Abstract
The selection of escape mutations has a major impact on immune control of infections with viruses such as human immunodeficiency virus (HIV). Viral evasion of CD8+ T-cell responses leaves predictable combinations of escape mutations, termed HLA “footprints.” The most clearly defined footprints are those associated with HLA alleles that are linked with successful control of HIV, such as HLA-B*57. Here we investigated the extent to which HLA footprint sites in HIV type 1 (HIV-1) are associated with viral evolution among and within clades. First, we examined the extent to which amino acid differences between HIV-1 clades share identity with sites of HLA-mediated selection pressure and observed a strong association, in particular with respect to sites of HLA-B selection (P < 10−6). Similarly, the sites of amino acid variability within a clade were found to overlap with sites of HLA-selected mutation. Second, we studied the impact of HLA selection on interclade phylogeny. Removing the sites of amino acid variability did not significantly affect clade-specific clustering, reflecting the central role of founder effects in establishing distinct clades. However, HLA footprints may underpin founder strains, and we show that amino acid substitutions between clades alter phylogeny, underlining a potentially substantial role for HLA in driving ongoing viral evolution. Finally, we investigated the impact of HLA selection on within-clade phylogeny and demonstrate that even a single HLA allele footprint can result in significant phylogenetic clustering of sequences. In conclusion, these data highlight the fact that HLA can be a strong selection force for both intra- and interclade HIV evolution at a population level.
Virus-specific CD8+ T cells play a central role in controlling infections with viruses such as human immunodeficiency virus (HIV) (15, 16). Cell surface presentation of viral peptides by HLA class I molecules allows recognition and killing of virus-infected cells by CD8+ T cells. This generates a strong selection pressure for viral escape mutations that reduce peptide recognition by CD8+ T cells and results in characteristic associations between particular escape mutations and the restricting HLA allele (5, 29, 30, 32, 38). These escape mutations can undermine the immune response (3, 14) but may also be detrimental to the virus by reducing infective or replicative capacity (29, 38). Indeed, HLA-mediated immune control of HIV may hinge critically upon the ability of particular CD8+ T cells to drive the selection of escape mutants that impose a significant fitness cost on the virus (2, 13, 27, 30).
Previous work has shown that HLA alleles, in particular those alleles associated with effective immune control of HIV, such as HLA-B*57 and HLA-B*27 (20, 21, 35), select a characteristic, predictable combination of escape mutations in the virus (8, 11, 25, 30, 38)—termed a “footprint” (31). The starting point for the present analysis is our observation, from previous studies of B-clade- and C-clade-infected cohorts (25, 29), that escape mutations selected in B-clade-infected individuals expressing HLA-B*57 frequently represented the consensus in C-clade sequences and that the consensus sequence in the AE clade bears many of the signature mutations of the HLA-B*57 footprint.
The aim of the present studies was to investigate the extent to which HLA footprints impact on HIV phylogeny, first by considering their relationship to sites of difference between clades and second by quantifying their influence on the clustering of sequences within a clade. We initially tested the hypothesis that amino acid differences between clades are commonly also those that represent escape variants associated with particular HLA alleles. To investigate this, we compared sites of interclade amino acid variability with sites at which HLA selection pressure had been previously identified from analysis of a C-clade-infected cohort of >700 study subjects in Durban, South Africa (30). The role of these variable sites in determining clade phylogeny was investigated by comparing the phylogenetic clustering of sequences in the presence and absence of the variable sites and also by swapping characteristic amino acid polymorphisms between clades.
Having established a potential role for HLA in driving amino acid sequence diversity between different clades, we next evaluated the extent to which HLA alleles might have an impact on viral evolution within a clade. We tested the hypothesis that characteristic combinations of CD8+ T-cell escape mutations might cause unrelated HIV nucleotide sequences of the same clade to cluster phylogenetically.
MATERIALS AND METHODS
Software and statistics.
We aligned nucleotide sequences, using Se-Al (tree.bio.ed.ac.uk/software/seal). We used PAUP* v.4.0 to construct neighbor-joining (NJ) phylogenetic trees from nucleotide sequences by using the HKY85 substitution model (17) and the parsimony algorithm implemented in MacClade v.4.0 (28) to quantify phylogenetic clustering. We used GARLI software (Genetic Algorithm for Rapid Likelihood Inference, available at www.nescent.org) to build maximum-likelihood (ML) trees from nucleotides and read bootstrap values from 100 bootstrap replicates in Bootscore (http://sourceforge.net/projects/bootscore). We used the Entropy One tool available at Los Alamos HIV databases (www.hiv.lanl.gov/content/sequence/ENTROPY) to calculate Shannon entropy. Statistical tests were undertaken with GraphPad Prism v.5.0.
Impact of HLA selection on amino acid polymorphisms between clades.
In order to investigate whether HLA footprints might account for some of the amino acid differences occurring between HIV type 1 (HIV-1) clades, we used clade A1, A2, AE, B, and C Gag, Pol, and Nef sequences published by the Los Alamos HIV database (www.hiv.lanl.gov). We selected these clades because A, B, and C account for the majority of infections worldwide and the most sequence data are available for these groups. In addition, we included analysis of CRF strain AE, in which Gag is derived from the A clade, to highlight the observation that these sequences show evidence of an HLA-B*57 footprint. We used the most recent (2004) consensus sequences for each clade to identify sites of amino acid differences between clade consensus sequences. We identified sites of HLA-mediated polymorphism by using previous analyses of C-clade Gag, Pol, and Nef sequences from 710 chronically infected adults (30). Fisher's exact test was used to test whether sites at which there is interclade variability are also sites of HLA-associated polymorphism. In order to determine the extent to which the results of our analysis are applicable despite the longitudinal HIV sequence changes that occur, we also compared ancestral HIV sequences for clades A1, B, and C (available at www.hiv.lanl.gov) to current consensus sequences for these three clades.
Impact of clade-specific amino acid polymorphisms on clade phylogeny.
To investigate the effect of the sites of interclade difference on the phylogenetic distinction between clades, we used Gag p24 sequences downloaded from www.hiv.lanl.gov, excluding clonal sequences. First, we sought to determine whether these variable sites are fundamental to defining clades. We randomly selected 20 taxa from clades A1, A2, AE, B, and C and used 12 taxa available for clade A2. We constructed NJ phylogenetic trees from nucleotides in the presence and absence of 27 codons at which we had identified amino acid differences between clades.
Second, we investigated whether amino acid polymorphisms characteristic of one clade alter phylogenetic clustering when superimposed onto sequences from another clade. We selected 20 sequences at random from clades A1, B, and C. Sequences were then modified at sites at which we had identified variability between clades (these sites and their HLA associations are shown in Fig. 1) by substituting the consensus codon for one clade for the same site in taxa selected from another clade. NJ phylogenetic trees were constructed by using the original nucleotide sequences for each clade plus the altered sequences bearing the characteristic codons of a different clade.
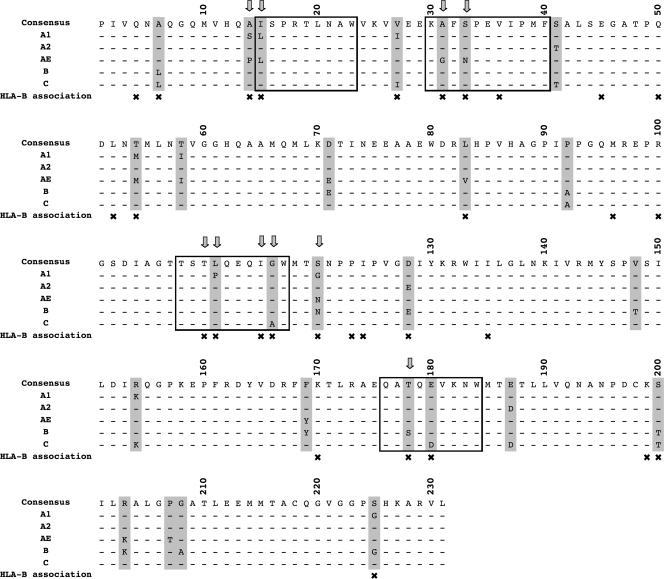
FIG. 1.
Sites of interclade variability and HIV-associated polymorphisms in p24 Gag. Consensus sequences (as of 2004) for HIV clades A1, A2, AE, B, and C are shown. Sites of interclade variability are marked with gray bars, and × indicates the sites of HLA-B-mediated selection pressure identified by analysis of subjects with C-clade infections in Durban (30). The four HLA-B*57/5801 epitopes in p24 Gag are enclosed in open boxes, with arrows indicating sites of HLA-B*57/5801-associated escape mutation that have been described in previous studies of B- and C-clade infections (6, 29, 30). There is a correlation between sites of interclade variability and sites of HLA-B-mediated selection; P < 10−6 (Fisher's exact test).
Study subjects for investigation of HLA footprint mutations with entropy and with phylogenetic clustering.
C-clade sequences were derived from a cohort of 710 chronically infected, treatment-naïve adult subjects from Durban, South Africa, as previously described (21, 22, 25, 30). The sequences used have the following GenBank (http://www.ncbi.nlm.nih.gov/) accession numbers: Gag, FJ198407 to FJ199088, Pol, FJ199532 to FJ199992; Nef, FJ199089 to FJ199531. B-clade sequences were derived from 149 subjects with acute HIV-1 infection recruited from Boston, MA, and Sydney, Australia (1, 26), and 234 subjects with chronic infection (cohort previously described by Schneidewind et al. [38]; sequences available at www.hiv.lanl.gov).
Relationship between sites of HLA selection within a clade.
We used C-clade population sequences from Gag, Pol, and Nef to investigate the relationship between entropy and HLA footprint sites. Sequences with gaps of more than five consecutive amino acids were removed from the analysis in order to avoid false overestimation of entropy due to missing data. Total sequence numbers for this analysis were as follows: p17 Gag, 584; p24 Gag, 646; p15 Gag, 421; protease, 402; reverse transcriptase (RT), 254; integrase, 244; Nef, 424. The Shannon entropy for each amino acid residue was calculated. We adopted a conservative approach by then excluding sites at which there is no amino acid variation (entropy score = 0) from further analysis. Sites of HLA-associated amino acid polymorphism were determined from our previously published analysis (30). We compared entropy at sites of HLA-A, -B, and -Cw selection to entropy at sites where no HLA selection pressure had been identified. Significant difference between the entropy scores of these two groups of sites was sought with a Mann-Whitney test.
Sequences used to asses phylogenetic clustering mediated by footprints of HLA-B*5703 and HLA-B*2705.
We used population sequences from C-clade Gag and Nef to evaluate the impact of the HLA-B*5703 footprint and from B-clade Gag to evaluate the impact of the HLA-B*2705 footprint. We selected C-clade taxa from a pool of 566 Gag sequences (p17 and p24, 1,080 nucleotides) and 443 Nef sequences (621 nucleotides). HLA B*5703 was present in 35 subjects with Gag sequences and 16 subjects with Nef sequences. This allele commonly selects five escape mutations in Gag (8, 11, 25) and two in Nef (30) (for the mutations and their frequencies, see Fig. S1 in the supplemental material).
In our B-clade analysis, 6 subjects from the acute infection cohort and 19 subjects with chronic infection had HLA-B*27. The HLA-B*27 footprint comprises three mutations selected in chronic infection, at Gag positions 173, 264, and 268 (37, 38). To contribute to a pool of HLA-B*27-negative subjects, we selected sequences from www.hiv.lanl.gov expressing the wild-type amino acid at HLA-B*27 footprint sites. Due to limited sequence availability in the chronic infection cohort (38), phylogenetic trees were restricted to taxa of 330 nucleotides length.
Quantification of phylogenetic clustering.
In order to quantify phylogenetic clustering, we adopted a maximum-parsimony approach. Our method calculates the minimum number of mutations required to produce an evolutionary history consistent with the specified amino acid pattern. We constructed NJ phylogenetic trees from nucleotide sequences and mapped all of the amino acid changes at the specified HLA footprint sites on this tree with the MacClade parsimony algorithm (28). The minimum number of mutations (parsimony score) for each of the footprint sites was then summed, giving a total minimum number of evolutionary changes. As a comparator, we calculated the equivalent parsimony score from the same sequences in the absence of the footprint sites. In order to assess the impact of the footprint mutations on clustering, we calculated the difference between the parsimony scores generated in the presence and absence of the footprint mutations for each pool of 100 sequences. A greater degree of phylogenetic clustering is reflected by a smaller parsimony score. Thus, as phylogenetic clustering accumulates, there is an increase in the difference between parsimony scores in the presence and absence of footprint sites; if no phylogenetic clustering is brought about by the footprint, we would expect this difference to be zero.
Phylogenetic clustering of sequences bearing an HLA-B*5703 footprint.
We evaluated the phylogenetic clustering of sequences bearing an HLA-B*5703 footprint by generating a data set containing 100 C-clade Gag sequences. These were selected at random to comprise 20 HLA-B*5703-positive and 80 HLA-B*5703-negative individuals from the aforementioned pool of sequence data. We then repeated the process of selecting 100 Gag sequences at random 100 times over, constructing an NJ tree for each data set, and quantifying phylogenetic clustering, as described above, in the presence and absence of the five HLA-B*5703 footprint sites. In order to account for the statistical variance arising from phylogeny estimation, we also used one set of these taxa (100 Gag sequences) to generate 100 bootstrap trees and quantified phylogenetic clustering in the presence and absence of the footprint sites.
To assess clustering mediated by HLA-B*5703 footprinting in Nef, we generated 50 randomized data sets, each containing 100 C-clade taxa. Each data set contained the same 16 sequences from HLA-B*5703-positive patients (due to limited data, we did not randomize these sequences) and 84 selected at random from HLA-B*5703-negative patients. We used the same methods as described for Gag to quantify phylogenetic clustering in the presence and absence of footprint sites.
Simulation of phylogenetic clustering according to a varying number of footprint mutations and an altered footprint frequency.
To further explore the phylogenetic impact of the HLA-B*5703 footprint, we used the methods described above to quantify the phylogenetic impact of varying the number of mutations per sequence and the proportion of sequences bearing mutations. To quantify the impact of between one and five footprint mutations, we used 100 Gag sequences selected at random from HLA-B*5703-negative subjects. We artificially superimposed a characteristic, conserved HLA-B*5703 footprint on 20 sequences from this pool of 100, starting with the mutation site with the strongest HLA-B*5703 association (defined by Fisher's exact test; see Fig. S1 in the supplemental material). We added the full footprint of five mutations, one site at a time, to each of the 20 selected taxa, quantifying phylogenetic clustering after the addition of each mutation and repeating this process of random sequence selection and addition of a conserved footprint 20 times. To explore how the proportion of sequences bearing the mutations impacts upon phylogenetic clustering, we repeated this analysis with all available HLA-B*5703-negative sequences (n = 526). We added sequential footprint mutations to 5%, 10%, and 20% of all taxa, selected at random, and repeated this process 10 times.
Phylogenetic clustering of Gag sequences bearing an HLA-B*27 footprint.
We investigated the impact of the HLA-B*27 footprint on viral phylogeny by the same methods described above. We modeled the phylogenetic impact of this footprint by using taxa from HLA-B*27-negative subjects from our C-clade cohort. We randomly selected 100 Gag sequences and superimposed the characteristic three-site HLA-B*27 footprint (37, 38) on 20 sequences selected at random, repeating this process 20 times.
We then assessed phylogenetic clustering among sequences from subjects from B-clade cohorts by using subjects truly expressing HLA-B*27. The HLA-B*27 footprint is selected late in infection (14, 38), so we compared clustering among subjects with acute and chronic infections. Due to limited sequence numbers, we did not perform randomizations but constructed a single ML tree with 100 nucleotide sequences. These taxa consisted of all sequences from subjects expressing HLA-B*27 (n = 6 in the acute infection cohort and n = 19 in the chronic infection cohort), with the remaining sequences selected at random from the pool of HLA-B*27-negative subjects.
RESULTS
HLA footprint sites overlap sites of interclade HIV amino acid polymorphism.
We addressed the potential impact of multiple HLA footprints on HIV evolution by investigating whether sites of HLA selection correspond to sites of variation in amino acid residues between clades in Gag, Pol, and Nef. In Gag sequences (p17, p24, and p15) from clades A1, A2, AE, B, and C (n = 1,230) downloaded from the Los Alamos HIV database, only 3.2% of the sites (16 of 500 residues) were invariant. We defined sites of interclade variability based on consensus sequences, identifying 103, 136, and 62 residues in Gag, Pol, and Nef, respectively, that vary among clades A1, A2, AE, B, and C. Consensus sequences from p24 Gag are shown (Fig. 1). We compared these sites with residues at which HLA-mediated immune selection pressure has been identified from a previous analysis of a C-clade cohort in which 84, 83, and 51 sites of HLA-associated polymorphism were described in Gag, Pol, and Nef, respectively (30).
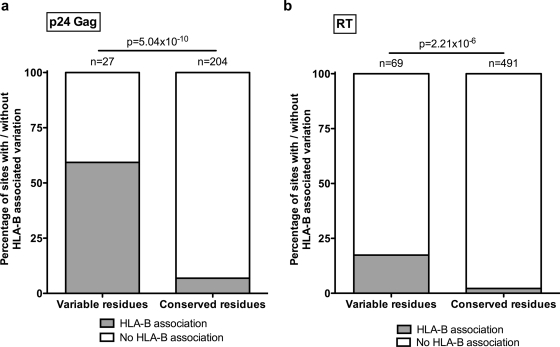
A strong association was seen between sites of interclade amino acid variability and sites of selection pressure mediated by HLA-A, -B, and -C (P = 4.1 × 10−7, P = 9.8 × 10−19, and P = 1.3 × 10−5, respectively; Table 1). Consistent with previous studies (21), the strongest association with interclade amino acid differences was for sites of selection pressure mediated by HLA-B in p24 Gag (P = 5.0 × 10−10) (Table 1 and Fig. 1 and 2a). This remained significant even when HLA-B*57/5801 was excluded from the analysis (P = 3.6 × 10−8), demonstrating that the association is not limited to these immunodominant alleles. Overall, 59% of the sites of interclade p24 Gag variability were also identified as sites of HLA-B-driven selection pressure (Fig. 1). For the Pol and Nef proteins, 18% of the variable residues were also sites of HLA-B-driven escape mutation (example shown in Fig. 2b).
TABLE 1.
Correlation between sites of HLA-mediated immune selection pressurea and sites of interclade variability in clades A1, A2, AE, B, and Cb
| Protein(s) | HLA-A | HLA-B | HLA-C |
|---|---|---|---|
| Gag | |||
| p17 | 0.15 | 8.68 × 10−3 | 1 |
| p24 | 4.27 × 10−3 | 5.04 × 10−10 | 0.62 |
| p15 | 1 | 0.47 | 1 |
| Pol | |||
| Protease | 0.076 | 1.92 × 10−3 | 0.19 |
| RT | 2.22 × 10−3 | 2.21 × 10−6 | 2.63 × 10−7 |
| Integrase | 0.14 | 1.19 × 10−3 | 1 |
| Nef | 0.032 | 0.018 | 0.24 |
| Gag + Pol + Nef | 4.13 × 10−7 | 9.77 × 10−19 | 1.34 × 10−5 |
q, <0.2.
Fisher's exact test was used to determine statistically significant associations (P < 0.001, corrected for multiple comparisons by the Bonferroni method), which are in bold and underlined.
FIG. 2.
Proportion of amino acid residues at which HLA-B-associated polymorphisms are detected. All amino acids in each protein are represented, divided into “variable residues” at which there are interclade differences in amino acids and “conserved residues” that are identical among consensus sequences for clades A1, A2, AE, B, and C. The proportion of each of these sites at which HLA-B selection has been previously identified (30) is shown in gray. (a) p24 Gag. (b) RT. P values were calculated with Fisher's exact test.
In comparing predicted ancestral sequences with contemporary (2004) consensus sequences, we identified differences in only 5.1% of the residues across Gag, Pol, and Nef, suggesting little overall longitudinal change in amino acids that characterize individual clades. The amino acids that differed in our comparison of ancestral and consensus sequences were largely the same as the amino acids that differed among modern consensus sequences for clades A1, A2, AE, B, and C (in 90%, 100%, and 96% of the cases in Gag, Pol, and Nef, respectively). This suggests that our strategy of comparing clade consensus sequences not only accounts for current differences among clades but also largely accounts for sites of amino acid diversity that have evolved longitudinally.
We also investigated whether HLA footprint sites are associated with sites of sequence variability within a clade. We used C-clade sequences to compare Shannon entropy for sites at which there is no known HLA selection to sites at which there is HLA-associated polymorphism (as identified previously [30]). As expected, we found significantly higher entropy scores at sites of HLA-mediated selection, compared to sites at which no HLA selection is detected (see Fig. S2 in the supplemental material); this relationship was most consistent for HLA-B (Table 2).
TABLE 2.
Relationship between Shannon entropy at sites of HLA-associated amino acid polymorphism compared to sites at which no HLA selection was detected in C-clade sequencesa
| Protein(s) | HLA-A | HLA-B | HLA-C |
|---|---|---|---|
| Gag | |||
| p17 | 0.0152 | 0.0005 | NAb |
| p24 | <0.0001 | <0.0001 | 0.0003 |
| p15 | NA | NA | NA |
| Pol | |||
| Protease | NA | 0.0024 | 0.0703 |
| RT | <0.0001 | <0.0001 | <0.0001 |
| Integrase | 0.0009 | <0.0001 | NA |
| Nef | <0.0003 | <0.0001 | 0.0123 |
| Gag + Pol + Nef | <0.0001 | <0.0001 | <0.0001 |
Invariant sites (Shannon entropy score = 0) were excluded from the analysis. Statistically significant associations (defined as P < 0.001 [Mann-Whitney test], corrected for multiple comparisons by the Bonferroni method) are in bold and underlined.
The NA (not applicable) designation applies where too few associations were detected to allow statistical analysis.
Synonymous and nonsynonymous nucleotide substitutions contribute to clade-specific phylogeny.
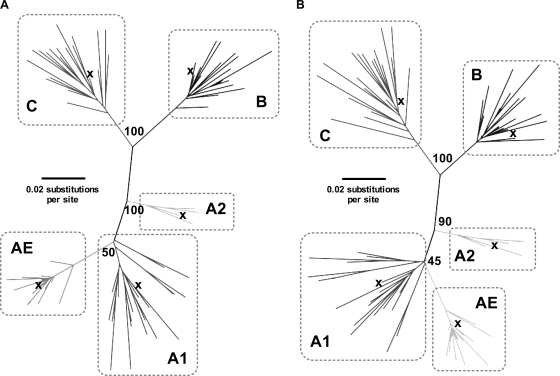
To investigate the extent to which sites of amino acid variability impact on phylogenetic distinction of HIV taxa into clades, we constructed phylogenetic trees in the presence and absence of the 27 codons that confer amino acid variability between p24 Gag clade consensus sequences. The majority, but not all, of these sites are also residues at which HLA selection has been shown to operate, as discussed above (Fig. 1 and 2). Not unexpectedly, the phylogenetic distinction between these clades persists in the absence of these variable sites (Fig. 3), showing that the distinction between clades exists as a result of synonymous nucleotide substitutions, as well as because of amino acid differences. We investigated the extent to which the presence of amino acid polymorphisms can affect phylogeny. Again not unexpectedly, by substituting the codons characteristic of one clade for the sequences of a second clade, we found evidence that changes at these sites can alter clade-specific phylogenetic clustering (characteristic examples shown in Fig. 4). Consensus sequences for each clade are shown to fall within their respective clade clusters (Fig. 3), demonstrating that the consensus sequences (also used elsewhere in our analysis) reliably reflect the random pool of sequences selected to represent each clade.
FIG. 3.
Phylogenetic trees illustrating the preservation of clade phylogeny in the presence and absence of sites of clade-specific difference. Sequences from clades A1, A2, AE, B, and C were selected at random from www.hiv.lanl.gov, and ML phylogenetic trees were constructed (midpoint rooted, bootstrap values based on 100 replicates). Each clade is enclosed within a box to show clustering in the presence and absence of sites that vary between clades. The 2004 consensus sequence for each clade is marked by ×. (A) Tree constructed from complete p24 sequences (693 nucleotides). (B) Tree constructed from p24 sequences in the absence of 27 codons at which there is amino acid difference between clade consensus sequences (612 nucleotides).
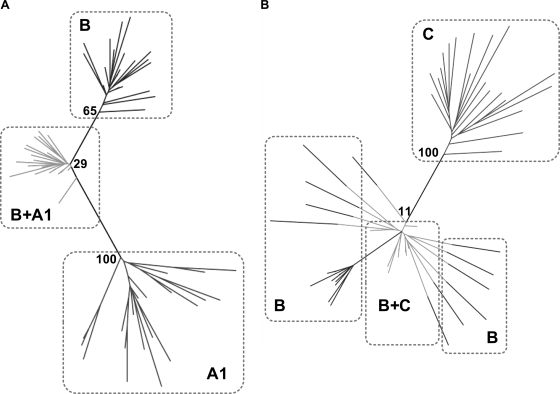
FIG. 4.
Phylogenetic trees illustrating altered distribution of taxa when codons determining clade-specific differences are swapped between clades. Twenty sequences were selected at random from the clades indicated within dashed boxes. ML trees were constructed from nucleotides (midpoint rooted, bootstrap values based on 100 replicates). (A) Codons defining amino acids characteristic of clade A1 (selected from the clade A1 consensus) were superimposed on 20 sequences from clade B. The clade B sequences are shown twice, once without alteration (marked B) and once with A1-clade amino acids superimposed (marked B+A1). (B) Codons defining amino acids characteristic of clade C (selected from the clade C consensus) were superimposed on the same 20 sequences from clade B. As before, the clade B sequences are shown twice, unchanged (marked B) and bearing the characteristic C-clade codons (marked B+C).
HIV subtype AE bears the HLA-B*57 footprint.
The AE subtype (CRF variant) dominates the HIV epidemic in Thailand. Despite the low frequency of HLA-B*57 in the Thai population (1.8%) (7), it is apparent that the AE consensus sequence closely resembles an HLA-B*5703 escape mutant, incorporating many components of the Gag HLA-B*5703 footprint (that is, A146P, I147L, A163G, and S165N; Fig. 1). The only omission is T242N, the escape mutation that reverts to the wild type following transmission (8, 25, 29) and hence does not persist at a population level. The AE subtype thus is a clear-cut example, consistent with the hypothesis that HLA selection may contribute significantly to founder strains of HIV.
Phylogenetic clustering can be mediated by an HLA-B*5703 footprint in Gag.
Previous analysis of C-clade sequences found that the HLA allele with the greatest number of associated HIV sequence polymorphisms was HLA-B*5703, with five strong associations in Gag alone (q, <0.05; P < 10−6; see Fig. S1 in the supplemental material) (30). Therefore, we initially focused on HLA-B*5703 to determine the potential impact of a single allelic footprint on viral evolution within a clade.
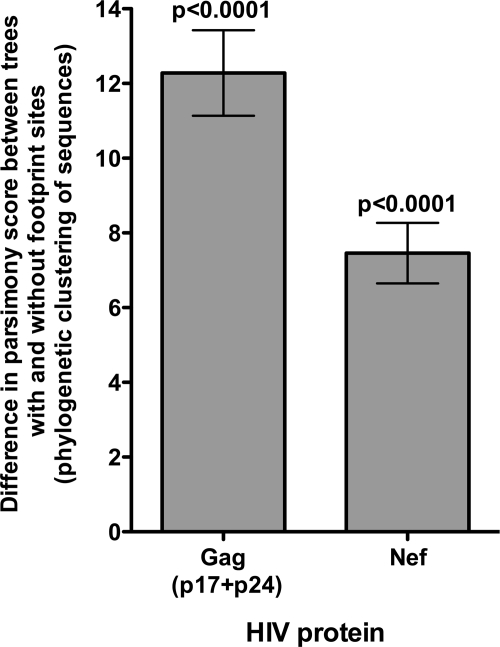
To determine whether HLA-B*5703-mediated selection pressure has a potential impact on viral evolution, we assessed the phylogenetic clustering of C-clade Gag and Nef sequences. Phylogenetic clustering among 100 Gag sequences, of which 20 were taken from subjects expressing HLA-B*5703, was quantified in the presence and absence of the five HLA-B*5703 footprint sites. In the absence of these five codons, phylogenetic clustering of the sequences was reduced. When this analysis was repeated with 100 randomized data sets, the clustering effect mediated by the HLA-B*5703 footprint sites was found to be highly statistically significant (P < 0.0001 [paired t test]; Fig. 5). In 100 bootstrap trees generated from a single data set of 100 taxa, this clustering effect remained equally significant (P < 0.0001 [paired t test]; data not shown). Such clustering occurs even though some HLA-B*5703-negative patients may also possess HLA-B*5703 footprint mutations as a result of transmission (2) or selection by non-HLA-B*5703 alleles (19; see Fig. S1 in the supplemental material).
FIG. 5.
Phylogenetic clustering in sequences from individuals with HLA-B*5703 with clustering analyzed in the presence and absence of the footprint sites. Phylogenetic clustering mediated by an HLA-B*5703 footprint was assessed in 100 NJ trees for Gag (with 20% of the sequences from subjects with HLA-B*5703) and 50 for Nef (with 16% of the sequences from subjects with HLA-B*5703). The mean difference in parsimony score is plotted, with error bars showing 95% confidence intervals. A significantly greater degree of phylogenetic clustering was observed in the presence of HLA-B*5703 footprint sites than in the absence of these sites for both proteins (P < 0.0001; paired t test).
To address the possibility that excluding any group of polymorphic codons might itself significantly affect phylogenetic clustering, we repeated the analysis of Gag sequences by using the same methods but excluding five randomly chosen polymorphic sites in place of the five footprint sites characteristic of HLA-B*5703. For each of five data sets, we chose five randomly selected polymorphic sites with Shannon entropy scores equivalent to those of the HLA-B*5703 footprint sites (P = 0.2, Mann-Whitney test). Comparison of phylogenetic clustering in the presence or absence of these sites had no significant effect on clustering (P = 0.5 [paired t test]; data not shown). Polymorphisms at these randomly selected sites occur independently, rather than arise in a distinct subset of taxa as a result of shared selection pressure, and therefore—as expected—have no significant impact on phylogenetic clustering.
Progressive phylogenetic clustering of Gag sequences occurs as sequential HLA-B*5703 footprint mutations are added.
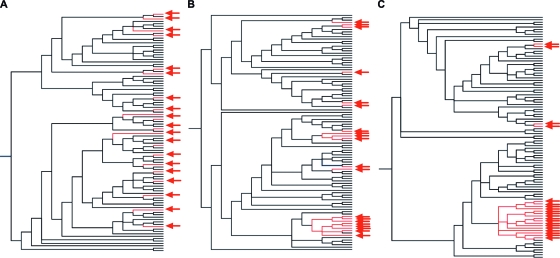
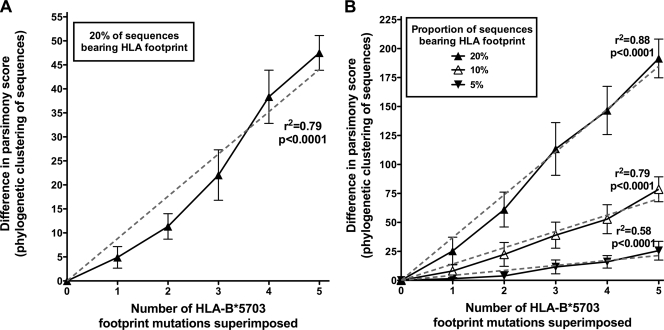
In order to quantify more specifically the impact of accumulating HLA-selected mutations, we conducted a simulation of HLA-B*5703 footprinting by using 100 sequences from HLA-B*5703-negative subjects and sequentially adding the five HLA-B*5703 footprint mutations to 20 of them selected at random. Representative phylogenetic trees constructed from these sequences are shown (Fig. 6), demonstrating progressive clustering between sequences bearing the footprint as more mutations were added. Quantifying this clustering by the methods described above, we found that progressively fewer mutations were required to explain the phylogeny as more footprint polymorphisms were added, reflecting increasing clustering of footprint-bearing taxa (Fig. 7A). Thus, significant phylogenetic clustering can arise as a consequence of imposing even a partial HLA-B*5703 footprint on randomly chosen Gag sequences.
FIG. 6.
Phylogenetic clustering as a consequence of artificial imposition of HLA-B*5703 mutations on HLA-B*5703-negative Gag sequences. ML phylogenetic trees constructed from 100 Gag sequences (selected at random from a pool of HLA-B*5703-negative individuals). The same 100 sequences are represented in each tree, and the same 20 are marked with arrows. (A) Twenty taxa were selected at random (marked with arrows). In this panel, no footprint mutations have been added. (B) The same sequences after the addition of three HLA-B*5703 footprint mutations to the arrowed sequences. (C) The same sequences after the addition of five footprint mutations. Progressive phylogenetic clustering among sequences bearing the HLA-B*5703 footprint is evident.
FIG. 7.
Model to show difference in phylogenetic clustering as five HLA-B*5703 footprint mutations were superimposed on Gag sequences. Each panel shows the mean difference in parsimony score between trees with no mutations and trees built from the same taxa with sequential HLA-B*5703 footprint mutations superimposed. Addition of mutations increases the difference in parsimony scores, reflecting progressive phylogenetic clustering (error bars show 95% confidence intervals; r2 from linear regression with the y intercept set to go through the origin). (A) One hundred Gag sequences were selected at random from HLA-B*5703-negative individuals and used to construct an NJ phylogenetic tree. Five characteristic HLA-B*5703 footprint mutations were added, one at a time, to 20 sequences in each tree. Twenty repetitions are shown. (B) All 526 taxa from HLA-B*5703-negative subjects were used. The footprint mutation was added to a varying proportion of sequences (individual sequences selected at random). Ten repetitions are shown.
With the same model, when we excluded footprint sites from the analysis, phylogenetic clustering was reduced (P = 0.0004; see Fig. S3 in the supplemental material); this reduction in clustering may relate to the presence of shared mutations at these sites selected by other closely related alleles (such as HLA-B*5702 and HLA-B*5801; see Fig. S1 in the supplemental material). However, the phylogenetic clustering we quantified in true sequences in the presence of the footprint sites (Fig. 5) is largely a consequence of the addition of progressive mutations rather than an artifact of removing footprint sites for the purpose of generating a comparator (see Fig. S3 in the supplemental material).
By quantification of phylogenetic clustering with a larger pool of sequences (n = 526) with a variable proportion bearing the footprint, we found that the degree of clustering increases as the proportion of footprint-bearing sequences is increased (Fig. 7B). Even with only 5% of the sequences bearing the footprint (comparable to the true population phenotypic frequency of HLA-B*5703 in Durban), there is still a significant increase in clustering as the footprint mutations accumulate (r2 = 0.58, P < 0.0001 [linear regression]).
Phylogenetic clustering can be mediated by an HLA-B*5703 footprint in Nef.
In order to examine the effect of a smaller HLA-B*5703 footprint in a more variable protein than Gag, we repeated the same analysis with Nef, which contains two polymorphisms associated with HLA-B*5703 (see Fig. S1 in the supplemental material) (30). We first investigated the true biological footprint, and subsequently imposed an artificial footprint of both mutations, by the same methods described above. Significant phylogenetic clustering of Nef sequences from B*5703-positive individuals was again seen as a consequence of shared footprint mutations (P < 0.0001 [paired t test]; Fig. 5).
Phylogenetic clustering can be mediated by an HLA-B*27 Gag footprint in chronic infection.
Having established the significant phylogenetic impact of a single HLA allele that imposes a substantial footprint on the virus, we sought evidence of phylogenetic clustering mediated by a second allele, HLA-B*2705. HLA-B*2705 is also associated with immune control of HIV but selects a smaller footprint of three mutations in Gag, arising late in the course of infection (14, 38). We observed significant phylogenetic clustering when we superimposed the characteristic footprint of three mutations (37, 38) on C-clade Gag sequences (P < 0.0001 [paired t test]; see Fig. S4a in the supplemental material). Analysis of the impact of the HLA-B*2705 Gag footprint arising as a result of natural selection showed, as expected, no clear evidence of phylogenetic clustering among HLA-B*2705 subjects in acute infection, since the escape mutations characteristically arise late (14; see Fig. S4b in the supplemental material). In contrast, in our analysis of sequence data from subjects with chronic infection (38), there is clustering of taxa from HLA-B*2705-positive subjects (see Fig. S4c in the supplemental material), indicating that the selection pressure imposed by this allele can also drive convergent evolution over time.
DISCUSSION
These studies set out to address the impact of HLA-selected mutations on HIV-1 evolution at the population level. We observed a strong association between sites of HLA selection and amino acid variability between and within clades, demonstrated that amino acid substitutions between clades can alter phylogenetic clustering, and showed that the footprint of even a single HLA allele can cause clustering within a clade.
Our observation that HIV amino acid differences between clades tend to be those that are also selected as CD8+ T-cell escape mutations has two possible explanations, (i) that these sites vary because of HLA selection or (ii) that sites of HLA escape are less constrained by a fitness cost to the virus. Both explanations may contribute to the observed findings. Analysis of the AE subtype that predominates in Thailand suggests that the former explanation may operate at least under some circumstances. The Gag mutations A163G and S165N are selected by the HLA-B*5703 CD8+ T-cell response to the epitope KAFSPEVIPMF (Gag 162 to 172) in C-clade infection (8). When the A163G mutant arises alone, it significantly reduces viral replicative capacity in vitro and reverts rapidly to the wild type in vivo in the absence of HLA-B*5703 (8). More commonly, A163G is found in association with the compensatory mutation S165N (8). The observation that T242N, the one mutation with an uncompensated fitness cost, does revert suggests that the other polymorphisms (presumably with compensatory mutations) do not revert because there is no fitness cost rather than as a consequence of ongoing selection pressure. We hypothesize that the founder strain may have been transmitted by an individual with HLA-B*5703, that is, that the AE subtype of HIV came to incorporate A163G/S165N as a consequence of HLA-B*5703 being the original driving force.
Similar observations can be made from the less extensive characterization that has been made of HCV-specific CD8+ T-cell responses: an escape mutation (Y1444F) within an immunodominant HLA-A*01 epitope in NS3 has been shown to accumulate in cohorts infected with genotypes 1 and 3 (23, 33). In the case of HLA B*27, the immunodominant epitope in NS5B in genotype 1 infection may undergo a double mutation which is associated with immune escape and loss of protection (34). These mutations form the consensus sequence of genotypes 2, 3, and 6 (Los Alamos Hepatitis C database; http://hcv.lanl.gov).
The contribution of HLA selection to amino acid sequence variation may be substantial, with 59% of the variation between Gag clade consensus sequences associated with sites of HLA-B selection. Moreover, these results are likely to be a considerable underestimate, as the HLA escape mutations we have considered here are limited to those identified in our Durban cohort (30). Sites that are under positive selection may also be identified by calculating the dN/dS ratio (9, 10), and this approach could be used to generate additional evidence that differences between clades relate to HLA selection. However, the results of dN/dS ratio calculations vary as a function of the frequency of the selecting allele, are affected by the rate of reversion, and are difficult to apply to large populations; for these reasons, this approach is outside the scope of the present analyses.
The substantial overlap between sites of amino acid variability and sites of HLA-driven escape mutation is of relevance to T-cell-based vaccines. These sites of amino acid variability are not simply “toggle” sites (12) of little significance to T-cell recognition. On the contrary, toggling between amino acid variants may be a function of positive selection (9), and our data suggest that vaccine constructs may need to be matched to the clade of virus prevailing in the target population.
We have demonstrated that sites of amino acid difference between clades are not required to distinguish clade specificity, suggesting that clades have been originally defined by nucleotide differences between founder sequences. Stripping sites of only nonsynonymous nucleotide substitutions is somewhat artificial since nonsynonymous and synonymous changes may arise in the same codon. However, the finding that clade clustering is preserved in the absence of synonymous changes is consistent with the previous observation that all HIV clades may be traced to ancestral sequences from the same region of Africa (39, 40), rather than occur subsequently as a consequence of HLA selection. Nevertheless, we have also shown that exchange of polymorphisms between clades does affect phylogeny, underscoring the potential for HLA selection to shape the future evolution of the epidemic.
We have shown that otherwise unrelated HIV sequences within a clade may cluster together phylogenetically as a consequence of selection pressure imposed by even a single HLA allele, both in true sequence data and in a model of serial mutations. This simulated approach is robust because the underlying sequences are altered only at footprint sites, and the mutations themselves are inferred from genuine sequence data. The model shows more phylogenetic clustering than that seen among the true sequences as a consequence of complete conservation of the mutations applied in the simulation compared to variations in the true sequences (number of mutations, sites of mutations, and nucleotide substitutions are all conserved in the model but vary in real sequences). Irrespective of this, we show that clustering mediated by the HLA-B*5703 footprint sites remains highly statistically significant in true biological sequences. The significance of this clustering effect is all the more striking when considered in the context of the enormous diversity of HIV and the potential for multiple HLA footprints to coexist.
This observation is of potential utility when considering the use of lineage-corrected approaches in the detection of HLA-mediated selection pressure on viruses. Statistical approaches to identify associations between HIV sequence polymorphisms and HLA alleles (21, 32) have been refined to account for a founder effect (4): lineage-based methods correct for similarities among taxa generated by their common ancestry, thus distinguishing bona fide HLA-escape mutations from artifactual associations mediated by founder effects (18, 36). Conversely, we show here that viral amino acid polymorphisms arising independently in individuals who share an HLA allele (29, 31, 38) may not be identified as independent mutations (homoplasies) in phylogenetic reconstruction but instead can mistakenly appear as shared ancestral mutations (synapomorphies). This may result in sequences that share common escape mutations being artificially grouped together during phylogenetic reconstruction. This phylogenetic bias has previously been addressed in the setting of HIV adaptation to neutralizing antibodies (18) and drug therapy (24) but not in the context of adaptation to CD8+ T-cell responses. This confounding effect can be minimized by excluding the nucleotides under analysis (footprint sites) when constructing the phylogenetic tree. However, this requires a priori knowledge of the sites of escape mutation. In the absence of this information, phylogenetic clustering is likely to be reduced by maximizing the length of the sequence analyzed. This is relevant to many studies that carry out phylogenetic analysis with short protein fragments (e.g., protease, 99 amino acids). Artifactual clustering is likely to be a particular problem for within-clade HIV data sets, which are characterized by many phylogenetically uninformative singleton polymorphisms.
In conclusion, these data support a role for both founder effects and HLA selection in establishing the epidemic and suggest that future HIV evolution—within and between clades—may be significantly shaped by the HLA alleles to which the virus is exposed. CD8+ T-cell vaccines may therefore need to be geared to the clade of virus affecting the target population and modified over time to keep pace with evolutionary changes in the virus driven by HLA.
Supplementary Material
Acknowledgments
Contributions to B-clade sequence data were made by Kristin O'Sullivan, Laura Reyor, Rafick Sekaly, and Marty Markowitz.
This work was funded by the National Institutes of Health (contracts NO1-A1-15422, 2RO1AI46995-06, and R01A1067073), the Wellcome Trust (A.L. and P.G.), the UK Medical Research Council (P.M. and A.P.), and the Mark and Lisa Schwartz Foundation. P.G. is an Elizabeth Glaser Pediatric AIDS Foundation Scientist.
Footnotes
Published ahead of print on 25 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, M. O'Sullivan, K., I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal, A., L. Yue, J. Conway, K. Yusim, J. Tang, J. Kappes, R. A. Kaslow, C. M. Wilson, and P. A. Goepfert. 2007. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS 212387-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 1024512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 5.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 8112608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brumme, Z. L., I. Tao, S. Szeto, C. J. Brumme, J. M. Carlson, D. Chan, C. Kadie, N. Frahm, C. Brander, B. Walker, D. Heckerman, and P. R. Harrigan. 2008. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS 221277-1286. [DOI] [PubMed] [Google Scholar]
- 7.Chandanayingyong, D., H. A. Stephens, R. Klaythong, M. Sirikong, S. Udee, P. Longta, R. Chantangpol, S. Bejrachandra, and E. Rungruang. 1997. HLA-A, -B, -DRB1, -DQA1, and -DQB1 polymorphism in Thais. Hum. Immunol. 53174-182. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 818346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delport, W., K. Scheffler, and C. Seoighe. 2008. Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Pathog. 4e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Oliveira, T., M. Salemi, M. Gordon, A. M. Vandamme, E. J. van Rensburg, S. Engelbrecht, H. M. Coovadia, and S. Cassol. 2004. Mapping sites of positive selection and amino acid diversification in the HIV genome: an alternative approach to vaccine design? Genetics 1671047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frahm, N., D. E. Kaufmann, K. Yusim, M. Muldoon, C. Kesmir, C. H. Linde, W. Fischer, T. M. Allen, B. Li, B. H. McMahon, K. L. Faircloth, H. S. Hewitt, E. W. Mackey, T. Miura, A. Khatri, S. Wolinsky, A. McMichael, R. K. Funkhouser, B. D. Walker, C. Brander, and B. T. Korber. 2007. Increased sequence diversity coverage improves detection of HIV-specific T cell responses. J. Immunol. 1796638-6650. [DOI] [PubMed] [Google Scholar]
- 13.Frater, A. J., H. Brown, A. Oxenius, H. F. Gunthard, B. Hirschel, N. Robinson, A. J. Leslie, R. Payne, H. Crawford, A. Prendergast, C. Brander, P. Kiepiela, B. D. Walker, P. J. Goulder, A. McLean, and R. E. Phillips. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 816742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4630-640. [DOI] [PubMed] [Google Scholar]
- 16.Goulder, P. J., and D. I. Watkins. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22160-174. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, E. C., L. Q. Zhang, P. Simmonds, C. A. Ludlam, and A. J. Brown. 1992. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc. Natl. Acad. Sci. USA 894835-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honeyborne, I., A. Prendergast, F. Pereyra, A. Leslie, H. Crawford, R. Payne, S. Reddy, K. Bishop, E. Moodley, K. Nair, M. van der Stok, N. McCarthy, C. M. Rousseau, M. Addo, J. I. Mullins, C. Brander, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple Gag-specific CD8+ T-cell epitopes. J. Virol. 813667-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2405-411. [DOI] [PubMed] [Google Scholar]
- 21.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432769-775. [DOI] [PubMed] [Google Scholar]
- 22.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 23.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 766104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemey, P., I. Derdelinckx, A. Rambaut, K. Van Laethem, S. Dumont, S. Vermeulen, E. Van Wijngaerden, and A. M. Vandamme. 2005. Molecular footprint of drug-selective pressure in a human immunodeficiency virus transmission chain. J. Virol. 7911981-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10282-289. [DOI] [PubMed] [Google Scholar]
- 26.Li, B., A. D. Gladden, M. Altfeld, J. M. Kaldor, D. A. Cooper, A. D. Kelleher, and T. M. Allen. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y., J. McNevin, H. Zhao, D. M. Tebit, R. M. Troyer, M. McSweyn, A. K. Ghosh, D. Shriner, E. J. Arts, M. J. McElrath, and J. I. Mullins. 2007. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J. Virol. 8112179-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison, D. R., and W. P. Maddison. 2000. MacClade. Analysis of phylogeny and character evolution, 4th edition. Sinauer Associates, Sunderland MA.
- 29.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 803617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews, P. C., A. Prendergast, A. Leslie, H. Crawford, R. Payne, C. Rousseau, M. Rolland, I. Honeyborne, J. Carlson, C. Kadie, C. Brander, K. Bishop, N. Mlotshwa, J. I. Mullins, H. Coovadia, T. Ndung'u, B. D. Walker, D. Heckerman, and P. J. Goulder. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 828548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMichael, A., and P. Klenerman. 2002. HIV/AIDS. HLA leaves its footprints on HIV. Science 2961410-1411. [DOI] [PubMed] [Google Scholar]
- 32.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 33.Neumann-Haefelin, C., D. N. Frick, J. J. Wang, O. G. Pybus, S. Salloum, G. S. Narula, A. Eckart, A. Biezynski, T. Eiermann, P. Klenerman, S. Viazov, M. Roggendorf, R. Thimme, M. Reiser, and J. Timm. 2008. Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J. Virol. 823438-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann-Haefelin, C., S. McKiernan, S. Ward, S. Viazov, H. C. Spangenberg, T. Killinger, T. F. Baumert, N. Nazarova, I. Sheridan, O. Pybus, F. von Weizsacker, M. Roggendorf, D. Kelleher, P. Klenerman, H. E. Blum, and R. Thimme. 2006. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 43563-572. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7379-381. [DOI] [PubMed] [Google Scholar]
- 36.Ridley, M. 1983. The explanation of organic diversity: the comparative method and adaptations for mating. Oxford University Press, New York, NY.
- 37.Schneidewind, A., M. A. Brockman, J. Sidney, Y. E. Wang, H. Chen, T. J. Suscovich, B. Li, R. I. Adam, R. L. Allgaier, B. R. Mothe, T. Kuntzen, C. Oniangue-Ndza, A. Trocha, X. G. Yu, C. Brander, A. Sette, B. D. Walker, and T. M. Allen. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 825594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 8112382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal, N., M. Peeters, C. Mulanga-Kabeya, N. Nzilambi, D. Robertson, W. Ilunga, H. Sema, K. Tshimanga, B. Bongo, and E. Delaporte. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 7410498-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worobey, M., M. Gemmel, D. E. Teuwen, T. Haselkorn, K. Kunstman, M. Bunce, J. J. Muyembe, J. M. Kabongo, R. M. Kalengayi, E. Van Marck, M. T. Gilbert, and S. M. Wolinsky. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.