VLP incorporation of a single vRNA segment, leaving in question why these viruses replicate poorly. A possible explanation is that the NP mutant facilitates virion incorporation of single, but not multiple, vRNA segment. In such a scenario, the NP would assemble vRNA segments into sets of eight (in the form of RNPs) that constitute a functional genome. To examine this possibility, we generated an MDCK cell line stably expressing wild-type NP and used the cell line for VLP infection. Since a helper virus was not used for this experiment, PA, PB1, and PB2 vRNA segments had to be provided by incoming VLPs to support the replication/transcription of HA-GFP vRNA. The number of GFP-expressing cells should, therefore, correspond to the number of VLPs possessing at least four vRNAs (i.e., PA, PB1, PB2, and HA-GFP vRNAs). As shown in Table 2, we found that the number of VLPs possessing at least four vRNAs produced from cells expressing R74A, R156A, R175A, R195A, R199A, or R361A was significantly lower than that from cells expressing wild-type NP. These results suggest that these mutations affect the efficient incorporation of multiple vRNA segments into VLPs, even though they had no or little effect on the virion incorporation of a single vRNA. As expected, the mutations D72A, K113A, R174A, and K325A completely inhibited VLP incorporation of four vRNAs.
DISCUSSION
In this study, we conducted a comprehensive mutational analysis to examine the roles of highly conserved amino acids in the influenza A virus NP for the virus life cycle. We found 17 amino acid substitutions (i.e., R19A, Y40A, L49A, R98A, G132A, S141A, G169A, R214A, V242A, P277A, F291A, N319A, C333A, K357A, E369A, G490A, and E495E) that had no or little effect on virus growth in vitro at either 33°C or 37°C, whereas other mutations tested significantly impaired viral replication efficiency. Using an in vitro replication assay, we identified 15 amino acid changes (i.e., R150A, R208A, R213A, E254A, A260R, K273A, A337R, E339A, R355A, A387R, Q405A, F412A, R416A, F488A, and F489A) that are crucial for viral-genome replication and/or transcription (Table 1). Several NP mutants supported efficient replication in an in vitro replication assay but were highly attenuated in their growth properties. Some of these mutants facilitated efficient virion incorporation of a single vRNA but not that of multiple vRNAs. This is the first report that suggests a role of an influenza virus protein in the assembly and/or virion incorporation of vRNA segments.
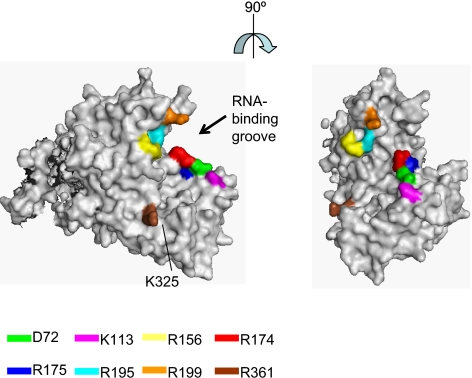
The genome of influenza A virus consists of eight RNA segments. Two packaging models have been proposed for the generation of infectious virions containing these eight vRNA segments: “random-packaging” and “selective-packaging” models (9, 10, 37). Recently, it has been suggested that each vRNA segment of influenza A viruses contains specific incorporation signals for the recruitment and/or packaging of vRNPs into virions (6, 7, 11-13, 20, 21, 28, 36). These data support the “selective-packaging” mechanism of vRNA recruitment into virions. However, the involvement of other factors (i.e., viral proteins and/or host proteins) in the selective incorporation of the eight vRNPs into progeny virions is still poorly understood. In this study, we found 10 mutations (D72A, R74A, K113A, R156A, R174A, R175A, R195A, R199A, K325A, and R361A) that impaired the efficient incorporation of a set of at least four vRNAs (PA, PB1, PB2, and HA-GFP vRNP) into VLPs, although they did not affect the VLP incorporation of a single vRNA, i.e., the HA-GFP vRNA (Table 2). As shown in Fig. 4, eight of these NP mutations (i.e., D72A, K113A, R156A, R174A, R175A, R195A, R199A, and R361A) cluster around a possible RNA-binding groove that lies between the head and body domains at the exterior of the NP oligomer and is lined with highly conserved basic residues (41). Two of these residues (R74 and K325) are located in an internal domain of NP. The architecture of RNP complexes in influenza A virus particles is such that the RNPs are organized in a distinct pattern of seven segments of different lengths surrounding a central segment. Close contact has been shown between the peripheral RNPs, as well as the central and peripheral RNPs (26). NP may be involved in this close contact among the peripheral RNPs, through the interaction of RNPs and/or other essential host cellular components, leading to the efficient incorporation of the eight vRNA segments into virus particles.
FIG. 4.
Crystal structure of influenza A virus NP. The images were created with the program PYMOL (W. L. Delane; http://www.pymol.org), and the NP structure was obtained from the Protein Data Bank (41) (accession number 2IQH). The locations of the amino acid residues that affect the efficient incorporation of multiple vRNA segments into VLPs are shown in color.
Influenza A virus NP has at least two NLS sequences. An unconventional NLS is located between residues 3 and 13 and is responsible for NP binding to karyopherins α1 and α2 (35). The second NLS is a bipartite signal located in the middle of NP (residues 198 to 216) (37). The bipartite NLS can function as an NLS to a limited extent when it is fused to a cytoplasmic reporter protein (37). Its contribution to NP nuclear import, however, is not as significant as that of NLS1 (37). In this study, we attempted to generate five viruses that possessed amino acid substitutions in the bipartite NLS (R199A, R208A, R213A, R214A, and R216A). The R208A and R213A mutant viruses were not recoverable because these mutations significantly reduced viral transcription, consistent with a previous report that the bipartite NLS was essential for vRNA transcription (31). In contrast, the single amino acid substitution R214A or R216A had no effect on viral-genome replication/transcription, NP localization, or virus replication. These results suggest that a single mutation at position 214 or 216 of NP may not be sufficient to affect NP function, although multiple amino acid changes in the bipartite NLS (positions 213, 214, and 216) completely disrupt vRNA transcription, NP nuclear accumulation, and virus replication (31). We also found that a mutation at position 199 did not affect viral-genome replication/transcription and NP nuclear transport.
In this study, we found that mutations A337R, E339A, Q405A, S407A, F412A, V414A, R416A, F488A, F489A, D491A, and E495A significantly decreased the transcription of influenza vRNA (Table 1). The crystal structure of influenza NP shows that each NP contains a tail loop, formed by residues 402 to 428 (41). This tail loop is inserted into the body domain of a neighboring molecule in a counterclockwise direction when viewed along the threefold axis from the side of the head domain. Mutagenesis revealed that the tail loop is essential for NP oligomerization, which in turn is necessary for vRNA transcription (8, 42). Since the mutations we tested (A337R, E339A, Q405A, S407A, F412A, V414A, R416A, F488A, F489A, D491A, and E495A) reside in or near the NP-NP-interacting region (8, 41), they may disrupt the NP-NP interaction, leading to inhibition of viral-genome replication and transcription.
Using coimmunoprecipitation, Biswas et al. showed that coexpression of the components of the polymerase protein complex (PB1, PB2, or PA) with NP either together or pairwise revealed that NP interacts with PB1 and PB2 but not with PA (3). These experiments implicated three NP regions—amino acids 1 to 160, 256 to 340, and 340 to 498—in binding to the PB2 subunit of the viral polymerase (3, 33). In our study, mutant viruses R150A, E254A, A260R, K273A, A337A, E339A, R355A, A387R, Q405A, F412A, R416A, F488A, and F489A were not viable, and their abilities to support vRNA transcription were decreased significantly. Our results suggest that these NP mutations (i.e., R150A, E254A, A260R, K273A, A337A, E339A, R355A, A387R, Q405A, F412A, R416A, F488A, and F489A, which lie in or near the PB2 binding domain) block the interaction of NP with the polymerase subunits (PB1 and PB2), leading to defective viral growth.
In summary, we have identified amino acids that alter the functionality of NP in viral-genome replication/transcription. We have also identified amino acids in NP that facilitate efficient incorporation of multiple vRNA segments into progeny virions, suggesting a role of these amino acid residues in the assembly of influenza A virus segments in the form of RNPs. Further analyses of the significance of NP should provide information essential to our understanding of influenza virus replication.
Supplementary Material
Acknowledgments
We thank Susan Watson for editing the manuscript and Krisna Wells and Martha McGregor for excellent technical assistance. We also thank Bill Sudgen for kindly providing plasmids.
This work was supported in part by U.S. National Institute of Allergy and Infectious Diseases Public Health Service research grants, by a grant-in-aid for Specially Promoted Research, and by a contract research fund for the Program of Funding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology; by ERATO (Japan Science and Technology Agency); and by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Published ahead of print on 18 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Albo, C., A. Valencia, and A. Portela. 1995. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J. Virol. 693799-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avalos, R. T., Z. Yu, and D. P. Nayak. 1997. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J. Virol. 712947-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 725493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng, T., O. G. Engelhardt, B. Thomas, A. V. Akoulitchev, G. G. Brownlee, and E. Fodor. 2006. Role of Ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 8011911-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Digard, P., D. Elton, K. Bishop, E. Medcalf, A. Weeds, and B. Pope. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 732222-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dos Santos Afonso, E., N. Escriou, I. Leclercq, S. van der Werf, and N. Naffakh. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 34134-46. [DOI] [PubMed] [Google Scholar]
- 7.Duhaut, S. D., and J. W. McCauley. 1996. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216326-337. [DOI] [PubMed] [Google Scholar]
- 8.Elton, D., L. Medcalf, K. Bishop, D. Harrison, and P. Digard. 1999. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 737357-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elton, D., M. Simpson-Holley, K. Archer, L. Medcalf, R. Hallam, J. McCauley, and P. Digard. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enami, M., G. Sharma, C. Benham, and P. Palese. 1991. An influenza virus containing nine different RNA segments. Virology 185291-298. [DOI] [PubMed] [Google Scholar]
- 11.Fujii, K., Y. Fujii, T. Noda, Y. Muramoto, T. Watanabe, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 793766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 1002002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gog, J. R., S. Afonso Edos, R. M. Dalton, I. Leclercq, L. Tiley, D. Elton, J. C. von Kirchbach, N. Naffakh, N. Escriou, and P. Digard. 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 351897-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda, A., K. Ueda, K. Nagata, and A. Ishihama. 1988. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J. Biochem. 1041021-1026. [DOI] [PubMed] [Google Scholar]
- 15.Huang, T. S., P. Palese, and M. Krystal. 1990. Determination of influenza virus proteins required for genome replication. J. Virol. 645669-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz, J. M., M. Wang, and R. G. Webster. 1990. Direct sequencing of the HA gene of influenza (H3N2) virus in original clinical samples reveals sequence identity with mammalian cell-grown virus. J. Virol. 641808-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 236901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 716706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, M., T. Toyoda, D. M. Adyshev, Y. Azuma, and A. Ishihama. 1994. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J. Virol. 688433-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, Y., Y. Hong, and T. G. Parslow. 2005. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 7910348-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, G. A., R. Hatami, and P. Palese. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 819727-9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67117-130. [DOI] [PubMed] [Google Scholar]
- 23.Mena, I., E. Jambrina, C. Albo, B. Perales, J. Ortin, M. Arrese, D. Vallejo, and A. Portela. 1999. Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J. Virol. 731186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 26.Noda, T., H. Sagara, A. Yen, A. Takada, H. Kida, R. H. Cheng, and Y. Kawaoka. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439490-492. [DOI] [PubMed] [Google Scholar]
- 27.Noton, S. L., E. Medcalf, D. Fisher, A. E. Mullin, D. Elton, and P. Digard. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 882280-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odagiri, T., and M. Tashiro. 1997. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J. Virol. 712138-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onishi, M., S. Kinoshita, Y. Morikawa, A. Shibuya, J. Phillips, L. L. Lanier, D. M. Gorman, G. P. Nolan, A. Miyajima, and T. Kitamura. 1996. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 24324-329. [PubMed] [Google Scholar]
- 31.Ozawa, M., K. Fujii, Y. Muramoto, S. Yamada, S. Yamayoshi, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 8130-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parvin, J. D., P. Palese, A. Honda, A. Ishihama, and M. Krystal. 1989. Promoter analysis of influenza virus RNA polymerase. J. Virol. 635142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portela, A., and P. Digard. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83723-734. [DOI] [PubMed] [Google Scholar]
- 34.Stegmann, T., J. M. White, and A. Helenius. 1990. Intermediates in influenza induced membrane fusion. EMBO J. 94231-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 711850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe, T., S. Watanabe, T. Noda, Y. Fujii, and Y. Kawaoka. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 7710575-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber, F., G. Kochs, S. Gruber, and O. Haller. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 2509-18. [DOI] [PubMed] [Google Scholar]
- 38.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol. 702743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, P. F., G. Neumann, and Y. Kawaoka. 2007. Orthomyxoviruses, p. 1691-1740. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 40.Yasuda, J., S. Nakada, A. Kato, T. Toyoda, and A. Ishihama. 1993. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology 196249-255. [DOI] [PubMed] [Google Scholar]
- 41.Ye, Q., R. M. Krug, and Y. J. Tao. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 4441078-1082. [DOI] [PubMed] [Google Scholar]
- 42.Ye, Z., T. Liu, D. P. Offringa, J. McInnis, and R. A. Levandowski. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 737467-7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.