Abstract
Hepatitis delta virus (HDV) is a subviral pathogen that increases the severity of liver disease caused by hepatitis B virus. Both the small circular RNA genome and its complement, the antigenome, form a characteristic unbranched rod structure in which approximately 70% of the nucleotides are base paired. These RNAs are associated with the sole virally encoded protein, hepatitis delta antigen (HDAg), in infected cells; however, the nature of the ribonucleoprotein complexes (RNPs) is not well understood. Previous analyses of binding in vitro using native, bacterially expressed HDAg have been hampered by a lack of specificity for HDV RNA. Here, we show that removal of the C-terminal 35 amino acids of HDAg yields a native, bacterially expressed protein, HDAg-160, that specifically binds HDV unbranched rod RNA with high affinity. In an electrophoretic mobility shift assay, this protein produced a discrete, micrococcal nuclease-resistant complex with an ∼400-nucleotide (nt) segment of HDV unbranched rod RNA. Binding occurred with several segments of HDV RNA, although with various affinities and efficiencies. Analysis of the effects of deleting segments of the unbranched rod indicated that binding did not require one or two specific binding sites within these RNA segments. Rather, a minimum-length HDV RNA unbranched rod approximately 311 nt was essential for RNP formation. The results are consistent with a model in which HDAg binds HDV unbranched rod RNA as multimers of fixed size rather than as individual subunits.
Hepatitis delta virus (HDV) is a unique human pathogen that increases the severity of liver disease in those infected with its helper virus, hepatitis B virus. HDV RNA replication is unique among known animal viruses in that it uses the DNA-dependent RNA polymerase activity of the host to replicate its genome without a DNA intermediate (21, 23, 24). The negative-stranded HDV genome is a single-stranded circular RNA predicted to form an unbranched rod structure in which ∼70% of the nucleotides form base pairs (15, 36). The RNA forms this structure in vitro (14), and mutational analyses have indicated that it not only occurs in cells but also is required for RNA replication (3, 27, 31). Genome replication involves a double rolling circle mechanism in which the antigenome, with a similar rod structure, serves as a replication intermediate (1, 16, 21, 33). Arranged along one side of a two-dimensional representation of the unbranched rod structure is the single HDV open reading frame, which encodes the hepatitis delta antigen (HDAg).
HDAg plays several critical functions in the HDV replication cycle: it has pivotal roles in both RNA replication and packaging (6, 17). The abilities of HDAg to form multimeric complexes and to bind HDV RNA are central to these functions (8, 9, 13, 25, 34, 38). In infected cells, both the genome and antigenome are associated with HDAg. However, the nature of the ribonucleoprotein (RNP) complex formed is not fully understood. For example, attempts to determine the relative amounts of HDAg and HDV RNA complexed in RNPs in viral particles and in cells have produced ratios ranging from 22 to 200 (12, 30).
Previous in vitro investigations of the interaction between HDAg and HDV RNA indicated that HDAg could specifically bind several segments of the HDV unbranched rod RNA (7, 20). However, these studies used bacterially expressed HDAg fusion proteins that were denatured prior to analysis of RNA binding and therefore did not provide quantitative binding data. Moreover, the particular binding determinants in the RNA have not yet been identified. More recent studies have demonstrated that native bacterially expressed HDAg can form complexes with HDV RNA in vitro and that these complexes can initiate HDV replication when transfected into cultured cells (10, 32). However, the structure of the complexes formed has not been fully addressed and it is not clear whether HDAg expressed in Escherichia coli binds HDV RNA specifically.
Here, we show that, in vitro, bacterially expressed, native HDAg forms heterogeneous complexes, possibly aggregates, with HDV RNAs as well as non-HDV RNAs. However, these obstacles to analysis of binding were overcome by using a truncated version of HDAg from which the C-terminal 35 amino acids (aa) of HDAg had been removed. In an electrophoretic mobility shift assay, this protein produced a discrete, micrococcal nuclease-resistant complex with an ∼400-nucleotide (nt) segment of HDV unbranched rod RNA. Binding was observed with several different segments of the unbranched rod with variable affinities, but no binding was observed with non-HDV RNAs. Deletional mapping indicated that binding did not require a particular site or a limited set of sites within the RNA; rather, a minimum-length HDV RNA unbranched rod of ∼311 nt was essential for complex formation. Our results are consistent with a model in which HDAg binds HDV RNA as a multimer of fixed size rather than as individual subunits.
MATERIALS AND METHODS
Plasmids.
pHDVΔApaI+LeftLoop is a nonreplicating expression plasmid in which the ApaI fragment of pHDVΔx1-NR (28) has been deleted and replaced with the sequence of the loop at the left end of the unbranched rod. This plasmid was used as a template for PCR amplification of templates for transcription of 384M, 207M, and g384M. pHDVΔSma is a nonreplicating expression plasmid in which the SmaI fragment of pHDVΔx1-NR has been deleted; this plasmid was used as a template for PCR amplification of the template for transcription of 405E. The plasmid pCMV-Ribo was created for expression of RNAs with defined 5′ and 3′ ends. This construct, which contains the cytomegalovirus (CMV) immediate-early promoter and the HDV antigenomic ribozyme, was made by inserting oligonucleotides containing sequences corresponding to the antigenomic ribozyme (nt 817 to 900) (nucleotide numbering according to reference 36) between the SacI and BamHI sites of pHDV·I(−)Ag(−) (4). An additional NotI site was added for cloning purposes. Plasmids for expression of 395L and 207L were created by insertion of PCR-amplified cDNA fragments for 395L and 207L between the SacI and NotI sites of pCMV-Ribo to make pCMV-Ribo-395L and pCMV-Ribo-207L. Upon transfection, pCMV-Ribo-395L transcribes nt 153 to 1441 and pCMV-Ribo-207L transcribes nt 59 to 1536.
In vitro transcription templates and T7 transcription.
RNAs were transcribed from PCR products into which the T7 promoter sequence was incorporated. RNAs were named according to length in nucleotides, and the location of the loop sequence (L for left, R for right, and NL for no loop). 395L, 311L, 253L, and 207L were transcribed from templates amplified from pCMV-DC1×1.2-AgS(−). 395L includes the left 395 nt from nt 153 (5′) to 1441 (3′). 311L includes nt 111 (5′) to 1484 (3′). 253L includes nt 81 (5′) to 1511 (3′). 207L includes nt 59 (5′) to 1536 (3′). The transcription templates for 384M, g384M, and 207M were amplified from the pHDVΔApaI+LeftLoop plasmid. 384M and g384M include nt 401 (5′) to 213, an ACAGA loop, and nt 1379 to 1193 (3′) while 207M includes nt 314 (5′) to 213, an ACAGA loop, and nt 1379 to 1283 (3′). The transcription template for 405E was amplified from the pHDVΔSma plasmid and includes nt 680 (5′) to 482 (at the SmaI site) and nt 1109 to 907 (3′).
In the template used to make 395R, 316R, and 298R, the sequence corresponding to the left-terminal loop of the HDV unbranched rod (5′-ACAGA-3′) was relocated such that the original location of the loop became the 5′ and 3′ ends (see Fig. 3). The 395R template was amplified by a two-step PCR procedure. Briefly, a first-round PCR amplification generated two PCR products (nt 153 to 1643 and nt 1441 to 1637). Each of these products included 20 bp of sequence that included the nucleotides of the left terminal loop. The second round of PCR used equal amounts of these two gel-purified PCR products as templates amplified with the outer pair of first-round primers. After gel purification, this second-round product was used as the transcription template for 395R which includes, from 5′ to 3′, nt 1637 to 1441, followed by the ACAGA loop, then nt 153 to 1643. PCR amplification, using the 395R template, was used to produce templates for transcription of 316R and 298R; 316R includes nt 1597 to 1441, followed by the ACAGA loop, and then nt 153 to 3; 298R includes nt 1589 to 1441, followed by the ACAGA loop, and then nt 153 to 13.
FIG. 3.
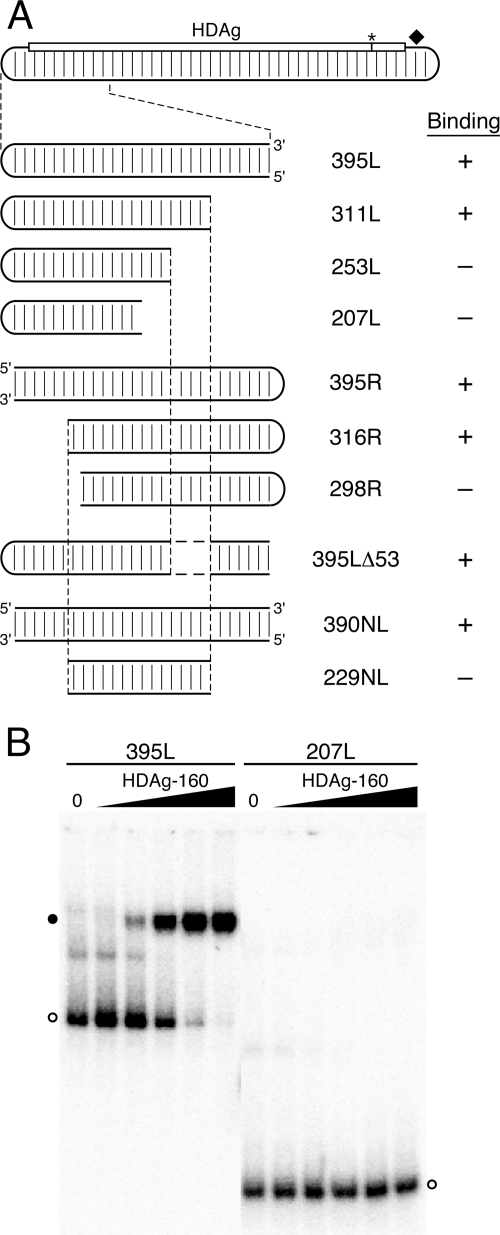
Effects of truncations and internal deletions of the unbranched rod on binding by HDAg-160. (A) Diagram of the HDV unbranched rod as in Fig. 1A. 395L and mutant RNAs are enlarged for clarity. Mutations that maintain the wild-type HDV left terminal loop and are truncated from the right are named 311L, 253L, and 207L. Mutations and truncated RNAs in which the wild-type HDV left terminal loop is moved to the right end of the RNA segment are named 395R, 316R, and 298R. 395LΔ53 contains an internal deletion from nt 110 to 82 and nt 1512 to 1484. Two RNAs have no loop: 390NL and 229NL. RNAs bound by HDAg-160 are indicated by +, and RNAs indicating no binding by HDAg-160 are indicated by −. (B) Electrophoretic mobility shift assays of 5.2 pM 395L and 207L with HDAg-160. The HDAg-160 concentrations were as in Fig. 1D. Free RNAs (open circles) and bound RNAs (closed circles) are indicated.
390NL and 229NL were each made by annealing two RNA transcripts. The two templates for 390NL were amplified to transcribe nt 153 to 1643 and nt 1637 to 1441, and the templates for 229NL were amplified to transcribe nt 111 to 3 and nt 1597 to 1484. In vitro RNA transcripts were purified by native polyacrylamide gel electrophoresis (PAGE). Equimolar amounts were annealed by overnight incubation at 45°C in a mixture of 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.7], 400 mM NaCl, and 1 mM dithiothreitol (DTT) after denaturation at 75°C for 5 min. Annealed RNAs were gel purified again.
The template for transcription of the hybrid 207L + 207M was generated by three-piece PCR. The first round amplified three products from pCMV-DC1×1.2-AgS(−). After gel purification, these three products acted as template for the second round using the outermost primers. This second-round product was used for T7 transcription. 207L plus 207M includes nt 314 (5′) to 213, nt 59 to 1536, and nt 1379 to 1283 (3′).
Transcription reactions included RNA polymerase buffer (New England Biolabs, Ipswich, MA); 10 mM DTT and 1 U/μl RNase inhibitor (Applied Biosystems, Foster City, CA); 500 μM (each) ATP, GTP, and UTP and 32 μM CTP (Invitrogen; Carlsbad, CA); DNA template and 0.8 μCi/μl [α-32P]CTP (Perkin Elmer, Waltham, MA); and 2.5 U/μl T7 RNA polymerase (New England Biolabs). Reaction mixtures were incubated at 37°C no less than 1 h and then chilled on ice no less than 1 h. Native transcripts were resolved on 6% native polyacrylamide gel (Bio-Rad, Hercules, CA) before column purification with Ultrafree-MC polyvinylidene difluoride 0.22-μm and Ultracel YM-30 molecular weight-30,000 cutoff (Millipore, Billerica, MA) filters.
Protein expression and purification.
Open reading frames of HDAg-195, HDAg-160 and HDAg-145 were cloned into the pET30 expression plasmid with an N-terminal His6 tag. Proteins were expressed in Rosetta(DE3) pLysS cells (Novagen, Darmstadt, Germany). A single colony from a newly transformed plate of cells was used to inoculate LB medium, including 10 μg/ml kanamycin and 34 μg/ml chloramphenicol. After growth at 37°C to an optical density at 600 nm of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at final concentration of 0.5 mM; growth was continued overnight at 30°C in a shaking incubator. Following growth, cells were pelleted and all subsequent procedures were conducted at 4°C. Cells were resuspended in native binding buffer (50 mM NaH2PO4, 0.5 M NaCl [pH 8.0], containing 1 mg/ml lysozyme and protease inhibitor cocktail [catalog no. P8340; Sigma, St. Louis, MO]). The cell suspension was incubated on ice for 30 min and then lysed by sonication on ice with six 10-s bursts at high intensity with a 10-s cooling period between each burst. After centrifugation at 3,000 × g for 15 min, the clarified lysate was mixed with Ni2+ ProBond resin (Invitrogen) with gentle agitation for 1 h. The resin was placed in a column and washed with several volumes of native binding buffer containing 100 mM imidazole. His-tagged HDAg was eluted with native binding buffer containing 250 mM imidazole. Purified proteins were dialyzed against 20 mM sodium phosphate buffer, pH 7.2 to 7.5. Protein concentrations were assessed by sodium dodecyl sulfate (SDS)-PAGE and SYPRO (Invitrogen) staining using bovine serum albumin as a standard. Proteins were determined to be greater than 95% pure by analysis of bands on SYPRO-stained SDS-NuPAGE 4 to 12% gradient gels (Invitrogen) (Fig. 1B).
FIG. 1.
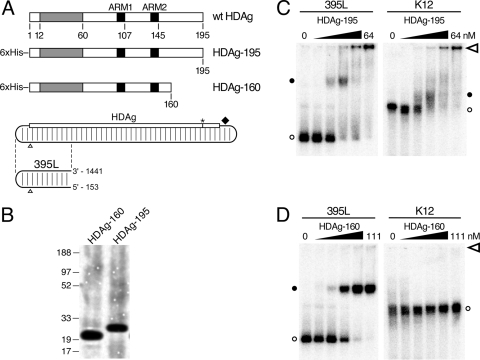
Binding of full-length and truncated HDAg to HDV and non-HDV RNAs. (A) Diagrams of wild-type (wt) HDAg and native bacterially expressed His-tagged HDAg-195 and HDAg-160. Dimerization and oligomerization domains (39) included within aa 12 to 60 are shaded in gray. The arginine-rich motifs (ARM1 and ARM2) are indicated in black (19). The HDV unbranched rod RNA is illustrated as a rounded rectangle with partial base pairing indicated by light vertical lines. The numbering origin is indicated by open triangles. The HDAg open reading frame reads from left to right on the top side of the rod with the amber/W editing site (2) (asterisk) and ribozyme (diamond) to the right. The 395-nt unbranched rod segment (395L) is indicated below the full-length HDV RNA. The genomic coordinates corresponding to the 5′ and 3′ ends of 395L RNA are shown. (B) SDS-PAGE of purified HDAg-195 and HDAg-160 protein preparations. (C) Electrophoretic mobility shift assays of HDV 395L RNA (left) and nonspecific K12 RNA (right) with HDAg-195. The RNA concentrations are each 52 pM. HDAg-195 concentrations increase fourfold from left to right: from 0 to 0.25, 1, 4, 16, and 64 nM. Six percent nondenaturing polyacrylamide gels were run at 250 V for 2 h (395L) or 3.5 h (K12) and are aligned from the top by the location of the well (open arrowhead). Free RNAs (open circles) and bound RNAs (closed circles) are designated. (D) Electrophoretic mobility shift assays of HDV 395L and K12 RNAs with HDAg-160. Conditions were as in panel C, except the HDAg-160 concentrations were 0, 0.04, 0.28, 2.1, 15, and 111 nM, respectively.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as follows. Binding reactions were done in 25 μl including 10 mM Tris-HCl (pH 7.0), 25 mM KCl, 10 mM NaCl, 0.1 μg/μl bovine serum albumin (New England Biolabs), 5% glycerol, 0.5 mM DTT, 0.2 U/μl RNase inhibitor (Applied Biosystems), and 1 mM phenylmethylsulfonyl fluoride solution. The radiolabeled RNA and protein concentrations were as indicated in the figure legends. Reaction mixtures were assembled on ice, incubated at 37°C for 1 h, and electrophoresed on 6% native polyacrylamide gels in 0.5× Tris-borate-EDTA at 240 V for 2.5 h unless otherwise noted. Levels of free and bound RNA were determined by radioanalytic scanning with a Molecular Dynamics Storm 475 phosphorimager. Binding was calculated as the intensity of unbound RNA relative to the intensity of the entire lane minus the background.
Cell transfection.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1 mM glutamine. A total of 3.8 × 105 cells were transfected in suspension with a total of 0.5 μg plasmid DNA and LipofectAMINE Plus (Invitrogen) according to the manufacturer's recommendations. Expression of HDAg was supplied in trans by including 0.02 μg of the HDAg-S-expression plasmid pCMV-AgS (28). All transfections were performed in duplicate. Transfection efficiencies varied by less than 50%.
Northern blot analysis.
Total cellular RNA was harvested 2 and 3 days posttransfection using QIAshredder and the RNeasy minikit (Qiagen, Valencia, CA) following the manufacturer's protocols. RNA (0.5 μg) was electrophoresed through 1.5% agarose gels containing 1.25 M glyoxal, transferred to positively charged nylon membranes, and hybridized with a genomic-sense 32P-labeled probe. RNA quality and loading uniformity were assessed by photography of ethidium bromide staining of rRNAs and by the hybridization signal from 28S RNA. To reduce interference from HDAg mRNA, the HDV probe was hybridized to the noncoding segment of the HDV RNA. Hybridization and washing were performed as described previously (4), except the hybridization temperature was 65°C and the high-stringency, posthybridization wash temperature was 70°C. Relative levels of HDV RNA were determined by radioanalytic scanning of blots with a Molecular Dynamics Storm 475 phosphorimager.
Micrococcal nuclease assay.
Binding reaction mixtures were assembled as described above using 5.2 pM radiolabeled RNA bound by 90 nM HDAg-160 or 300 nM HDAg-160, as indicated. Binding was achieved by incubation at 37°C for 30 min. A total of 0.2 to 200 U micrococcal nuclease (New England Biolabs) prepared in nuclease buffer was added to each reaction mixture, and incubation continued for 15 min. Digestion was stopped by immediate electrophoresis on 6% polyacrylamide gels at 240 V for 2.5 h.
RESULTS
C-terminally truncated HDAg binds a segment of the HDV unbranched rod RNA with high affinity and specificity in vitro.
We characterized the interaction between HDV RNA and HDAg using bacterially expressed N-terminally His-tagged HDAg (Fig. 1A; HDAg-195) in an electrophoretic mobility shift assay. The N-terminal tag does not interfere with HDAg function; Dingle et al. showed that bacterially expressed N-terminally His-tagged HDAg forms complexes with HDV RNA in vitro that can initiate HDV replication following transfection into cultured cells (10). To simplify the analysis of complexes formed, we used a 395-nt RNA, referred to here as 395L, derived from the left end of the antigenomic unbranched rod, rather than the full-length 1,679-nt RNA (Fig. 1A; 395L). Analysis of this RNA sequence with the RNA secondary structure folding algorithm mfold (22, 40) predicts that it forms an unbranched rod structure; after gel purification, greater than 95% migrates as a single band on a nondenaturing polyacrylamide gel. Natively purified HDAg-195 was incubated with 52 pM 32P-labeled 395L RNA for 1 h at 37°C, prior to electrophoresis for 2 h at room temperature in a 6% polyacrylamide gel. We observed that increasing concentrations of HDAg-195 led to the disappearance of unbound 395L RNA and the appearance of more slowly migrating RNA-protein complexes (Fig. 1C, left panel). At a concentration of 1 nM HDAg-195, we observed the appearance of a complex with about one-third the mobility of free 395L RNA. That the intensities of the shifted complexes did not equal those seen with the unbound 395L could indicate that the majority of the RNA is not bound in discrete complexes by HDAg-195. With higher HDAg-195 concentrations (≥16 nM), a high-molecular-weight complex formed that barely entered the gel. The apparent dissociation constant (Kd), determined by nonlinear regression (GraphPad Prism) analysis of the disappearance of unbound 395L, was found to be 1 to 2 nM.
To examine the specificity of HDAg-195 for HDV RNA, we analyzed binding to an RNA derived from the K12 gene of human herpesvirus 8 (11). This RNA is similar in size (393 nt) to 395L, but is not known or predicted to form a rod-like structure. Consistent with the different structure, the migration of this RNA in a native polyacrylamide gel in the absence of protein was significantly slower than that of 395L. We observed that HDAg-195 bound this non-HDV RNA (Fig. 1C, right panel) with affinity indistinguishable from that observed for 395L. Moreover, similar to the behavior of 395L RNA, K12 RNA formed high-molecular-weight complexes with limited or no mobility at HDAg-195 concentrations ≥16 nM. We have observed comparable results with other non-HDV RNAs. Chao et al. observed a similar lack of specificity of HDAg for HDV RNA in a mobility shift assay using a bacterially expressed HDAg-fusion protein (7). Overall, these results indicate that HDAg-195 binds HDV RNA; however there is little or no specificity to the binding, and much of the bound RNA is present in large heterogeneous complexes.
The failure of complexes to migrate into the gel suggests the formation of aggregates at high HDAg-195 concentrations; such aggregate formation could also affect the specificity of binding and complex heterogeneity. We therefore examined the effects of adding various detergents to the binding assay. We found that low concentrations of SDS (e.g., 0.02%) eliminated the formation of complexes that did not enter the gel but did not abolish complex formation. Furthermore, this concentration of SDS resulted in increased specificity of HDAg-195 for HDV RNA (data not shown). These results could indicate that the formation of aggregates promotes nonspecific binding of HDAg-195 to RNA. Possibly, the proline/glycine-rich C-terminal 35 aa of HDAg, which are not known to have any RNA binding activity, could be contributing to aggregate formation. In an attempt to improve the quality of the in vitro RNA binding activity of bacterially expressed HDAg, we removed these C-terminal 35 residues from the coding sequence to express HDAg-160 (Fig. 1A).
We observed that this C-terminal truncation of HDAg substantially improved the binding characteristics of the protein in the electrophoretic mobility shift assay. Unlike HDAg-195, HDAg-160 bound 395L RNA as a single discrete complex, which migrated with about half the mobility of free 395L RNA and slightly faster than the complex formed by HDAg-195 and 395L RNA (Fig. 1D). Greater than 95% of 395L RNA was bound by HDAg-160 in this complex at the highest protein concentration tested (111 nM). No high-molecular-weight complexes with limited or no mobility were observed, even at HDAg-160 concentrations as high as 1.1 μM (data not shown). Binding was highly specific for the unbranched rod structure of HDV RNA; in contrast to full-length HDAg-195, HDAg-160 showed no evidence of binding K12 RNA. Furthermore, we also observed that fully double-stranded RNA formed by two annealed complementary segments of HDV RNA did not bind, nor did segments of HDV RNA that could not form the unbranched rod structure (data not shown). The apparent Kd for HDAg-160 binding to 395L RNA was determined to be 2.46 ± 0.31 nM, similar to that of HDAg-195. Thus, the C-terminal truncation substantially improved the quality of the binding observed without reducing the binding activity of the protein for HDV RNA.
HDAg-160 binds several subgenomic segments of the HDV unbranched rod.
Previous analyses have indicated that HDAg binds to different segments of the HDV genome and antigenome that have the ability to form the unbranched rod structure (7, 20). However, these studies used bacterially expressed fusion proteins that had been denatured prior to binding analysis. Thus, it was not possible to quantitatively compare binding of different RNAs or to characterize the nature of any complexes formed. We therefore analyzed the binding of native HDAg-160 to two additional segments of the antigenomic RNA unbranched rod, 384M and 405E, which are similar in size to 395L (Fig. 2). We also analyzed binding to the genomic RNA segment complementary to 384M, named g384M.
FIG. 2.
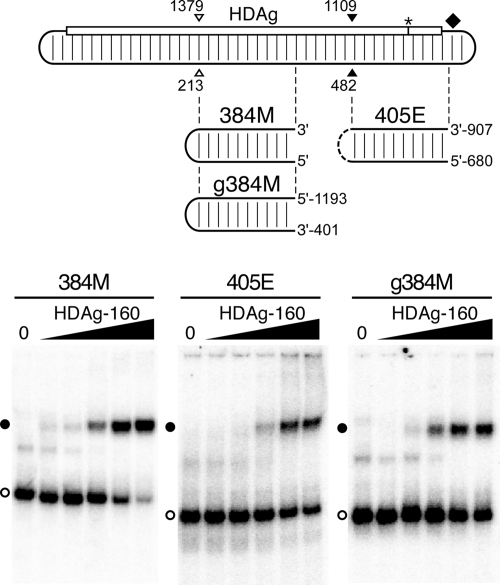
HDAg-160 binds several segments of HDV unbranched rod RNA. (Top) Diagram of the unbranched rod antigenomic RNA, as in Fig. 1A. Open and closed arrowheads indicate the locations of the ApaI and SmaI restriction site sequences, respectively. Locations of 384M, g384M, and 405E RNAs are shown graphically; the genome coordinates of the 5′ and 3′ ends of the g384M and 405E RNAs are shown. (Bottom) Mobility shift assays of 5.2 pM 384M, 405E, and g384M RNAs with HDAg-160. Results are presented as in Fig. 1D, except the HDAg-160 concentrations for 384M are 0, 0.2, 0.5, 2.9, 18, and 111 nM, respectively. Free RNAs (open circles) and bound RNAs (closed circles) are indicated.
We observed that, consistent with the results of Chao et al. and Lin et al. (7, 20), HDAg-160 bound each of these unbranched rod RNAs (Fig. 2). Moreover, all three RNAs formed discrete complexes with HDAg-160 that migrated with mobilities similar to that of the complex formed by 395L and HDAg-160. Nevertheless, in terms of both the affinity and the amount of RNA bound at the highest HDAg-160 concentration, these RNAs exhibited distinct binding behaviors. 395L and g384M (apparent Kds of 2.46 nM and 1.53 nM, respectively) bound about two- to fourfold more tightly than 384M and 405E. Also, while most of the 395L and 384M RNAs were bound by HDAg-160 at the highest concentrations tested, less than half of the 405E and g384M RNAs formed complexes, even at protein concentrations considerably higher than the apparent Kd. Overall, considering both affinity and the maximum amount of RNA bound, 395L, derived from the left end of the antigenome, was the best binder.
The length of the HDV unbranched rod RNA plays a critical role in HDAg-160 binding.
To characterize the minimal determinants within 395L for binding, we created truncated RNAs by shortening the transcription templates from both the 5′ and 3′ ends, such that the predicted unbranched rod structure of the RNA transcripts was preserved. We observed that 207L RNA, 207 nt in length, was not bound by HDAg-160, even at the highest protein concentration tested, 111 nM, which bound >95% of 395L RNA (Fig. 3B). We therefore examined binding of HDAg-160 to a panel of RNAs progressively shortened from the right or the left to identify the structures required for binding. Analysis of RNAs truncated on the right indicated that HDAg-160 efficiently bound 311L RNA, but not 253L (Fig. 3A). In order to examine truncations from the left end of the 395L unbranched rod structure, we first created 395R RNA, in which the HDV terminal loop sequence was moved from the left side of the RNA segment to the right side. Levels of binding of HDAg-160 to RNAs 395L and 395R were indistinguishable, indicating that the context of the loop does not affect protein binding. Mobility shift assays performed on RNAs truncated on the left indicated that HDAg-160 forms a complex with 316R but not with 298R.
The binding results obtained for 311L and 253L indicate that a critical binding feature within 311L has been deleted in 253L. Similarly, binding results for RNAs truncated on the left show that 298R has lost a critical binding feature that is present in 316R. However, the sequences deleted from 253L are present in 298R, and, conversely, the sequences deleted from 298R are present in 253L; yet, neither 253L nor 298R is bound by HDAg-160. Thus, there is no single unique feature that serves as a binding site for HDAg-160 within this HDV segment. Perhaps two binding features are present within 395L RNA—one that includes sequences between the ends of 311L and 253L, and another that includes sequences between the ends of 316R and 298R—and neither one alone is sufficient for binding. Contradicting this explanation, an RNA (395LΔ53) with the same 5′ and 3′ ends as 395L, but lacking the sequences deleted from 311L to create 253L, exhibited high-affinity binding similar to 395L RNA. The determinants of binding were further examined by creating an RNA substrate (229NL) that includes just the 229-nt region common to the two shortest truncations that bind, 311L and 316R. The 229NL RNA was made by annealing two ∼115-nt RNA strands and contains no loop. As a control, we analyzed binding of a 390-nt RNA, 390NL, which also contains two annealed RNA strands, but no loop. While 390NL was bound by HDAg-160 with the same affinity as 395L and 395R, 229NL exhibited no binding by HDAg-160. These data indicate that features critical for binding of 311L and 316R by HDAg-160 are, alone, not sufficient for binding.
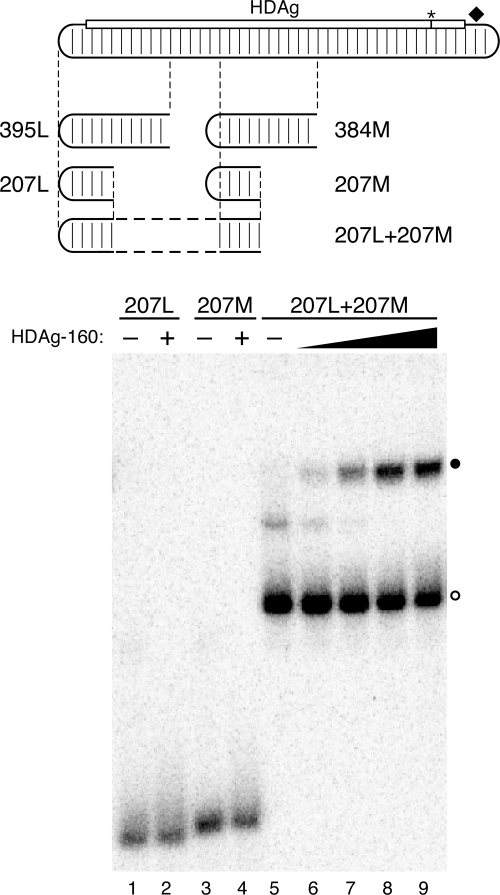
What features, then, determine the high-affinity specific binding of HDAg-160 to HDV RNA? Inspection of the HDV unbranched rod RNAs analyzed in Fig. 3 reveals that all of the RNAs longer than 311 nt were bound by HDAg-160, and all RNAs shorter than 298 nt were not bound. We tested the possibility that binding requires HDV RNA substrates with a minimum length of unbranched rod structure by analyzing the binding of an RNA formed by joining two short, noncontiguous segments of the unbranched rod that are not bound by HDAg-160 (Fig. 4). The short segments used were 207L, which is derived from 395L, and 207M, which is derived from 384M; neither of these 207-nt RNAs formed complexes with HDAg-160 (Fig. 3 and 4). The lack of binding by 207M indicates that the dependence of binding on the size of the RNA is not restricted to the left end of the unbranched rod structure. We made a 405-nt hybrid RNA, 207L+207M, by joining the sequences of 207L and 207M. We observed that this RNA forms a complex with HDAg-160 in the mobility shift assay (Fig. 4). While this binding was not as complete as that of 395L, it was comparable to that shown by other similarly sized unbranched rod RNAs analyzed (Fig. 2). The observed binding of 207L+207M supports the conclusion that HDAg-160 requires a minimum-size unbranched rod structure for binding. Based on the RNA truncations analyzed in Fig. 3, this minimum size is between 298 nt and 311 nt, or approximately 300 nt. Thus, the results presented in Fig. 3 and 4 indicate that binding by HDAg-160 requires an HDV unbranched rod structure of at least about 311 nt.
FIG. 4.
A minimum length of the HDV RNA unbranched rod is required for binding HDAg-160 in vitro. The diagram indicates locations of the 395L, 384M, 207L, and 207M RNAs. Regions of 207M and 207L combined in the 207L+207M hybrid are indicated. Results of electrophoretic mobility shift assays of 5.2 pM 207L, 207M, and 207L+207M RNAs are shown. The HDAg-160 concentrations in lanes 2 and 4 are 1.1 μM. HDAg-160 concentrations increase sevenfold from left to right in lanes 5 to 9: 0, 0.5, 3.3, 23, and 159 nM. Free RNAs (open circles) and bound RNAs (closed circles) are indicated.
Binding of HDAg-160 to HDV RNA in vitro correlates with RNA accumulation in cells expressing HDAg.
To examine the biological relevance of RNP complexes formed by bacterially expressed HDAg-160 in vitro, we analyzed the size-dependent interaction between HDV RNA and HDAg in cells. For this analysis, we relied on the observation that nonreplicating segments of HDV RNA accumulate to higher levels in cells expressing HDAg in trans (17). This accumulation is most likely due to protection of the RNA from cellular nucleases by HDAg binding. Whereas Lazinski et al. studied the effects of HDAg expression on the accumulation of circular RNAs (17), we chose to study whether HDAg could affect the accumulation of linear RNAs, which are less stable in the cell than circular RNAs (29). We used full-length HDAg—without the His6 tag—rather than HDAg-160 in order to examine the dependence of RNA stabilization on RNA size in cells using the most biologically relevant form of HDAg. HEK293T cells were transfected with expression constructs for either 395L or 207L RNA, with or without an expression construct for full-length HDAg. Cellular RNA levels were assessed by Northern blot analysis on days 2 and 3 posttransfection (Fig. 5). We observed that 395L RNA accumulated to approximately three- to fourfold higher levels in cells cotransfected with HDAg (Fig. 5). In contrast, no increase in the amount of 207L RNA was observed in the presence of HDAg. The accumulation of 395L in the presence of HDAg is consistent with the interpretation that this RNA is bound by HDAg in cells and is thereby protected against cellular RNase activity. The lack of any observed accumulation of 207L in the presence of HDAg could indicate that this RNA is rapidly degraded in the cell because it is not bound by HDAg. This interpretation is consistent with the observation that HDAg-160 did not bind this RNA in vitro (Fig. 3). Thus, binding of HDAg-160 to these RNAs in vitro was correlated with their accumulation in the presence of HDAg in cells. This correlation underscores the biological significance of the RNA binding activity of native bacterially expressed HDAg-160.
FIG. 5.
In vitro RNA binding by HDAg-160 is correlated with RNA accumulation in cells expressing HDAg. (A) Northern blot analysis of total cellular RNA harvested 2 and 3 days posttransfection. HEK293T cells were transfected with pCMV-Ribo (empty vector), pCMV-Ribo-395L (395L), or pCMV-Ribo-207L (207L) with or without cotransfection of the HDAg expression plasmid. (A) A representative blot shows RNA harvested 3 days posttransfection. In vitro-transcribed 395L and 207L (lanes 7 and 8) are shown for comparison. (B) Graphic representation of the effect of HDAg expression on the levels of 395L and 207L RNAs in transfected cells. Ratios with and without HDAg were determined by dividing the lane-normalized intensity (using 28S rRNA) obtained in the presence of HDAg by that in the absence of HDAg, after subtraction of background intensity from cells not expressing these RNAs. Results are averages from two independent experiments, each involving duplicate transfections; error bars indicate standard deviations.
The complex formed by HDAg-160 and HDV RNA is resistant to micrococcal nuclease digestion.
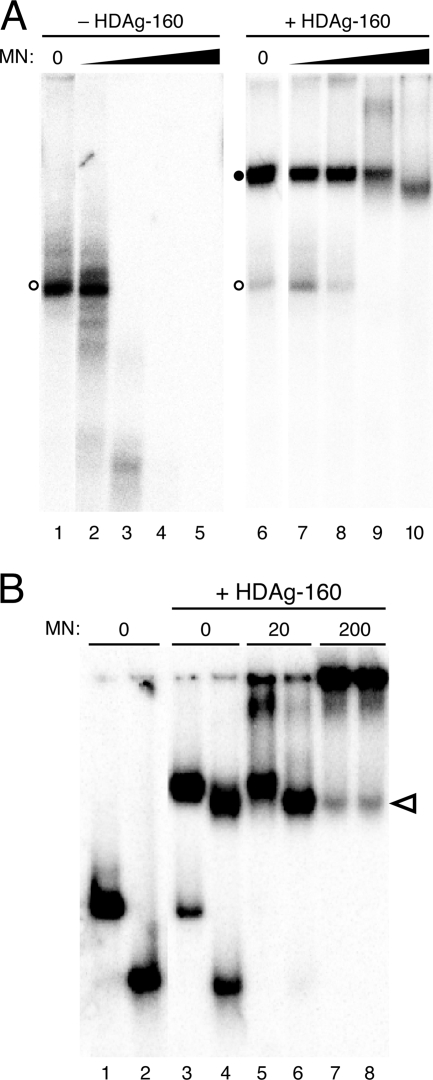
As noted above, the effect of HDAg on the accumulation of 395L RNA in cells could indicate that the complex formed by HDAg and 395L RNA protects the RNA from nuclease activity (17). To examine this possibility in vitro, we examined the micrococcal nuclease sensitivity of the complex formed by HDAg-160 and HDV RNA. HDAg-160 and 395L RNA were allowed to bind and then were incubated with increasing amounts of micrococcal nuclease prior to electrophoresis on a native polyacrylamide gel. We observed that the RNA-protein complex was highly resistant to micrococcal nuclease. Whereas 2 U of nuclease extensively degraded 395L RNA in the absence of HDAg-160, there was no effect on the complex formed by HDAg-160 and 395L (Fig. 6A, lanes 3 and 8). A considerable amount of complexed RNA was still observed in the presence of 20 and even 200 U of the nuclease, whereas unbound RNA was completely degraded. The nuclease resistance of the complex formed by HDAg-160 and 395L is consistent with the conclusion that the accumulation of 395L RNA in cells in the presence of HDAg is due to the protection of bound RNA against cellular nuclease activities. The increased mobility of the complex following treatment with 200 U of enzyme (Fig. 6A, lane 10) is consistent with digestion of portions of the RNA that are not in direct contact with HDAg-160.
FIG. 6.
(A) HDAg-160 protection of 395L RNA from micrococcal nuclease digestion. 395L RNA in the absence (left panel) or presence (right panel) of HDAg-160 was treated with increasing amounts of micrococcal nuclease (MN): lanes 1 and 6, 0 U; lanes 2 and 7, 0.2 U; lanes 3 and 8, 2 U; lanes 4 and 9, 20 U; and lanes 5 and 10, 200 U. Free RNAs (open circles) and bound RNAs (closed circles) are indicated. (B) Micrococcal nuclease digestion of 395L and 311L RNAs complexed with HDAg-160. Shown are the results of mobility shift assays of 395L (odd lanes) and 311L (even lanes) RNAs in the absence (lanes 1 and 2) or presence (lanes 3 to 8) of 300 nM HDAg-160. Amounts of micrococcal nuclease are indicated. The change in migration of 395L complexed with HDAg-160 in the presence of 200 U of micrococcal nuclease (lane 7) is indicated by the open arrowhead.
To more directly examine how HDAg is assembled on the RNA, we compared the mobilities of complexes formed by HDAg-160 and either 395L or 311L, both before and after treatment with increasing concentrations of nuclease. In the absence of micrococcal nuclease, the complex formed by HDAg-160 and 311L migrated just ahead of that formed by 395L (Fig. 6B, lanes 4 and 3, respectively). Similar to the results shown in Fig. 6A, 395L and 311L RNAs that remained unbound were degraded by micrococcal nuclease, but HDAg-160 complexes formed by both RNAs were protected against digestion (Fig. 6B, lanes 5 and 6). With 200 U of nuclease, the intensities of the bands due to both 395L and 311L complexes decreased, possibly due to some degradation of the RNA in these complexes or to the formation of larger complexes with the nuclease (37), as suggested by the increased intensity in the well. Most notable, however, was that the mobility of the complex formed by 395L in the presence of 200 U of micrococcal nuclease increased such that it was identical to that of 311L (Fig. 6B, lanes 7 and 8). Multiple independent repeats of this experiment consistently showed increased mobility of the complex formed by HDAg-160 and 395L with this high concentration of micrococcal nuclease. The mobility of the 311L-HDAg-160 complex was unaffected by the nuclease (Fig. 6B, lanes 4, 6, and 8). Thus, for both RNAs, the size of the complex protected from nuclease digestion is the same as that formed by the smallest RNA bound by HDAg-160. Furthermore, these results suggest that the amounts of HDAg-160 bound are the same for RNAs between 311 nt and 395 nt in length.
DISCUSSION
We have found that a C-terminally truncated form of HDAg, HDAg-160, expressed in E. coli and purified under native conditions, exhibits specific binding to unbranched rod segments of HDV RNA, as indicated by the formation of a discrete complex in an electrophoretic mobility shift assay. The use of bacterially expressed HDAg-160 in binding assays is a significant improvement over bacterially expressed full-length protein, which produced heterogeneously migrating complexes, bound to non-HDV RNAs, and formed aggregates (Fig. 1). Similar to our results with HDAg-195, Chao et al. observed that nondenatured full-length HDAg, expressed as a fusion protein in bacteria, exhibited no clear specificity for HDV RNA (7). Previous studies that have shown specific binding of full-length HDAg to the HDV unbranched rod structure used HDAg expressed in bacteria as a fusion protein that was denatured and renatured prior to RNA binding (7, 20). These previous analyses were limited by potential effects of large fusion protein partners on HDAg structure, the sensitivity of binding to renaturation conditions, and the inability to evaluate the extent or quality of the renatured protein. The nonspecific RNA binding activity of native, full-length, bacterially expressed HDAg is most likely an effect of the bacterial expression. Secondary structure prediction analyses performed on the segment of the protein removed to create HDAg-160 indicated no defined structures. Possibly, in bacterially expressed HDAg-195, this proline/glycine-rich 35-aa region contributes to structural variability and/or aggregate formation that contributes to nonspecific RNA binding activity. Removal of this region is unlikely to directly affect RNA binding; it is not among those regions of the protein implicated in RNA binding by other studies that have used either site-directed mutagenesis or deletional analysis (18-20, 26). An HDAg protein created by a 50 aa C-terminal truncation (HDAg-145) yielded binding results similar to those of HDAg-160, except that the discrete complex formed migrated slightly faster (not shown).
Binding of HDAg-160 was specific for the HDV RNA unbranched rod structure; fully double-stranded RNAs and single-stranded RNAs incapable of forming this structure, even if derived from HDV, were not bound (data not shown). Our deletional analysis of the RNA requirements for binding indicated that binding is not determined by just one or two unique local structural features in the RNA; rather, complex formation in vitro required that the unbranched rod be greater than 298 nt (Fig. 3). We also observed a size-dependent relationship in cells via analysis of RNA stabilization by HDAg that verified the biological relevance of the complexes formed by HDAg-160 (Fig. 5). Previous studies that were able to demonstrate specific binding of HDAg to HDV RNA in vitro indicated that binding required the HDV unbranched rod structure (7, 20), but the RNAs analyzed in those studies were larger than those analyzed here and the dependence of binding on the size of the RNA was not investigated. The length requirement that we have observed for HDV RNA binding to HDAg could be significant for the virus in that it may provide a means by which HDAg discriminates between binding to its cognate RNA rather than shorter similarly structured RNAs (i.e., extended hairpins) in infected cells.
Cells infected by HDV contain both genomic and antigenomic RNAs, as well as two isoforms of HDAg that differ by 19 or 20 aa at the C terminus. Our deletional analysis was conducted using a segment of the antigenomic RNA, but it seems probable that similar size-dependent binding will be obtained for the genome. Chang et al. observed that segments of genomic HDV RNA 311 nt or longer were efficiently packaged into virus-like particles from cells expressing the long isoform of HDAg, while a 258-nt segment was not (5). The nature of the difference in packaging was not further explored at the time, but could be explained by our in vitro binding results: the failure of the shorter 258-nt RNA to be efficiently packaged is likely due to its inability to be bound by HDAg. It is not yet clear whether RNPs with varying HDAg isoform compositions differ structurally. HDAg-160 contains all of the known RNA-binding regions within both isoforms of HDAg.
The results presented in Fig. 2 indicate that HDAg-160 bound several different unbranched RNA segments derived from the HDV antigenome and genome. Previous binding studies, which used denatured bacterially expressed HDAg, were qualitative, and could not distinguish binding characteristics of different RNAs. With natively expressed HDAg-160, we found that binding to different unbranched RNAs was not identical, in terms of either affinity or the maximum levels of binding (Fig. 2). Variations in affinity could be due to subtle structural differences in the RNAs that modulate binding activity. It is not clear why some RNAs failed to be completely bound during the assay. Perhaps, structural heterogeneity in the RNA that affects binding, without affecting gel mobility, is responsible. Another possibility is that binding requires conformational changes—in the RNA, the protein, or both—and the time required for these changes varies among the different RNA-protein interactions. Consistent with these models, we have observed that binding is strongly dependent on both the temperature and time of incubation (D. A. Defenbaugh and J. L. Casey, unpublished data). Regardless of the mechanistic explanation, the variable binding to different segments of the RNA suggests that binding in the context of the full-length unbranched rod RNA might occur preferentially at certain sites. In cells, such preferential binding could lead to an ordered assembly of HDAg on the RNA that is likely to play a role in RNA transcription of both the genome and antigenome as well as packaging of the genome.
HDAg is known to form dimers, and several reports have indicated that the protein assembles into higher-order structures, possibly octamers (9, 35, 38, 39). The results of our electrophoretic mobility shift assays are most consistent with the formation of an HDV RNA-HDAg complex containing a large HDAg multimer, rather than an RNP consisting of either an HDAg monomer or dimer. The mobility of the complex formed by HDAg-160 and 395L, a 395-nt RNA derived from the left end of the antigenomic unbranched rod, was approximately half that of unbound 395L RNA (Fig. 1). The molecular mass of an HDAg-160 monomer is about 19 kDa; complexes formed with 395L and either a single monomer or dimer of HDAg-160 would have molecular masses 15% (149 kDa) to 30% (168 kDa) greater than that of the RNA alone, respectively. On the other hand, a complex involving 395L RNA and an HDAg-160 octamer (152 kDa), for example, would produce an RNA-protein complex with a mass more than twice the size of the RNA alone (282 kDa versus 130 kDa), consistent with the large shift in mobility observed. Of course, in addition to effects of the mass of the protein on RNP mobility, conformational changes in the RNA that might occur upon protein binding could also alter the mobility in a native gel. However, two additional findings support the conclusion that a larger mass of protein is bound. Comparison of the mobility of RNA-protein complexes formed by HDAg-195 and HDAg-160 (Fig. 1) and HDAg-145 (not shown) on a given RNA demonstrates readily detectable changes in mobility, consistent with a large contribution of the protein mass to the mobility of the complex. Furthermore, the resistance of RNA-protein complexes to nuclease digestion (Fig. 6) is more consistent with binding of a large amount of protein to the RNA. Because no RNPs of intermediate mobility were observed as the level of HDAg-160 increased (Fig. 1), the protein either assembles on the RNA with a high degree of cooperativity or binds as a preassembled protein complex. While others have suggested that HDAg exists as an octameric structure in cells and in vitro, even in the absence of HDV RNA (25, 39), our results presented here cannot distinguish between these two possibilities, nor can we determine the number of HDAg subunits involved.
The complex formed between HDAg-160 and 395L RNA was resistant to digestion with micrococcal nuclease. Interestingly, high concentrations of nuclease reduced the size of the complex formed by 395L RNA, which is greater than the minimum-length RNA required for binding, to the same as that formed by 311L RNA, which is the smallest RNA bound by HDAg-160 (Fig. 6). These results indicate that (i) most of the bound RNA is tightly associated with HDAg-160, and (ii) the 395-nt RNA does not bind more protein than the 311-nt RNA. This result, together with the minimum length requirement for binding and our conclusion that HDAg binds as a large multimer, suggests that binding of the multimeric unit occurs in discrete amounts. Thus, given that the minimum length for binding of HDAg-160 to HDV unbranched rod RNA is ∼300 nt, we expect five HDAg-160 multimers are able to bind per full-length HDV RNA. If the multimeric unit is an octamer (as suggested by Zuccola et al. [39]), then we would expect the RNA/protein ratio to be 1:40. This number is near the low end of the range of RNA/protein ratios determined by previous analyses of HDV RNA-protein complexes in cells and in virions (12, 30). In future studies, determination of the exact size of the HDAg multimer involved in binding, as well as the number of such complexes bound to progressively larger RNAs, will permit a more precise evaluation of the composition of the HDV RNA-protein complex.
Acknowledgments
This work was supported in part by grant R01-AI42324 from the National Institutes of Health.
We extend our thanks to Alex Israel for protein preparation.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223450-455. [DOI] [PubMed] [Google Scholar]
- 2.Casey, J. L. 2006. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 30767-89. [DOI] [PubMed] [Google Scholar]
- 3.Casey, J. L. 2002. RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure. J. Virol. 767385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey, J. L., and J. L. Gerin. 1998. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 722806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, M.-F., C.-H. Chen, S.-L. Lin, C.-J. Chen, and S.-C. Chang. 1995. Functional domains of delta antigens and viral RNA required for RNA packaging of hepatitis delta virus. J. Virol. 692508-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 645066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, M., S.-Y. Hsieh, and J. Taylor. 1991. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J. Virol. 654057-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Q., G. C. Jayan, and J. L. Casey. 2003. Differential inhibition of RNA editing in hepatitis delta virus genotype III by the short and long forms of hepatitis delta antigen. J. Virol. 777786-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornillez-Ty, C. T., and D. W. Lazinski. 2003. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 7710314-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle, K., V. Bichko, H. Zuccola, J. Hogle, and J. Taylor. 1998. Initiation of hepatitis delta virus genome replication. J. Virol. 724783-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandy, S. Z., S. D. Linnstaedt, S. Muralidhar, K. A. Cashman, L. J. Rosenthal, and J. L. Casey. 2007. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 8113544-13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudima, S., J. Chang, G. Moraleda, A. Azvolinsky, and J. Taylor. 2002. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 763709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeng, K.-S., P.-Y. Su, and M. M. C. Lai. 1996. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J. Virol. 704205-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323558-560. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, M. Y. P., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 621855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai, M. M. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64259-286. [DOI] [PubMed] [Google Scholar]
- 17.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 682879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 672672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C.-Z., J.-H. Lin, M. Chao, K. McKnight, and M. M. C. Lai. 1993. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 672221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, J.-H., M.-F. Chang, S. C. Baker, S. Govindarajan, and M. M. C. Lai. 1990. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J. Virol. 644051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. C. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 763920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 23.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 206030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 7510161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Malley, B., and D. W. Lazinski. 2005. Roles of carboxyl-terminal and farnesylated residues in the functions of the large hepatitis delta antigen. J. Virol. 791142-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poisson, F., P. Roingeard, and A. Goudeau. 1995. Direct investigation of protein RNA-binding domains using digoxigenin-labelled RNAs and synthetic peptides: application to the hepatitis delta antigen. J. Virol. Methods 55381-389. [DOI] [PubMed] [Google Scholar]
- 27.Polson, A. G., B. L. Bass, and J. L. Casey. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380454-456. [DOI] [PubMed] [Google Scholar]
- 28.Polson, A. G., H. L. Ley III, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 181919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puttaraju, M., A. T. Perrotta, and M. D. Been. 1993. A circular trans-acting hepatitis delta virus ribozyme. Nucleic Acids Res. 214253-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu, W.-S., H. J. Netter, M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 673281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, S., C. Cornillez-Ty, and D. W. Lazinski. 2004. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J. Virol. 788120-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheu, G. T., and M. M. Lai. 2000. Recombinant hepatitis delta antigen from E. coli promotes hepatitis delta virus RNA replication only from the genomic strand but not the antigenomic strand. Virology 278578-586. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, J. M. 2006. Structure and replication of hepatitis delta virus RNA. Curr. Top. Microbiol. Immunol. 3071-23. [DOI] [PubMed] [Google Scholar]
- 34.Wang, H.-W., P.-J. Chen, C.-Z. Lee, H.-L. Wu, and D.-S. Chen. 1994. Packaging of hepatitis delta virus RNA via the RNA-binding domain of hepatitis delta antigens: different roles for the small and large delta antigens. J. Virol. 686363-6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, J.-G., and S. M. Lemon. 1993. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J. Virol. 67446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature 323508-514. [DOI] [PubMed] [Google Scholar]
- 37.Wang, M. J., and P. Gegenheimer. 1990. Substrate masking: binding of RNA by EGTA-inactivated micrococcal nuclease results in artifactual inhibition of RNA processing reactions. Nucleic Acids Res. 186625-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia, Y.-P., and M. M. C. Lai. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 666641-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuccola, H. J., J. E. Rozzelle, S. M. Lemon, B. W. Erickson, and J. M. Hogle. 1998. Structural basis of the oligomerization of hepatitis delta antigen. Structure 6821-830. [DOI] [PubMed] [Google Scholar]
- 40.Zuker, M., and A. B. Jacobson. 1995. “Well-determined” regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res. 232791-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]