Abstract
Resting CD4+ T cells restrict human immunodeficiency virus (HIV) infection at or before reverse transcription, resulting in slower kinetics of reverse transcription. In a previous study, we showed that, despite this restriction at reverse transcription, HIV integration occurs in resting CD4+ T cells, albeit with slower kinetics. In that study, the resting T cells were a mixture of memory and naïve cells. Here we asked whether the more quiescent naïve cell subset could be directly infected by HIV and, if so, whether the level of integration in naïve cells was comparable to that in memory cells. We found that HIV integrates in the naïve subset of resting CD4+ T cells without prior activation of the cells. The level of integration (proviruses/cell) in naïve cells was lower than that in memory cells. This difference between naïve and memory cells was observed whether we inoculated the cells with R5 or X4 HIV and could not be explained solely by differences in coreceptor expression. The presence of endogenous dendritic cells did not change the number of proviruses/cell in memory or naïve cells, and deoxynucleoside pools were equally limiting. Our results instead indicate the existence of a novel restriction point in naïve T cells at viral fusion that results in reduced levels of fusion to naïve CD4+ T cells. We conclude that HIV can integrate into both naïve and memory cells directly. Our data further support our hypothesis that integrated proviral infection of resting T cells can be established without T-cell activation.
Resting CD4+ T cells are the major, best-understood reservoir of latent human immunodeficiency virus (HIV) infection and are a significant barrier to cure because, upon stimulation of the resting cells, they are a source of viremia when antiretroviral therapy is interrupted (29, 37). At the same time, resting CD4+ T cells are considered to be relatively resistant to HIV infection in vitro (35, 58, 61, 69, 70); thus, it is unclear how latent proviruses are established in these cells. The apparent resistance of resting CD4+ T cells to HIV infection in vitro led to the prevailing belief that an activation step is required in order for HIV to integrate into resting cells and therefore that an activation step is required in order for proviruses, when not defective, to establish latency (29, 37).
Resting CD4+ T cells can be subdivided phenotypically into naïve and memory subsets as defined by the expression of multiple surface markers, including CD45RA and CD62L (3, 17). Memory resting CD4+ T cells differ from naïve resting CD4+ T cells in that they have a lower threshold for activation (3, 12, 16, 19, 34). In addition, a subset of memory resting CD4+ T cells express higher levels of the HIV-1 coreceptor CCR5 than do naïve resting CD4+ T cells while naïve cells express slightly higher levels of CXCR4 than do memory cells (6, 38, 67). These differences between these two CD4+ T-cell subsets may have important implications for HIV pathogenesis (56).
Memory cells comprise a greater portion of latent proviral infection than do naïve cells (52), as memory cells contain higher levels of total HIV DNA (7, 18) and integrated HIV DNA (14, 51) than do naïve resting cells in vivo. It is unclear why memory cells are more infected than naïve cells. One favored explanation is that memory cells are infected while in an activated state before returning to quiescence (29, 37) and that the occasional naïve cells infected with HIV in vivo are cells that have reverted from a memory phenotype (3). This explanation has been favored because it was thought until recently that HIV could not directly infect cells in the G0/G1a stage of the cell cycle (29, 37, 60).
Alternatively, direct infection of resting CD4+ T cells may occur, which may lead to latent infection when the provirus is not defective. Recent data suggest that this is possible given that HIV can directly infect and integrate into G0/G1a CD4+ T cells, albeit with slower kinetics (2, 53, 62, 64). Furthermore, massive infection of “apparently” resting memory CD4+ T cells is detected during acute infection (39, 40, 55), suggesting that direct infection of resting cells may occur in vivo. In addition, infected naïve T-cell subsets have been identified in HIV+ individuals, again suggesting that direct infection of resting T cells may occur in vivo (51, 71).
If latent proviral infection can be established in naïve cells without activation, then an alternative mechanism for why memory cells are more infected is required. One possible mechanism is that resting memory cells are inherently more susceptible to direct infection with HIV. This alternative mechanism should be considered given that a subset of memory CD4+ T cells express higher levels of CCR5 (6, 38, 67) and given that memory cells are considered to be in a more active state than naïve cells (3).
While, in our previous studies, we showed that resting CD4+ T cells could be directly infected, we did not separate the cells into naïve and memory subsets and so did not address whether HIV could directly infect the more quiescent naïve cells. It is important to answer these questions because proviral persistence in naïve cells may be regulated by factors different from those in memory cells. Therefore, therapeutic strategies to eliminate infection in naïve cells may be different from strategies that target infection in memory cells.
Here we used an in vitro system in which we infected resting CD4+ T cells with HIV by spinoculation, cultured these cells for 3 days, and then sorted the cells into naïve and memory resting CD4+ T cells and assayed discrete steps in viral replication. We present two novel findings. First, we show that naïve resting CD4+ T cells can be directly infected by X4 HIV without prior activation. Second, we show that memory resting CD4+ T cells are more susceptible to HIV infection than naïve cells in large part because viral fusion occurs more efficiently in memory cells.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses.
The CD4+ T-lymphoblastoid cell line CEM-ss (23, 46) was maintained at 1 × 105 to 5 × 105 cells/ml. The culture medium used was RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 25 mm HEPES, and 100 μg/ml penicillin-streptomycin (Invitrogen Life Technologies). Virions were collected 48 h after calcium-phosphate transfection of 293 T cells. Viral supernatants with peak reverse transcription activity were treated with DNase I at 30 μg/ml (Roche Molecular Biochemicals, Indianapolis, IN) for 1 h at 37°C. pNL4-3 transfection supernatants (1) (donated to the AIDS Reagents Program by Malcolm Martin), pRF1 transfection supernatants, or the CCR5-tropic viral isolate BaL (26) (purchased from the Center for AIDS Research core facility at the University of Pennsylvania) was used to generate the data for Fig. 3 to 5, 7, and 8. pRF1 (a gift from Michael Malim) is equivalent to pYU2 except that a mutation in Vpu is corrected (24). pNL4-3 and pNL-AD8 (25) (donated to the AIDS Reagents Program by Eric O. Freed) were cotransfected with BlaM-Vpr (10, 63) (donated by Michael Miller to the AIDS Reagents Program) to generate virus for Fig. 6. pNL4-3 and pRF1 were cotransfected with vesicular stomatitis virus glycoprotein (VSV-G) (pHIT) (24) to generate data for Fig. 9. Viral potency was based on the level of reverse transcription (second-strand transfer [SST]) in CEM-ss cells 24 h after infection in a single-cycle infection using saquinavir to prevent spreading infection.
FIG. 3.
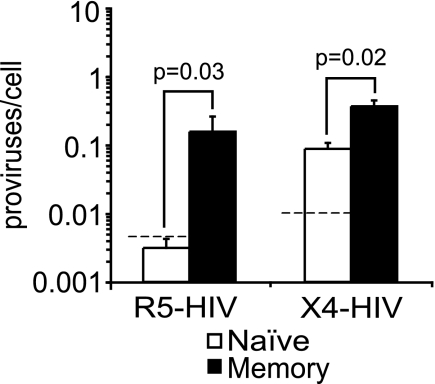
X4 HIV integration is detected in naïve resting CD4+ T cells but at lower levels than in memory resting CD4+ T cells. CD4+ T cells were infected by spinoculation with R5 HIV (n = 2 for pRF1 and n = 2 for BaL) or X4 HIV (pNL4-3), cultured for 3 days, and sorted into naïve and memory resting cells. Integration (proviruses/cell) was measured by Alu-PCR. The dashed line represents the expected level of integration from contaminating memory cells (3% contaminants). The bars represent the averages of four independent R5 experiments and five independent X4 experiments. Error bars represent the standard errors. Integration was measured two to four times in each independent experiment. The P values were calculated with the Wilcoxon rank-sum test.
FIG. 5.
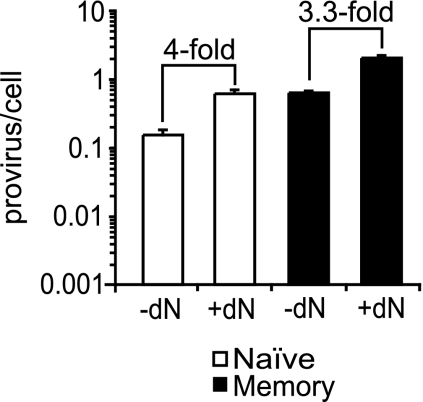
Limiting dNs are not responsible for lower susceptibility of naïve resting cells to HIV integration compared to memory resting CD4+ T cells. CD4+ T cells were infected with pNL4-3 (X4 HIV) by spinoculation, followed by culture for 3 days and then sorting into naïve and memory resting CD4+ T cells. The inoculation and culture of the cells were conducted in the presence or absence of 50 μM of dNs. Integration (proviruses/cell) was measured by Alu-PCR. Error bars represent the standard deviations of three measurements of integration.
FIG. 7.
Both R5 HIV and X4 HIV bind equally to naïve and memory resting CD4+ T cells. Sorted naïve (CD45RA+ CD62L+) and memory (CD45RA− and/or CD62L−) resting CD4+ T cells were spinoculated with BaL (R5 HIV) and pNL4-3 (X4 HIV) for 2 h at 25°C. The level of viral binding was estimated by measuring the level of cell-associated p24Gag by ELISA immediately after inoculation and after removal of unbound virions. The graph represents the averages of a total of six measurements of viral binding from three independent inoculations. Error bars represent the standard errors. The P values were calculated with the Wilcoxon rank-sum test.
FIG. 8.
HIV reverse transcription is less efficient in naïve than in memory resting CD4+ T cells. The efficiencies of integration (proviruses/SST) are similar in both naïve and memory resting CD4+ T cells. CD4+ T-cell and DC mixtures were infected with an R5 HIV (BaL for two experiments and pRF1 for two experiments) or an X4 HIV (pNL4-3) (five experiments) by spinoculation, cultured for 3 days, and then sorted into naïve and memory resting CD4+ T cells. (A) Late reverse transcription was measured by quantitative PCR, using primers that detect the SST of reverse transcription. (B) The ratios of integration to total DNA (proviruses/SST) were obtained by dividing the data in Fig. 3 by the data in panel A. Error bars represent the standard errors of four (R5) and five (X4) independent experiments. The P values were calculated with the Wilcoxon rank-sum test.
FIG. 6.
Fusion of R5- and X4-tropic HIV to naïve cells is less efficient than that to memory resting CD4+ T cells. CD4+ T-cell and DC mixtures were spinoculated with either pNL-AD8 (R5 HIV) (B) or pNL4-3 (X4 HIV) (D) particles that carry BlaM-Vpr. After inoculation the cells were loaded with the dye CCF2-AM. Uninfected cells (A and C) loaded with CCF2-AM were used as a negative control for each inoculation. After infection of the cells with R5 HIV (B) or X4 HIV (D), naïve and memory resting CD4+ T cells were gated for cells that had taken up the green dye and fusion was measured based on the percentage of cells that fluoresced blue. Fusion was measured independently among the naïve (CD45RA+ CD62L+) and memory (CD45RA− and/or CD62L−) subsets of resting CD4+ T cells (CD69, CD25, and HLA-DR negative). This experiment is representative of four.
FIG. 9.
VSV-G-pseudotyped HIV infects both naïve and memory resting CD4+ T cells with similar efficiencies. CD4+ T cells were infected with RF1 (R5 HIV) or pNL4-3 (X4 HIV) particles pseudotyped with VSV-G by spinoculation, followed by culture and sorting into naïve and memory resting CD4+ T cells. Integration was measured as described for Fig. 3 to 5. Error bars represent the standard errors of two independent experiments. The P values were calculated with the Wilcoxon rank-sum test. The X4 HIV(VSV-G) used for these experiments is five times more potent than the R5 HIV(VSV-G) as assessed by the level of reverse transcripts present in CEM-ss cells 24 h after a single-round infection.
Reagents.
The following phycoerythrin (PE)-, fluorescein isothiocyanate-, peridinin chlorophyll protein-, allophycocyanin (APC)-, or APC-Cy7-conjugated monoclonal antibodies were purchased from Becton Dickinson (Sunnyvale, CA): CD45RA, CD3, CD62L, HLA-DR, CD69, CD25, CXCR4, CCR5, and Ki67. PE-conjugated BDCA-1, BDCA-4, and anti-PE magnetic beads were obtained from Miltenyi Biotec Inc. (Auburn, CA). Saquinavir was used at 1.25 μM (Roche US Pharmaceuticals). Deoxynucleosides (dNs) were purchased from Sigma-Aldrich (St. Louis, MO). The CCF2-AM staining kit used for measuring HIV fusion was purchased from Invitrogen.
Preparation and purification of CD4+ cells and T-cell subsets.
CD4+ cells were isolated by negative selection from leukapheresis-enriched peripheral blood mononuclear cells (PBMC) using the RosetteSep kit (Stem Cell Technologies, Inc.) as recommended by the manufacturer. Briefly, leukapheresis-enriched PBMC were labeled with antibodies recognizing glycophorin A, CD8, CD16, CD19, CD36, CD56, CD66b, and T-cell receptor γ/δ. A secondary anti-mouse antibody was added to cross-link labeled PBMC to labeled red blood cells. Following these labeling steps, the cells were applied to a Ficoll-Paque (GE Healthcare) density gradient to remove the cross-linked cell complexes. This purification step normally yields a CD4+ T-cell and dendritic cell (DC) mixture. DC-depleted CD4+ cells were then isolated by negative selection with saturating concentrations of PE-labeled antibodies specific for BDCA-1 and BDCA-4 and anti-PE magnetic beads as recommended by the manufacturers. Immediately after DC depletion, the levels of DCs present were determined using antibodies against BDCA-1, BDCA-4, and CD3. After 3 days of culture of the spinoculated cells, T-cell subsets were isolated from the cultures with or without DC depletion. To do this, the cells were labeled with FITC-labeled CD45RA, PE-labeled HLA-DR, CD25, CD69, APC-labeled CD62L, and APC-Cy7-labeled CD3. Naïve and memory T cells were sorted, using a BD FACS-Vantage cell sorter, based on CD3, CD62L, and CD45RA expression and lack of activation markers (HLA-DR, CD69, and CD25).
Flow cytometry.
To determine the level of the coreceptors CCR5 and CXCR4 and Ki67, we sort purified an aliquot of naïve and memory cells on the day of inoculation. The coreceptor levels were determined by surface staining in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline with 1% bovine serum albumin and 2 mM EDTA) and by blocking nonspecific binding with mouse immunoglobulin G. For cell proliferation marker Ki67 staining, the purified T-cell subsets were first fixed with 70% ethanol, followed by intracellular staining with PE-labeled Ki67 as recommended by the manufacturer. The results were analyzed with FlowJo software (Treestar).
HIV infection of human primary cells.
All the cells were cultured in RPMI 1640 medium containing 10% heat-inactivated FBS, supplemented with 2 mM l-glutamine, 100 μg/ml penicillin-streptomycin, and 25 mM HEPES prior to infection. Cells diluted at 1 × 107 cells/ml in viral supernatant were spinoculated at 1,200 × g for 2 h at 25°C as similarly described (49). When testing the effect of dNs on integration, cells were resuspended with both the appropriate virus and a dN mixture at 50 μM (at equal contents of each dN) and spinoculated. After spinoculation, the cells were washed three times to remove unbound virions, resuspended in medium in the presence of 1.25 μM saquinavir and 50 μM of dNs (when indicated), and incubated at 37°C.
Real-time PCR analysis for viral DNA intermediates.
DNA was prepared after HIV infection using the QIAmp blood and cell culture kit (Qiagen, Valencia, CA). Real-time PCR was performed to detect long reverse transcripts (completed SST), integrated HIV DNA, and β-globin as described previously (2, 48, 62).
Fusion assay.
Transfection supernatants of pNL4-3 or pNL-AD8 viruses containing BlaM-Vpr were utilized to inoculate pure resting CD4+ T cells by spinoculation at 1,200 × g for 2 h at 4°C. After inoculation, the cells were washed twice with cold phosphate-buffered saline plus 10% FBS to remove unbound virions. Cells were then resuspended in RPMI plus 10% FBS and incubated at 37°C for 30 min to induce viral fusion. After fusion, cells were washed twice with CO2-independent medium (Invitrogen) without serum and without antibiotics. A stock CCF2-AM loading solution was prepared for staining the cells: 2 μl of CCF2 at 200 μg/ml and 8 μl of 100-mg/ml Pluronic-F127 surfactant in dimethyl sulfoxide and 0.1% acetic acid, diluted to a total of 1 ml with CO2-independent medium without antibiotics or serum. One million cells were resuspended in 100 μl of CCF2 loading solution and incubated at room temperature for 1 h to allow dye uptake. Cells were subsequently washed with CO2-independent medium, resuspended in CO2-independent medium containing 10% FBS and 2.5 mM of probenecid (Sigma-Aldrich), and incubated at 25°C for ∼12 h. After incubation at 25°C, the cells were washed once with FACS buffer and stained for the following markers: activation markers (HLA-DR, CD69, and CD25), CD4, CD62L, and CD45RA. Stained cells were then analyzed by multicolor flow cytometry using a BD LSR II flow cytometer.
Measuring viral binding.
Viral binding was estimated by measuring the level of cell-associated p24Gag immediately after inoculation using a p24-specific enzyme-linked immunosorbent assay (ELISA; Perkin-Elmer). The approximate number of viral particles bound per cell was calculated as previously described (2, 49).
RESULTS
Purification of memory and naïve cells.
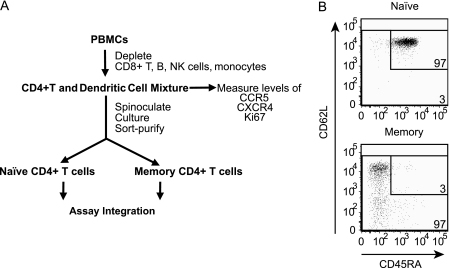
In this study, our approach to determining the relative susceptibilities of memory and naïve cells differed from our prior approaches, in that we sort purified the cells 3 days after inoculation. Starting with PBMC, we isolated a mixed population of CD4+ T cells and DCs, spinoculated this mixed population with HIV, cultured cells for 3 days, and then sort purified naïve and memory resting CD4+ T-cell subsets and assayed for intermediates of HIV infection (Fig. 1A). Specifically, we obtained PBMC from healthy donors using an institutional review board-approved protocol and negatively selected against CD8+ and T-cell receptor γ/δ T cells, CD36+ monocytes, CD56+ and CD16+ NK cells, CD19+ B cells, and CD66b+ granulocytes using a rosette method (RosetteSep kit; Stem Cell Technologies, Inc.). This negative selection protocol left us with a CD4+ T-cell and DC mixture. This cell mixture was spinoculated with either X4 or R5 HIV and cultured for 3 days. After 3 days of culture, the CD4+ T-cell and DC mixture was sort purified into memory and naïve resting CD4+ T cells by selecting against activation markers (HLA-DR, CD69, and CD25) and by CD45RA and CD62L expression patterns. Our approach sorted naïve and memory cells, as demonstrated by the postsort analysis (Fig. 1B): 3% of the sorted naïve cells (CD45RA+ and CD62L+) were memory cells, and 3% of the sorted memory cells (CD45RA− and/or CD62L−) were naïve cells.
FIG. 1.
(A) Outline of purification, infection, and sorting strategy of memory and naïve resting CD4+ T cells. (B) After resting CD4+ T cells were sorted, naïve cells contained 3% contaminating memory cells and memory cells contained 3% contaminating naïve cells. The CD4+ cells were sorted by selecting for CD3+ cells that lacked activation markers (HLA-DR, CD69, and CD25) and had the pattern of expression of CD45RA and CD62L as shown. Fluorescence-minus-one controls were used to place the L-shaped gate that measures the purity of naïve and memory resting cells. The fluorescence-minus-one gates were placed so that 1% of unstained cells appeared as stained. Less than 1% of the cells were endogenous activated (not shown) as defined in our prior papers (2, 62).
We chose the approach of sorting on day 3 postinoculation based on CD62L and CD45RA expression for several reasons. First, by omitting positive selection of memory and naïve T cells before inoculation, we prevented unnecessary stimulation of the cells, which could influence the susceptibility of the cells to HIV infection. Second, this strategy allowed us to ask later whether the presence of DCs enhances infection of naïve or memory CD4+ T cells. Lastly, in the two studies which showed that in vivo memory cells have higher levels of integrated HIV DNA than naïve cells, CD62L and CD45RA expression was used to define memory and naïve CD4+ T-cell subsets (14, 51).
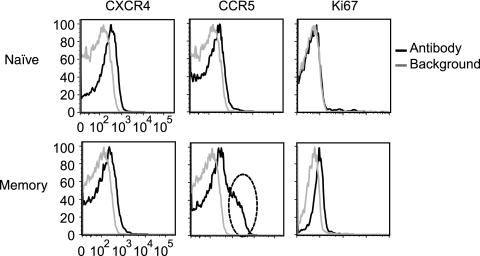
We also wanted to determine the expression levels of CXCR4, CCR5, and Ki67 on naïve and memory cells at the time of inoculation to determine the levels in our system. We were also interested to determine if significant levels of Ki67 were expressed in our purified naïve and memory resting CD4+ T cells, since this has been reported as a marker for actively cycling cells (8, 21, 22). To do this, we removed an aliquot of the CD4+ T-cell and DC mixture prior to spinoculation and sort purified the cells into naïve and memory resting CD4+ T cells (using CD62L and CD45RA, CD3, and activation markers). We then stained the sorted aliquot with antibodies against surface CXCR4 and CCR5 and intracellular Ki67 and analyzed the level of expression in memory and naïve resting CD4+ T cells (Fig. 1A). Consistent with other reports (6, 27, 38), we found that the level of CXCR4 was slightly higher on naïve cells (Fig. 2). We also found that a subset of memory cells expressed higher levels of CCR5 as previously described (5, 6, 38, 65) (Fig. 2). Ki67 expression was undetectable on naïve cells and present at very low levels in memory cells (Fig. 2).
FIG. 2.
Expression levels of CCR5, CXCR4, and Ki67 in memory and naïve resting CD4+ T cells. The level of expression of the HIV coreceptors CCR5 and CXCR4 and the level of expression of the cellular activation markers Ki67 were assessed by flow cytometry. A subset of memory cells (dashed oval) expressed higher levels of CCR5 than did most memory cells. Minimal expression of the cell cycle marker Ki67 was detected in the memory subset, and no expression was detected in the naïve cells. Unstained cells were used to show the background level for CXCR4 and CCR5 staining. An isotype control was used to determine the background level for the Ki67 intracellular staining.
Higher levels of integration in memory cells than in naïve cells.
In comparing naïve and memory resting cells, the first step of the viral life cycle that we investigated was integration. As described for Fig. 1A, we inoculated cells with HIV, cultured them for 3 days, sort purified naïve and memory cells, and then quantified the level of integration (2, 53, 62). We assayed integration after the inoculated cells were cultured for 3 days because we previously showed that the level of integration plateaus at that time point (2, 53, 62). We found that integration was detectable in naïve cells inoculated with X4-tropic HIV (pNL4-3) (Fig. 3). The level of integration in R5- and X4-inoculated memory cells was higher than that in naïve cells (Fig. 3). When we averaged the ratio of memory to naïve cell integration levels, we found on average 36-fold-higher integration with R5 HIV (but the ratio ranged from 10- to 73-fold higher) and 4-fold-higher integration with X4 HIV. We cannot determine conclusively whether R5 HIV can directly integrate into naïve cells, because the signal is close to the signal expected from contaminating memory cells. Nonetheless, we can conclude that X4 HIV directly integrates into naïve cells since 25% contaminating memory cells would be required to account for the signal obtained in the naïve subset.
Since integration occurred at higher levels in memory cells, we wanted to test two possible explanations. One is that DCs have been reported to enhance CD4+ T-cell infection (66) and, thus, DCs present during the inoculation and culture might preferentially activate the memory cells. Furthermore, it is known that the requirements for activation of memory cells differ from the requirements of naïve cells (16). The other is that nucleotide levels may be more limiting in naïve cells than in memory cells (53).
Next, we asked whether the difference in integration levels between memory and naïve cells was altered by coculturing CD4+ T cells with DCs. To test this, we modified the protocol described for Fig. 1A in the following ways: after preparing a CD4+ T-cell and DC mixture, we either depleted or did not deplete DCs (defined as BDCA-1+ and BDCA-4+ cells). Before depletion, DCs comprised 0.89% of the CD4+ T-cell and DC mixture; after depletion DCs comprised ∼0.02% of the mixture (Fig. 4A). The two cell preparations were then spinoculated with HIV, cultured for 3 days, and sort purified into memory and naïve resting subsets, and the level of integrated DNA was measured. We found that depletion of DCs had no apparent effect on the integration levels of R5 or X4 virus in either naïve or memory CD4+ T cells (Fig. 4B). Therefore, we conclude that the presence of endogenous, unstimulated DCs does not affect the susceptibility of naïve or memory resting CD4+ T cells and does not account for the differences in the susceptibility of memory and naïve cells to HIV infection.
FIG. 4.
The presence of endogenous, unstimulated DCs does not influence the susceptibility of resting naïve or memory cells to HIV integration. (A) FACS analysis of spinoculated cells before and after DC depletion. CD4+ T-cell and DC mixtures were exposed to anti-BDCA-1 and anti-BDCA-4 PE-labeled antibodies followed by depletion of BDCA-1+ and BDCA-4+ DCs with anti-PE beads. The cells were then analyzed for the presence of DCs and T cells by staining with anti-BDCA-1, anti-BDCA-4, and anti-CD3 antibodies. (B and C) CD4+ cells depleted or not depleted of endogenous DCs were infected with BaL (R5 HIV) (B) or pNL4-3 (X4 HIV) (C) by spinoculation. At 3 days postinoculation, resting naïve and memory cells were sorted and integration (proviruses/cell) was measured by Alu-PCR. Error bars represent the standard deviations of three to four measurements of integration. The P values were calculated with the Wilcoxon rank-sum test. Similar results were obtained when the DCs were depleted with anti-HLA-DR instead of BDCA-1 and BDCA-4 (not shown).
Because we had previously demonstrated that the addition of dNs enhanced integration in resting CD4+ T cells (53), here we asked whether adding dNs enhanced integration in naïve cells only or in both memory and naïve resting CD4+ T cells. In a modification of the protocol shown in Fig. 1A, we either added or did not add dNs to the CD4+ T-cell and DC mixture at the time of spinoculation. After 3 days of culture, the level of integration was assayed in sort-purified naïve and memory resting CD4+ T cells. We observed that the addition of dNs increased the level of integration in both populations similarly, about fourfold (3.3-fold for memory and 4.0-fold for naïve cells) (Fig. 5). We concluded that the relative difference between naïve and memory cells was not altered by the addition of dNs and that the difference in susceptibility cannot be attributed to nucleotide pools (Fig. 5).
Higher levels of fusion to memory cells than to naïve cells.
Given that neither dN pools nor the presence of DCs could explain the higher levels of HIV integration in memory than in naïve cells (Fig. 3), we next asked which steps were more restricted in naïve cells. We decided to start from the beginning of the life cycle and test the level of fusion and then the level of reverse transcription in both cell types. To compare HIV fusion in memory and naïve cells, we used the BlaM-Vpr fusion assay (10, 63). In this assay, virions are loaded with β-lactamase that is covalently linked to Vpr and the cells are loaded with the β-lactamase substrate CCF2-AM, a dye that fluoresces green. When β-lactamase-containing virions fuse to CCF2-dyed cells, β-lactamase cleaves CCF2, causing a change in cellular fluorescence from green to blue that can be detected and quantitated by FACS (10). Notably, this assay measures fusion and not viral uncoating, since CCF2 can still be cleaved in intact viral cores (11). In other words, BlaM-Vpr will be able to cleave CCF2 after viral fusion regardless of whether or not the virus has uncoated. To validate the assay, we measured viral fusion to resting CD4+ T cells in the presence or absence of the fusion inhibitor enfurvitide (T-20) (see the figure in the supplemental material). When we compared viral fusion between memory cells and naïve cells using R5 HIV, we found that virus fused to 0.8% of naïve cells and fused to 5.3% of memory cells (Fig. 6). Percent fusion was calculated by subtracting the background percent blue cells in uninfected cells from the percent blue cells in infected cells (e.g., for naïve cells, 1.3 − 0.5 = 0.8%, and for memory cells, 5.8 − 0.5 = 5.3%). When we performed this experiment using X4 HIV, we found that 5.8% of naïve cells fused with the HIV virions and 21.9% of memory cells fused with the HIV virions (Fig. 6).
Because viral binding precedes viral fusion, the differences that we observed in viral fusion between naïve and memory cells could have been due to differences in viral binding, although this is unlikely since the two cell types express similar levels of CD4. To address this possibility, we estimated the level of viral binding to naïve and memory resting CD4+ T cells by measuring the level of cell-associated p24Gag immediately after inoculation. Similar to the data for Fig. 1B, naïve and memory resting CD4+ T cells were sort purified from CD4+ T cells based on the expression of CD45RA and CD62L and the absence of the activation markers CD25, CD69, and HLA-DR. Following sort purification, naïve and memory resting cells were spinoculated with R5 or X4 HIV. After inoculation, the cells were washed to remove unbound virions and the level of cell-associated p24Gag was measured by ELISA. We found that the level of viral binding to naïve cells was equal to the level of binding to memory cells for both R5 and X4 HIV (Fig. 7), suggesting that the difference in viral fusion that we see between naïve and memory cells cannot be attributed to differences in viral binding. We note that fusion may occur during our spinoculation procedure at 25°C, especially to memory CD4+ T cells, and that this may affect the level of viral binding that we detect. However, given that we saw similar viral binding to both memory and naïve cells, any effect of viral fusion during spinoculation on binding is likely to be below the detection limit of our binding assay.
In summary, compared to naïve cells, we detected sevenfold-more fusion in R5-inoculated memory cells and fourfold-more fusion in X4-inoculated memory cells. Given that naïve cells express slightly more CXCR4 than memory cells, we were surprised to find that X4 viruses fused more efficiently to memory cells than to naïve cells. From these data, we concluded that fusion of both X4 and R5 HIV occurs more readily to memory than to naïve resting CD4+ T cells. In other words, it appears that naïve cells are relatively restricted at fusion.
Higher levels of reverse transcription in memory cells than naïve cells.
Finally, we wanted to assess if there are differences in HIV reverse transcription efficiency between naïve and memory resting CD4+ T cells that could contribute to the differences in susceptibility of these cell subsets to HIV infection. To do this, we compared the efficiency of HIV reverse transcription between memory cells and naïve cells, by measuring the level of reverse transcription that has completed the SST and by calculating the fraction of those reverse transcripts that integrated (proviruses/SST or integrated HIV DNA/total HIV DNA). We found on average 20-fold-more R5 reverse transcripts and fourfold-more X4 reverse transcripts in memory cells than in naïve cells (Fig. 8A). This difference paralleled that seen with integration (Fig. 3).
Reverse transcription may also occur less efficiently in naïve than in memory cells. However, the differences in reverse transcription efficiency between memory and naïve cells, if present, are modest and so are difficult to quantitate.
When we calculated the fraction of reverse transcripts that integrated in the naïve and memory subsets (proviruses/SST, by dividing the data in Fig. 3 by the data in Fig. 8A), we found that the fractions were comparable in the two subsets with both X4 and R5 HIV infection (Fig. 8B). There was no statistical difference between memory and naïve resting CD4+ T cells when we averaged experiments (Fig. 8B, n = 4 for R5 and n = 5 for X4). This finding suggests that the susceptibility to infection of memory and naïve CD4+ T cells is not determined at the level of integration and that integration is not impaired in naïve T cells.
VSV-G-pseudotyped HIV integrates into the genome of naïve and memory resting CD4+ T cells to similar extents.
We then hypothesized that if memory cells are “more infected” because naïve cells are restricted at fusion, then pseudotyping HIV with VSV-G envelope should overcome this restriction. This reasoning was based on the expectation that VSV-G-pseudotyped HIV would fuse to naïve and memory cells to similar extents given the ubiquitous nature of the VSV-G receptor (15). First, we tested if pseudotyping HIV with VSV-G enhanced infection of naïve cells. To do this, we spinoculated the CD4+ T-cell and DC mixture with R5 HIV(VSV-G) or X4 HIV(VSV-G), cultured the cells for 3 days, sorted the cells into subsets, and then assayed integration. We found that the levels of integration of R5 or X4 HIV(VSV-G) in naïve and memory cells were comparable and not statistically different (Fig. 9) (P = 0.4 for R5 virus and P = 0.2 for X4 virus). The observation that pseudotyping HIV with VSV-G resulted in comparable levels of integrated DNA strongly suggests that naïve cells, relative to memory cells, are largely restricted at HIV fusion and possibly modestly restricted at reverse transcription. Stated another way, memory cells are inherently more susceptible to HIV fusion by both X4 and R5 viruses.
DISCUSSION
In the present study, we show for the first time that X4 HIV can directly integrate into the genomes of naïve resting CD4+ T cells without prior activation of the cells. Although prior work demonstrated that resting CD4+ T cells are susceptible to HIV integration (2, 53, 62, 64) and although it is known that naïve cells represent 30 to 70% of CD4+ T cells in blood, it was possible that all the integrated DNA was in the memory subset. Thus, the susceptibility of naïve cells to HIV integration compared to memory cells was unclear. Consistent with in vivo data, the memory resting CD4+ T cells are inherently more susceptible to HIV infection than naïve resting CD4+ T cells. We also show for the first time that the enhanced susceptibility of memory cells is due, in large part, to more efficient HIV fusion to memory than to naïve resting CD4+ T cells (Fig. 6). Reverse transcription may also occur slightly more efficiently in memory resting cells, but this effect is modest. Integration, on the other hand, occurs with similar efficiencies in the two cell types with both R5 and X4 viruses based on ratios of proviruses to SST (Fig. 8B), suggesting that integration is not impaired in naïve resting cells compared to memory resting cells. In addition, we show that dNs enhance infection in the two cell types to similar extents, suggesting that nucleotide pools are equally limiting in the two cell types. Finally, we show that coculture of endogenous unstimulated blood DCs does not enhance infection of either naïve or memory CD4+ T cells.
Our approach was to sort resting CD4+ T cells into naïve and memory subsets 3 days after inoculation and then to assay integration in naïve and memory cells. This approach prevented unnecessary stimulation of the resting cells by cross-linking antibodies prior to infection. By carefully accounting for the contaminating memory cells in the purified naïve cell fractions, in Fig. 3, we were able to show that X4 and R5 HIVs were able to directly integrate into naïve resting CD4+ T cells. We acknowledge that, in some experiments (for example, Fig. 4), the lower levels of integration of R5 in the naïve cells could be due to residual contaminating memory cells. However, our data clearly demonstrate that X4 viruses were able to directly integrate into naïve resting CD4+ T cells and that this signal was not due to contaminating memory cells.
Role of DCs.
We also showed that coinoculation of endogenous blood DCs does not enhance the level of HIV integration in naïve or memory cells. This is in agreement with studies by Hladik et al. (33) but appears to contradict prior studies (66). However, in many of the studies reviewed (67), trans-infections rather than coinfections were performed. In other words, the DCs were usually first isolated and then activated, followed by exposure to HIV, and finally cocultured with T cells. Our studies and the studies by Hladik et al. (33) looked at the role of DCs during coinoculation without prior activation of the DCs and found no enhancement of infection of the T cells through the presence of DCs. Thus, cell-mediated stimuli are not required for infection of resting naïve cells.
Role of nucleotide levels.
We also explored the possibility that the reduced level of integration seen in naïve cells could have been due to lower nucleotide pools in these cells, given that naïve cells are considered to be more quiescent (3). We previously showed that dN addition enhances the level of integration (53). However, we found that supplying dNs exogenously enhanced integration of the two cell types equally. This suggests that nucleotides are limiting in both cell types and does not explain the difference in susceptibility between naïve and memory T cells.
Role of activation status.
We were interested in determining whether Ki67 levels on cells correlated with susceptibility to HIV infection. We found that HIV can integrate into the genomes of Ki67-negative, noncycling naïve cells. Low levels of Ki67 were detected on resting memory cells, consistent with the idea that these cells are slightly more activated than naïve cells or at a slightly different stage in the cell cycle. It has been reported that memory cells are in G1a and naïve cells in G0 based on levels of RNA (3). We speculate that the level of Ki67 may correlate with a T cell's susceptibility to HIV, although the significance is unclear.
Fusion.
We also showed that the difference in infection between memory and naïve cells was largely due to fusion efficiency (Fig. 6 to 9). We did this by measuring viral fusion immediately after inoculation. For example, X4 viruses fused four times more efficiently to memory cells than to naïve cells, while R5 viruses fused seven times more efficiently to memory cells than to naïve cells. Importantly, one would expect X4 viruses to fuse to naïve cells more than memory cells given that naïve cells express slightly more CXCR4 than do memory cells (6, 38, 67) (also Fig. 2), but the opposite occurs. Finally, we confirmed that the higher susceptibility of memory cells to HIV infection is mostly due to more efficient viral fusion compared to naïve cells, by pseudotyping HIV particles with VSV-G envelope. We found that the difference in HIV integration level between memory and naïve cells was greatly reduced when particles carried VSV-G envelope, which is thought to have a ubiquitous receptor.
Potential mechanism for enhanced fusion to memory cells.
In the case of R5 infection, the increased susceptibility of memory cells could simply be due to higher CCR5 levels, but in the case of X4 infection, other explanations are required since the level of CXCR4 is slightly higher on naïve cells. One explanation could be that naïve cells may express different conformations of the chemokine receptor (28) that may not allow fusion as efficiently as the conformations found in memory cells. Another explanation is that some form of virus-induced cellular stimulation could enhance fusion and naïve cells may require a higher threshold of virus-mediated stimulation (3, 12, 16, 19). For example, more HIV receptors or coreceptors may need to be engaged for fusion to naïve cells to occur. Finally, engaging the coreceptors may enhance fusion through signaling to a greater extent in naïve cells than in memory cells. Most studies suggest that coreceptor signaling is not necessary for HIV fusion (4, 9, 50, 54, 68), but one recent study suggests that signaling can play a role in one model of HIV envelope-mediated cell-to-cell fusion (30).
Here we addressed the relative susceptibility of naïve and memory resting CD4+ T cells to HIV infection and which replication step determines susceptibility. Prior studies compared the relative susceptibilities of naïve and memory cells only after activating the cells to divide and studied only relative levels of reverse transcription and production (20, 36, 44, 57, 59). We found that, consistent with simian immunodeficiency virus and HIV in vivo infection (39, 40, 42, 43), memory cells are more susceptible to fusion with both X4 and R5 viruses, but HIV can directly infect and integrate into the genomes of both cell types. These experiments support our previous finding that cells in G0/G1a can be infected by HIV (2, 62). We emphasize that our results do not disagree with prior work showing restrictions at reverse transcription and integration within 24 h of infection (13, 35, 36, 61, 69, 70). We detect significant levels of integration only 2 to 3 days after infection (47, 53). In other words, the restriction in resting CD4+ T cells alters the tempo of infection but not the final outcome—integration of HIV into the host genome. Our results provide further support for an alternative mechanism for how HIV proviruses establish latent infection—by direct infection of resting CD4+ T cells.
Our results are consistent with the model that proviruses may establish latent infection by direct infection of resting CD4+ T cells (2, 53, 62, 64). Thus, proviruses may establish latent infection in the absence of an activation step. One argument for a model of direct infection of resting CD4+ T cells is that the ratios of infected memory and naïve cells in the blood of HIV-infected individuals and in our in vitro model are similar: approximately ∼16-fold-more integrated DNA is found in memory cells from HIV-infected individuals (51). Another argument for direct infection is that resting cells have a much longer half-life than activated cells (31, 32, 41, 45). Notably, naïve resting CD4+ T cells have a longer half-life than memory resting CD4+ T cells, which in turn have a longer half-life than activated CD4+ T cells. Infected resting CD4+ T cells are likely to continue to have a survival advantage over infected activated CD4+ T cells, since activated CD4+ T cells produce more infectious virus than resting CD4+ T cells, which likely shortens the life span of the infected activated cells to a greater extent than that of the infected resting cells (32, 55, 71, 72). Furthermore, the predominance of infected “apparently” resting memory cells in acute infection suggests that direct infection of resting cells occurs in vivo (39, 43). Thus, our data, taken together with T-cell turnover data and in vivo studies of acute infection, suggest that direct infection of naïve and memory resting CD4+ T cells by HIV may be an important mechanism whereby HIV proviruses establish latent infection in vivo.
Supplementary Material
Acknowledgments
We thank Richard Carroll, Avinash Bhandoola, and Liz Colston for critical reading of the manuscript and Wei Peter Yang for his helpful advice on statistical analysis. We thank the flow cytometry core facility at Penn for sorting uninfected samples and Paul Hallberg at the cell sorting core facility at the Children's Hospital of Philadelphia for sorting infected samples. We thank Penn CFAR for providing the R5 strain BaL and for p24 testing. We thank the AIDS Research and Reference Reagent Program for providing pNL4-3, pNL-AD8, and BlaM-Vpr.
This work was supported by the NIH grants R01-AI058862-06 and R01AI058862-04S1.
Footnotes
Published ahead of print on 11 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agosto, L. M., J. J. Yu, J. Dai, R. Kaletsky, D. Monie, and U. O'Doherty. 2007. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 36860-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berard, M., and D. F. Tough. 2002. Qualitative differences between naive and memory T cells. Immunology 106127-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Ann. Rev. Immunol. 17657-700. [DOI] [PubMed] [Google Scholar]
- 5.Biancotto, A., J. C. Grivel, S. J. Iglehart, C. Vanpouille, A. Lisco, S. F. Sieg, R. Debernardo, K. Garate, B. Rodriguez, L. B. Margolis, and M. M. Lederman. 2007. Abnormal activation and cytokine spectra in lymph nodes of people chronically infected with HIV-1. Blood 1094272-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 941925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, J. Yazdani, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 781160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullwinkel, J., B. Baron-Luhr, A. Ludemann, C. Wohlenberg, J. Gerdes, and T. Scholzen. 2006. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J. Cell. Physiol. 206624-635. [DOI] [PubMed] [Google Scholar]
- 9.Busillo, J. M., and J. L. Benovic. 2007. Regulation of CXCR4 signaling. Biochim. Biophys. Acta 1768952-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 201151-1154. [DOI] [PubMed] [Google Scholar]
- 11.Cavrois, M., J. Neidleman, W. Yonemoto, D. Fenard, and W. C. Greene. 2004. HIV-1 virion fusion assay: uncoating not required and no effect of Nef on fusion. Virology 32836-44. [DOI] [PubMed] [Google Scholar]
- 12.Chandok, M. R., and D. L. Farber. 2004. Signaling control of memory T cell generation and function. Semin. Immunol. 16285-293. [DOI] [PubMed] [Google Scholar]
- 13.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 14.Chun, T.-W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y.-H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387183-188. [DOI] [PubMed] [Google Scholar]
- 15.Coil, D. A., and A. D. Miller. 2004. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 7810920-10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft, M. 1994. Activation of naive, memory and effector T cells. Curr. Opin. Immunol. 6431-437. [DOI] [PubMed] [Google Scholar]
- 17.De Rosa, S. C., L. A. Herzenberg, and M. Roederer. 2001. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 7245-248. [DOI] [PubMed] [Google Scholar]
- 18.Douek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 41795-98. [DOI] [PubMed] [Google Scholar]
- 19.Dutton, R. W., L. M. Bradley, and S. L. Swain. 1998. T cell memory. Annu. Rev. Immunol. 16201-223. [DOI] [PubMed] [Google Scholar]
- 20.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 15671-682. [DOI] [PubMed] [Google Scholar]
- 21.Endl, E., and J. Gerdes. 2000. The Ki-67 protein: fascinating forms and an unknown function. Exp. Cell Res. 257231-237. [DOI] [PubMed] [Google Scholar]
- 22.Endl, E., C. Hollmann, and J. Gerdes. 2001. Antibodies against the Ki-67 protein: assessment of the growth fraction and tools for cell cycle analysis. Methods Cell Biol. 63399-418. [DOI] [PubMed] [Google Scholar]
- 23.Foley, G. E., H. Lazarus, S. Farber, B. G. Uszman, B. A. Boone, and R. E. McCarthy. 1965. Continuous culture of human lymphoblasts from periphral blood of a child with acute leukemia. Cancer 18522-529. [DOI] [PubMed] [Google Scholar]
- 24.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 164531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 693949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233215-219. [DOI] [PubMed] [Google Scholar]
- 27.Groot, F., T. M. van Capel, J. Schuitemaker, B. Berkhout, and E. C. de Jong. 2006. Differential susceptibility of naive, central memory and effector memory T cells to dendritic cell-mediated HIV-1 transmission. Retrovirology 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta, S. K., and K. Pillarisetti. 1999. Cutting edge: CXCR4-Lo: molecular cloning and functional expression of a novel human CXCR4 splice variant. J. Immunol. 1632368-2372. [PubMed] [Google Scholar]
- 29.Han, Y., M. Wind-Rotolo, H. C. Yang, J. D. Siliciano, and R. F. Siliciano. 2007. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 595-106. [DOI] [PubMed] [Google Scholar]
- 30.Harmon, B., and L. Ratner. 2008. Induction of the Gαq signaling cascade by the human immunodeficiency virus envelope is required for virus entry. J. Virol. 829191-9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellerstein, M., M. B. Hanley, D. Cesar, S. Siler, C. Papageorgopoulos, E. Wieder, D. Schmidt, R. Hoh, R. Neese, D. Macallan, S. Deeks, and J. M. McCune. 1999. Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat. Med. 583-89. [DOI] [PubMed] [Google Scholar]
- 32.Hellerstein, M. K., R. A. Hoh, M. B. Hanley, D. Cesar, D. Lee, R. A. Neese, and J. M. McCune. 2003. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Investig. 112956-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hladik, F., P. Sakchalathorn, L. Ballweber, G. Lentz, M. Fialkow, D. Eschenbach, and M. J. McElrath. 2007. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity 26257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inaba, K., J. P. Metlay, M. T. Crowley, M. Witmer-Pack, and R. M. Steinman. 1990. Dendritic cells as antigen presenting cells in vivo. Int. Rev. Immunol. 6197-206. [DOI] [PubMed] [Google Scholar]
- 35.Korin, Y. D., and J. A. Zack. 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J. Virol. 723161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreisberg, J. F., W. Yonemoto, and W. C. Greene. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10525-531. [DOI] [PubMed] [Google Scholar]
- 38.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 965215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 4341148-1152. [DOI] [PubMed] [Google Scholar]
- 40.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 4341093-1097. [DOI] [PubMed] [Google Scholar]
- 41.McLean, A. R., and C. A. Michie. 1995. In vivo estimates of division and death rates of human T lymphocytes. Proc. Natl. Acad. Sci. USA 923707-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehandru, S., M. A. Poles, K. Tenner-Racz, V. Manuelli, P. Jean-Pierre, P. Lopez, A. Shet, A. Low, H. Mohri, D. Boden, P. Racz, and M. Markowitz. 2007. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J. Virol. 81599-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mengozzi, M., M. Malipatlolla, S. C. De Rosa, L. A. Herzenberg, and M. Roederer. 2001. Naive CD4 T cells inhibit CD28-costimulated R5 HIV replication in memory CD4 T cells. Proc. Natl. Acad. Sci. USA 9811644-11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michie, C. A., A. McLean, C. Alcock, and P. C. L. Beverley. 1992. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360264-265. [DOI] [PubMed] [Google Scholar]
- 46.Nara, P. L., and P. J. Fischinger. 1988. Quantitative infectivity assay for HIV-1 and -2. Nature 332469-470. [DOI] [PubMed] [Google Scholar]
- 47.O'Doherty, U. 2005. Mechanisms of human immunodeficiency virus-1 latency. Transfusion 4588S-91S. [DOI] [PubMed] [Google Scholar]
- 48.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative, assay for human immunodeficiency virus type 1 integration. J. Virol. 7610942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 7410074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oppermann, M. 2004. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell. Signal. 161201-1210. [DOI] [PubMed] [Google Scholar]
- 51.Ostrowski, M. A., T.-W. Chun, S. J. Justement, I. Motola, M. A. Spinelli, J. Adelsberger, L. A. Ehler, S. B. Mizell, C. W. Hallahan, and A. S. Fauci. 1999. Both memory and CD45RA+/CD62L+ naive CD4+ T cells are infected in human immunodeficiency type 1-infected individuals. J. Virol. 736430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierson, T., T. L. Hoffman, J. Blankson, D. Finzi, K. Chadwick, J. B. Margolick, C. Buck, J. D. Siliciano, R. W. Doms, and R. F. Siliciano. 2000. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 747824-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plesa, G., J. Dai, C. Baytop, J. L. Riley, C. H. June, and U. O'Doherty. 2007. Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J. Virol. 8113938-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popik, W., and P. M. Pitha. 2000. Exploitation of cellular signaling by HIV-1: unwelcome guests with master keys that signal their entry. Virology 2761-6. [DOI] [PubMed] [Google Scholar]
- 55.Reilly, C., S. Wietgrefe, G. Sedgewick, and A. Haase. 2007. Determination of simian immunodeficiency virus production by infected activated and resting cells. AIDS 21163-168. [DOI] [PubMed] [Google Scholar]
- 56.Ribeiro, R. M., M. D. Hazenberg, A. S. Perelson, and M. P. Davenport. 2006. Naive and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J. Virol. 80802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riley, J. L., B. L. Levine, N. Craighead, T. Francomano, D. Kim, R. G. Carroll, and C. H. June. 1998. Naive and memory CD4 T cells differ in their susceptibilities to human immunodeficiency virus type 1 infection following CD28 costimulation: implications for transmission and pathogenesis. J. Virol. 728273-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 692877-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spina, C. A., H. E. Prince, and D. D. Richman. 1997. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J. Clin. Investig. 991774-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9853-860. [DOI] [PubMed] [Google Scholar]
- 61.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 91551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swiggard, W. J., C. Baytop, J. J. Yu, J. Dai, C. Li, R. Schretzenmair, T. Theodosopoulos, and U. O'Doherty. 2005. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J. Virol. 7914179-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tobiume, M., J. E. Lineberger, C. A. Lundquist, M. D. Miller, and C. Aiken. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 7710645-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vatakis, D. N., G. Bristol, T. A. Wilkinson, S. A. Chow, and J. A. Zack. 2007. Immediate activation fails to rescue efficient human immunodeficiency virus replication in quiescent CD4+ T cells. J. Virol. 813574-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veazey, R. S., and A. A. Lackner. 2005. HIV swiftly guts the immune system. Nat. Med. 11469-470. [DOI] [PubMed] [Google Scholar]
- 66.Wu, L., and V. N. KewalRamani. 2006. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat. Rev. Immunol. 6859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 1851681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoder, A., D. Yu, L. Dong, S. R. Iyer, X. Xu, J. Kelly, J. Liu, W. Wang, P. J. Vorster, L. Agulto, D. A. Stephany, J. N. Cooper, J. W. Marsh, and Y. Wu. 2008. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61213-222. [DOI] [PubMed] [Google Scholar]
- 70.Zack, J. A., A. M. Haislip, P. Krogstad, and I. S. Y. Chen. 1992. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J. Virol. 661717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Z.-Q., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 2861353-1357. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Z. Q., S. W. Wietgrefe, Q. Li, M. D. Shore, L. Duan, C. Reilly, J. D. Lifson, and A. T. Haase. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 1015640-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.