Abstract
One characteristic of all positive-strand RNA viruses is the necessity to assemble viral RNA replication complexes on host intracellular membranes, a process whose molecular details are poorly understood. To study viral replication complex assembly we use the established model system of Flock House virus (FHV), which assembles its replication complexes on the mitochondrial outer membrane. The FHV RNA-dependent RNA polymerase, protein A, is the only viral protein necessary for genome replication in the budding yeast Saccharomyces cerevisiae. To examine the host components involved in protein A-membrane interactions, an initial step of FHV RNA replication complex assembly, we established an in vitro protein A membrane association assay. Protein A translated in vitro rapidly and specifically associated with mitochondria isolated from yeast, insect, and mammalian cells. This process was temperature dependent but independent of protease-sensitive mitochondrial outer membrane components or the host mitochondrial import machinery. Furthermore, lipid-binding studies revealed that protein A preferentially bound to specific anionic phospholipids, in particular the mitochondrion-specific phospholipid cardiolipin. These studies implicate membrane phospholipids as important host determinants for FHV RNA polymerase membrane association and provide evidence for the involvement of host phospholipids in positive-strand RNA virus membrane-specific targeting.
Positive-strand RNA viruses make up a large and diverse group of microbial pathogens responsible for significant clinical diseases in humans. The number of available vaccines and antiviral therapies targeted toward these viruses is limited, and thus the study of common molecular mechanisms involved in positive-strand RNA virus pathogenesis is important for the identification of novel candidate antiviral targets. One universal characteristic of all positive-strand RNA viruses is the necessity to assemble viral RNA replication complexes on host intracellular membranes. Viral replication complexes have been found associated with a variety of intracellular membranes, including the endoplasmic reticulum (ER), endosomes, peroxisomes, chloroplasts, and the mitochondria (32). To facilitate the formation of viral RNA replication complexes, individual viruses encode replicase proteins containing membrane-specific targeting signals, several of which have been characterized in detail (6, 29, 31, 45, 47). However, to date the host components present on organelle-specific membranes that facilitate targeting and assembly are poorly defined, and only a limited number of cellular membrane components have been shown to be important in viral RNA replication complex assembly (16, 35, 37). The identification of host factors necessary for this essential step in the positive-strand RNA virus life cycle will enhance our understanding of the molecular mechanisms of replication complex assembly and may highlight possible targets for future antiviral therapies.
To investigate the role of host factors in viral RNA replication complex assembly, we use the established and versatile model alphanodavirus, Flock House virus (FHV). FHV is the best-studied member of the Nodaviridae family and has been used extensively to investigate positive-strand RNA virus replication (7, 14, 15, 30, 31, 40, 41), host innate immunity (9, 24), and virus assembly (26, 48). FHV virions contain a copackaged bipartite genome consisting of RNA1 (3.1 kb) and RNA2 (1.4 kb), which encode protein A, the viral RNA-dependent RNA polymerase, and the capsid protein precursor, respectively. Additionally, during active RNA replication a small subgenomic RNA3 (0.4 kb) colinear with the 3′ end of RNA1 is produced which encodes protein B2, a potent inhibitor of the innate immunity RNA interference pathway (9, 24). FHV RNA replication is supported in cells derived from a wide variety of genetically tractable hosts, including Drosophila melanogaster (30), Caenorhabditis elegans (24), and the budding yeast Saccharomyces cerevisiae (31, 40, 41), which makes FHV a unique model virus to study the molecular mechanisms of host-pathogen interactions that control RNA replication complex assembly.
FHV protein A is the only viral protein necessary for RNA replication complex assembly, a process that takes place on the mitochondrial outer membrane (29-31). Protein A is targeted to the mitochondrial outer membrane via an amino-terminal targeting signal (29). This signal resembles those present in signal-anchored proteins that are targeted to the mitochondrial outer membrane (4, 20, 27, 56), suggesting that FHV may use established mitochondrial targeting and trafficking machinery for assembly. Mitochondrial protein import has been studied extensively in yeast in part due to the facile genetics of this model organism and the development of an easily manipulated in vitro import system (23, 46, 54, 58). The majority of mitochondrial proteins are translated from nuclear-encoded genes within the cytosol and therefore must be properly trafficked and targeted to their final mitochondrial compartments. The import of most proteins into mitochondria is facilitated by the translocase of the mitochondrial outer membrane (TOM) complex, a large macromolecular structure consisting of the main import component, Tom40, three receptors, Tom20, Tom22, and Tom70, and three accessory proteins, Tom5, Tom6, and Tom7 (8, 38, 42). However, it has recently been shown that some mitochondrial outer membrane proteins containing a signal anchor amino-terminal domain, including the NADH-cytochrome b5 reductase isoform encoded by the yeast MCR1 gene, can use an undefined TOM complex-independent mechanism for membrane targeting and insertion (27).
In this report, we investigated the role of host mitochondrial outer membrane components in the binding and insertion of protein A to mitochondria, an initial step of FHV RNA replication complex assembly. To facilitate these studies we established an in vitro protein A translation and membrane association system with which we were able to recapitulate many of the in vivo biochemical characteristics of protein A. We found that protein A rapidly and specifically associated with yeast, insect, and mammalian mitochondria after in vitro translation and furthermore acquired resistance to alkaline extraction, consistent with its characteristic as an integral membrane protein in vivo (29, 30). Protein A membrane insertion was independent of protease-sensitive outer membrane components and the main component of the TOM complex, Tom40. However, we found that protein A associated efficiently with anionic phospholipids, and in particular the mitochondrion-specific anionic phospholipid cardiolipin. These studies provide insight into the early steps of FHV RNA replication complex assembly and implicate organelle membrane lipids in the specific intracellular targeting of positive-strand RNA virus replication.
MATERIALS AND METHODS
Yeast strains, Drosophila cells, and culture conditions.
Yeast strains used in this study are listed in Table 1. For isolation of mitochondria, yeast were grown in YPG (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 3% glycerol) or YPD (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% dextrose) medium. Drosophila S2 cells were grown in Schneider's Drosophila medium with 10% heat-inactivated fetal bovine serum at 25°C as previously described (18). Mammalian baby hamster kidney (BHK) cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 5% bovine growth serum, 10 units per ml penicillin, and 10 μg per ml streptomycin.
TABLE 1.
S. cerevisiae strains used in the study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4743 (WT) | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0 | ATCC |
| Δtom70 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0 tom70Δ/tom70Δ | ATCC |
| YPH499 (WT) | MATα ade2-101 his3Δ200 leu2Δ1 ura3-52 trp1-Δ63 lys-801 | 58 |
| MM112-C (Δtom20) | MATα ade2-101 his3Δ200 leu2Δ1 ura3-52 trp1-Δ63 lys-801 tom20::URA3 (Yep13-TOM22) | 58 |
| KKY3.7 (WT) (TK8) | MATα his3-Δ200 leu2-3,112 ade2-101 suc2-Δ9 trp1-Δ901 ura3-52 tom40::HIS3 (pRS314-Tom40) | 58 |
| KKY3.3 (TK4) tom40-3 | MATα his3-Δ200 leu2-3,112 ade2-101 suc2-Δ9 trp1-Δ901 ura3-52 tom40::HIS3 (pRS314-Tom40-3) | 2 |
| KKY3.6 (TK7) tom40-6 | MATα his3-Δ200 leu2-3,112 ade2-101 suc2-Δ9 trp1-Δ901 ura3-52 tom40::HIS3 (pRS314-Tom40-6) | 58 |
Plasmids and antibodies.
Standard molecular biology techniques were used for the production of all plasmids. The protein A in vitro expression plasmid pIVT-PA was generated by placing the open reading frame from pS2FA (18) into the EcoRI/XbaI sites of pCMV-TnT (Promega, Madison, WI). The β-galactosidase (β-Gal) in vitro expression plasmid pIVT-LacZ was generated by placing the SpeI/AgeI fragment from pMT/V5/LacZ (BD Biosciences, San Jose, CA) into the AvrII/BspEI sites of pIVT-PA. The protein A-green fluorescent protein (GFP) chimera in vitro expression plasmid pIVT-PA1-46/GFP was generated by placing the PstI/BamHI fragment from pPA1-46/GFP (29) into the same sites of pGEM-4Z (Promega). The GFP in vitro expression plasmid pIVT-GFP was generated by first placing the PstI/HindIII fragment from pGal-YFP into the same sites of pGEM-4Z to create pIVT-YFP, followed by placing the BspEI/HindIII fragment from pPA1-46/GFP (29) into the same sites of pIVT-YFP. The in vitro expression plasmid encoding the ATP/ADP carrier protein (AAC) was generously provided by F. Ulrich Hartl (Max-Planck-Institute of Biochemistry, Germany). In vitro expression plasmids encoding Tom20, porin, or His6-tagged pSu9-dihydrofolate reductase (DHFR) have been previously described (2, 27). We expressed and purified pSu9-DHFR from Escherichia coli BL21 cells by nickel affinity chromatography as previously described (2, 27). Mouse monoclonal antibodies against yeast porin were purchased from Molecular Probes (Carlsbad, CA). Rabbit polyclonal antibodies against yeast Tim44 or Tom40 were generously provided by Donna M. Gordon (University of Pennsylvania) or Nikolaus Pfanner (University of Freiburg, Freiburg, Germany), respectively. Rabbit polyclonal antibodies against yeast Tom20 and Tom70 have been previously described (2, 27).
Mitochondrial isolation and in vitro membrane association.
Yeast mitochondria were isolated as previously described (28) and stored in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM 3-morpholinopropanesulphonic acid [MOPS]-KOH, pH 7.2) at −80°C. Drosophila S2 cell and BHK mitochondria were isolated by mechanical disruption and differential centrifugation as previously described (11, 18). mRNAs were generated by in vitro transcription with SP6 or T7 RNA polymerase (Epicentre, Madison, WI) from pGEM-4Z- or pCMV-TnT-based vectors, respectively. Initial in vitro translation experiments with wheat germ, S. cerevisiae, and D. melanogaster cell-free lysates failed to produce detectable full-length protein A (data not shown), and thus we used commercially available nuclease-treated rabbit reticulocyte lysates (RRL; Promega) for all subsequent experiments. Proteins for in vitro assays were translated using RRLs in the presence of 400 μCi/ml [35S]methionine-cysteine (Amersham) with optimized amounts of in vitro-transcribed mRNAs for 1 h at 25°C. Translation mixtures (5 μl per 100-μl reaction mixture) were incubated with isolated mitochondria (50 μg total protein per 100-μl reaction mixture) in import buffer (250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 10 mM MOPS-KOH [pH 7.2], 0.25 mg/ml bovine serum albumin, 2 mM ATP, 2 mM NADH) at 25°C. Import reaction mixtures were diluted with 4 volumes of SEM buffer, and mitochondria were pelleted by centrifugation at 12,000 × g for 15 min, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% [wt/vol] glycerol, 0.001 mg/ml bromophenol blue, 5% 2-mercaptoethanol), and heated to 95°C for 5 min. For carbonate extractions, pelleted membranes were resuspended in 0.1 M sodium carbonate (pH 11.5) and incubated on ice for 30 min. Extracted membranes were reisolated by centrifugation at 55,000 × g for 30 min at 4°C and resuspended in sample buffer. Porin and AAC protein import were analyzed by incubating mitochondria with 100 μg per ml proteinase K for 5 min on ice followed by the addition of 1 mM phenylmethanesulfonyl fluoride. Mitochondria were reisolated by centrifugation at 12,000 × g for 15 min and resuspended in sample buffer. Denatured samples were separated by SDS-PAGE, and gels were fixed in 25% isopropanol-10% acetic acid, incubated in 1 M sodium salicylate for 1 h, dried under vacuum, and exposed to autoradiography film. Digital images were obtained using an Alpha Innotech Fluorchem 8900 and quantified with AlphaEase FC software, and final images were prepared with Adobe Photoshop software. All contrast adjustments to the final images were done prior to cropping.
Flotation of membranes and liposomes.
In vitro membrane association reaction mixtures were brought to 300 μl with flotation buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 50 mM KCl, 2 mM MgCl2, pH 7.5] and mixed with 900 μl 50% Nycodenz solution to yield a final concentration of 37.5% Nycodenz. Samples were loaded under 3.2 ml of 35% and 0.2 ml of 5% Nycodenz solutions and centrifuged at 134,000 × g for 20 h at 4°C in a Beckman MLS-50 swing bucket rotor. Fractions of equal volumes were collected and analyzed by SDS-PAGE and fluorography as described above. E. coli and porcine brain total lipid extracts as well as purified 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC), 1,2-dioleoyl-sn-glycero-3-[phospho-l-serine] (PS), l-α-phosphatidylinositol (PI), 1,2-dioleoyl-sn-glycero-3-phosphate (PA), 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (PG), 1,1′,2,2′-tetraoleoyl cardiolipin (CL), and 1-oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl]-sn-glycero-3-phosphocholine (NBD-PC) were purchased from Avanti Polar Lipids (Alabaster, AL). Chloroform solvent was removed under vacuum centrifugation for 1 h and lipids were resuspended in HEPES buffer (20 mM HEPES-KOH, pH 7.2) by vortexing at 4°C for 1 h. Lipids (250 μg per 100-μl reaction mixture) were incubated with in vitro translation mixtures in protein import buffer for 30 min at 25°C, diluted to 1 ml with 85% sucrose in HEPES buffer to a final sucrose concentration of 71.25%, overlaid with 2.5 ml 65% sucrose and 1 ml 10% sucrose in HEPES buffer, and centrifuged at 134,000 × g for 20 h at 4°C in a Beckman MLS-50 swing bucket rotor. Gradients were fractionated and analyzed as described above.
Statistics.
We used a two-tailed Student's t test, assumed equal variances for comparative statistical analyses, and considered a P value of <0.05 significant. Unless otherwise indicated all quantitative data represent results from at least three independent experiments and are presented as the means ± standard errors of the means.
RESULTS
In vitro-translated FHV protein A associates specifically with mitochondria.
FHV assembles RNA replication complexes on the mitochondrial outer membrane (29, 30). To investigate the initial steps of replication complex assembly and identify host components responsible for protein A membrane association we developed an in vitro mitochondrial binding assay, similar to the in vitro system used to study the import and assembly of endogenous mitochondrial proteins (23, 46, 58). We initially attempted to purify full-length FHV RNA polymerase produced in E. coli for in vitro experiments, but protein A produced with this heterologous system was highly insoluble (D. Miller, unpublished data). Thus, we explored an in vitro translation system, in which FHV protein A was synthesized from a synthetic mRNA template in nuclease-treated RRL. Initial studies with a carboxy-terminal epitope-tagged construct and subsequent immunoblotting with epitope tag-specific antibodies demonstrated that full-length protein A was produced (data not shown). For all subsequent experiments 35S-radiolabeled protein was used to enhance detection sensitivity.
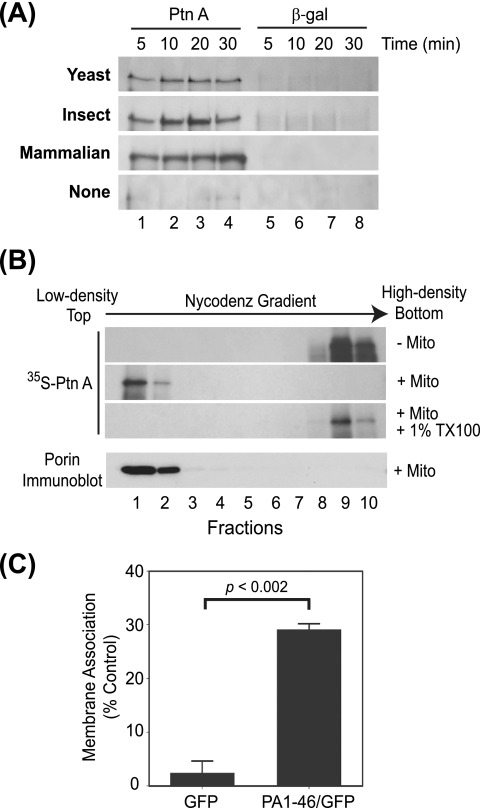
We incubated in vitro translation mixtures with mitochondria isolated from cells capable of supporting robust FHV RNA replication, including wild-type YPH499 yeast cells (31, 40, 41), Drosophila S2 cells (30), or mammalian BHK cells (15). Mitochondria were subsequently pelleted by centrifugation and membrane association was analyzed by SDS-PAGE and fluorography of pelleted fractions (Fig. 1). We found protein A rapidly sedimented in the presence of mitochondria from yeast, insect, or mammalian cells, whereas β-Gal, a similarly sized (∼100-kDa) control protein that was translated as efficiently as protein A, showed negligible mitochondrial association and sedimentation (Fig. 1A). In the absence of mitochondria neither protein A nor β-Gal was found in significant amounts in the pellet fractions, indicating that the in vitro-translated proteins were not forming aggregates that rapidly sedimented with centrifugation (Fig. 1A, bottom gel). Maximal binding of protein A was 70 to 80% of input protein and was seen after 10 to 20 min. Protein A association with mitochondria was independent of ATP levels, and extended incubations past 30 min did not significantly increase the fraction of bound protein A (data not shown). Furthermore, 60% of protein A translated in vitro remained associated with mitochondrial membranes even after alkaline extraction with 0.1 M sodium carbonate, suggesting that protein A behaved as an integral membrane protein after in vitro translation and mitochondrial association, similar to results obtained in vivo with both yeast and cultured Drosophila cells (29, 30). Based on the comparable results with yeast, insect, and mammalian mitochondria and the ready availability of yeast strains with deletions or mutations in specific mitochondrial outer membrane components, we used yeast mitochondria for all subsequent studies unless otherwise indicated.
FIG. 1.
FHV protein A specifically associates with mitochondrial membranes in vitro. (A) Equal amounts of in vitro-translated 35S-labeled protein A (PtnA) (lanes 1 to 4) or β-Gal (lanes 5 to 8) were incubated in the absence of mitochondria or with isolated yeast (S. cerevisiae), insect (D. melanogaster), or mammalian (BHK cell) mitochondria at 25°C for the indicated time period. The mitochondrion source is shown in bold on the left. Mitochondria were pelleted by centrifugation and membrane association was analyzed by SDS-PAGE and fluorography. (B) In vitro-translated protein A was incubated in the presence or absence of purified yeast mitochondria and detergent and subjected to equilibrium centrifugation in Nycodenz gradients, and equal-volume fractions were analyzed as described above. The bottom image represents an immunoblot for porin to identify membrane-containing fractions from yeast mitochondria. (C) In vitro-translated GFP or a GFP chimera fused to the first 46 amino acids of protein A (PA1-46/GFP) was incubated with isolated wild-type yeast mitochondria for 20 min at 25°C and separated into pellet and supernatant fractions by centrifugation, and membrane association was quantified as the percentage of total in vitro-translated protein present in the pellet fraction.
To examine the biochemical characteristics of in vitro-translated protein A and further exclude the possibility of aggregation due to the multiple hydrophobic regions within the protein (29), we used membrane flotation assays (Fig. 1B). In vitro translation or membrane association reaction mixtures were combined with Nycodenz, subjected to equilibrium centrifugation, and fractionated to analyze protein A distribution in low-density and high-density regions, which correspond to membrane-associated and soluble or aggregated proteins, respectively (29, 30). Protein A was recovered in high-density fractions in the absence of mitochondria (Fig. 1B, first gel), whereas with the addition of mitochondria protein A became membrane associated and floated to the low-density fractions (Fig. 1B, second gel), indicating that protein A was not aggregating after in vitro translation and mitochondrial association. Similar results were obtained when mitochondria were extracted with 0.1 M sodium carbonate prior to flotation, further confirming the protein A was inserted as an integral membrane protein (data not shown). As a control, the addition of the detergent Triton X-100 shifted protein A distribution back to the high-density fractions (Fig. 1B, third gel). Immunoblotting against the mitochondrial outer membrane protein porin was used as a control to identify membrane-containing fractions (Fig. 1B, bottom blot).
To explore membrane specificity of in vitro-translated protein A, we initially used constructs with defined amino-terminal deletions that disrupt mitochondrial targeting and membrane association in vivo (29). Deletions of amino acids 9 to 45 or 9 to 135 reduced protein A membrane association with mitochondria in vitro by 20% or 40%, respectively, and there was a direct correlation between the levels of membrane association in vitro with purified mitochondria and in vivo with intact yeast (R = 0.999, P < 0.02). We also conducted gain-of-function studies and found that the amino-terminal region containing 46 amino acids from protein A, which includes a mitochondrion-targeting signal (29), was sufficient to increase the in vitro mitochondrial association of a GFP fusion construct 10-fold (Fig. 1C). To further test membrane specificity, we examined the ability of wild-type protein A to associate with canine microsomes in vitro. As a control for these experiments, we used a chimeric protein A in which the amino-terminal mitochondrion-targeting signal was replaced with a hepatitis C virus NS5B ER-targeting signal (31) and performed the assays cotranslationally to mimic protein import in the ER. We found that 51.9 ± 6.0% of the protein A-ER chimera associated with canine microsomes in vitro compared to only 26.8 ± 6.2% of wild-type protein A (P < 0.004). We concluded from these data that FHV protein A translated in vitro recapitulates important membrane specificity and biochemical characteristics of protein produced in vivo.
The in vitro membrane insertion of protein A is independent of Tom20, Tom70, and the general import pore.
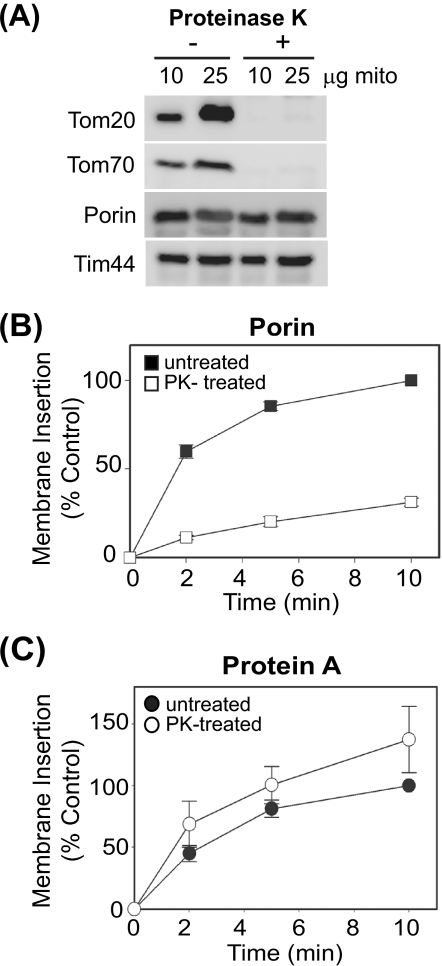
The majority of host mitochondrial proteins utilize the TOM complex for transport to their appropriate mitochondrial compartments (8, 38, 42). Thus, we examined the role of this macromolecular complex in FHV protein A mitochondrial membrane association and insertion using both biochemical and genetic approaches. Several TOM components, such as the primary receptors Tom20 and Tom70, are exposed to the cytosol (5, 19, 23) and therefore susceptible to protease digestion. We treated mitochondria isolated from YPH499 yeast with proteinase K to degrade accessible outer membrane components and analyzed protein A membrane association and subsequent bilayer insertion in vitro by assessing resistance to alkaline extraction (Fig. 2). Immunoblot examination of protease-treated mitochondria revealed the near-complete loss of Tom20 and Tom70, whereas a pore-forming protein wholly embedded in the mitochondrial outer membrane (porin) and an inner membrane protein (Tim44) were relatively unaffected by protease treatment (Fig. 2A). Protease-treated mitochondria showed a marked defect in the insertion of porin (Fig. 2B), an endogenous host protein previously shown to use the Tom20 receptor for proper membrane insertion (23). In contrast, protein A showed a slight increase in insertion efficiency upon the removal of protease-sensitive outer membrane components (Fig. 2C). These results suggested that protease-sensitive components were not required for efficient protein A mitochondrial membrane association and insertion.
FIG. 2.
Protease-sensitive outer membrane components are not required for protein A mitochondrial membrane association and insertion in vitro. (A) Immunoblot analysis of yeast mitochondrial membrane proteins after proteinase K digestion of purified mitochondria. (B) In vitro-translated porin was incubated with untreated or proteinase K (PK)-treated yeast mitochondria for the indicated times. Mitochondria were isolated by centrifugation and porin membrane insertion was analyzed by resistance to protease digestion followed by SDS-PAGE and fluorography. Data are presented as percentages of porin insertion at 10 min in untreated mitochondria. (C) In vitro-translated protein A (PtnA) was incubated with untreated or proteinase K (PK)-treated mitochondria for the indicated times. Mitochondria were isolated by centrifugation and protein A membrane insertion was analyzed by resistance to alkaline extraction followed by SDS-PAGE and fluorography. Data are presented as percentages of protein A insertion at 10 min in untreated mitochondria.
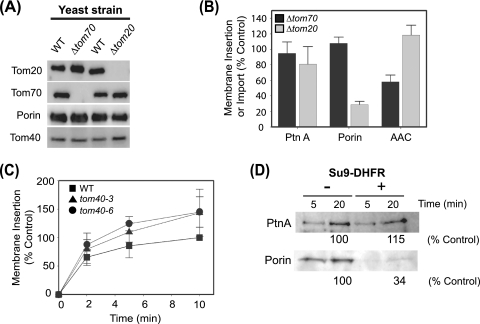
To further examine the role of specific mitochondrial outer membrane components in protein A insertion, we conducted in vitro assays with mitochondria purified from yeast strains with deletions or mutations in individual TOM components (Table 1 and Fig. 3). We verified the genotype of deletion strains by PCR (data not shown) and immunoblotting (Fig. 3A) and used purified mitochondria for protein A insertion assays. Protein A insertion was not significantly reduced using mitochondria from Δtom20 or Δtom70 compared to wild-type yeast, whereas insertion or import of porin or AAC, which have previously been shown to depend primarily on Tom20 or Tom70, respectively (23, 61), was reduced in mitochondria from the appropriate deletion strain (Fig. 3B). We also examined the role of Tom40, the main component of the general import pore, in protein A membrane insertion. Since Tom40 is an essential protein, we made use of two temperature-sensitive (ts) strains, tom40-3 and tom40-6. Mitochondria from tom40-3 have a defect in Tom20 import (2), whereas mitochondria from tom40-6 have not been shown to have any defects in protein import but do have reduced levels of assembled TOM complex (58). Wild-type and ts Tom40 strains were grown at 25°C for mitochondrial isolation, and prior to insertion assays mitochondria were incubated at 37°C for 15 min to induce the ts phenotype through temperature-dependent destabilization of Tom40. Mitochondria from tom40-3 or tom40-6 yeast showed no reduction in protein A insertion but rather showed a trend toward increased insertion efficiency (Fig. 3C), similar to results with protease-treated mitochondria (Fig. 2C). To further examine the role of the TOM complex in protein A mitochondrial insertion we conducted competition experiments with purified pSu9-DHFR, a chimeric protein that competes for protein insertion through the main import pore (2, 23, 27). Protein A insertion into wild-type mitochondria was not inhibited by Su9-DHFR, whereas porin insertion was substantially reduced (Fig. 3D). Taken together, these data suggested that protein A association and insertion into the mitochondrial membrane did not require Tom20, Tom70, and the general import pore.
FIG. 3.
The mitochondrial import machinery is not required for protein A membrane association and insertion in vitro. (A) Immunoblot analysis of purified mitochondria from Δtom20, Δtom70, and corresponding wild-type (WT) yeast strains (Table 1). (B) In vitro-translated protein A (PtnA), porin, or AAC was incubated with mitochondria from Δtom70 and Δtom20 yeast for 10 min at 25°C. Protein A and porin membrane insertion were analyzed as described for Fig. 2. AAC import was analyzed by resistance to protease digestion followed by SDS-PAGE and fluorography. Data are presented as percentages of mitochondrial insertion or import compared to control wild-type yeast. (C) Protein A insertion into mitochondria from wild-type (WT), tom40-3, or tom40-6 yeast cells. Mitochondrial membrane insertion was analyzed as described for Fig. 2 and data are presented as percentages of control mitochondria from wild-type yeast at 10 min. (D) Wild-type yeast mitochondria were incubated with excess recombinant Su9-DHFR for 5 min prior to incubation with in vitro translation mixtures. Protein A and porin insertion were analyzed at 5 and 20 min as described for Fig. 2. Quantitative data are presented as the percentages of protein A or porin inserted at 20 min in untreated samples and are the averages of two independent experiments.
Protein A is a lipid-binding protein with affinity for specific anionic phospholipids.
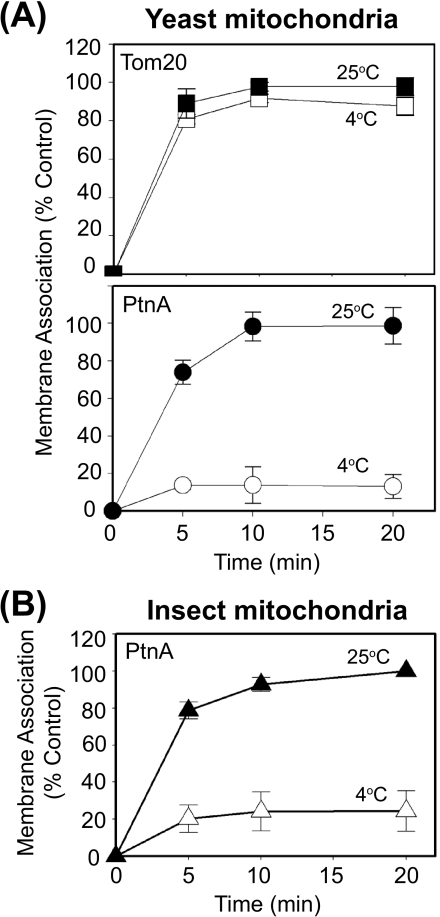
The observation that protein A membrane association and insertion were independent of protease-sensitive outer membrane components and Tom40 (Fig. 2 and 3) suggested that nonprotein components, such as membrane lipids, may be involved. To initially test this hypothesis we examined the temperature dependence of protein A-membrane interactions in vitro (Fig. 4). Protein-protein interactions are often temperature independent, and this characteristic is frequently used to study processes such as initial receptor-ligand binding at low temperatures to avoid receptor internalization or remodeling after ligand-mediated activation. We found that Tom20 binding to purified mitochondria occurred equally well at 4°C and 25°C (Fig. 4A, upper graph), consistent with its known interaction with the import pore Tom40 during mitochondrial membrane binding and insertion (2, 4). We obtained similar temperature-independent results with porin (data not shown). In contrast, protein A membrane association with purified wild-type yeast (Fig. 4A, bottom graph) and Drosophila (Fig. 4B) mitochondria was significantly reduced at 4°C to less than 20% of the levels seen at 25°C.
FIG. 4.
Protein A membrane association is temperature dependent. (A) In vitro-translated Tom20 (upper graph) or protein A (lower graph) was incubated with purified yeast mitochondria at 4°C or 25°C for the indicated time periods. Mitochondria were isolated by centrifugation and protein membrane association was analyzed as described for Fig. 1. Data are presented as the percentages of control membrane association at 20 min and 25°C. (B) In vitro-translated protein A was incubated with purified Drosophila mitochondria at 4°C or 25°C for the indicated time periods and membrane association was analyzed as described above.
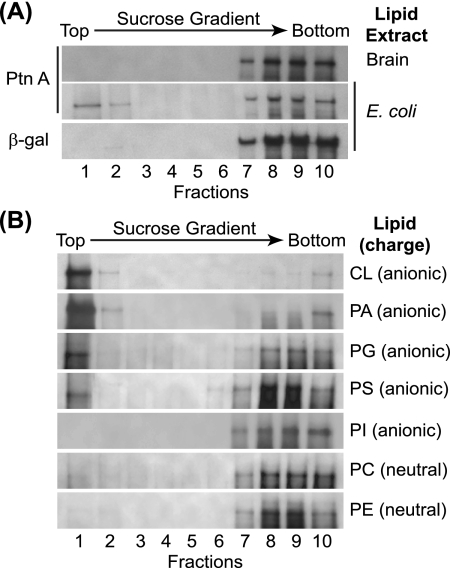
To directly examine potential protein A-lipid interactions, we used lipid extracts from E. coli and brain with in vitro binding assays. We selected these extracts due in part to their differences in phospholipid composition. For example, E. coli lipid extracts contain a lipid composition similar to that of eukaryotic mitochondria with large quantities of the anionic phospholipids PG (15.1%) and CL (9.8%) (Avanti Polar Lipids product data). In contrast, brain extracts contain a more diverse group of phospholipids with low concentrations of anionic phospholipids and a larger quantity of neutral phospholipids. We incubated in vitro-translated protein A or β-Gal with liposomes generated from individual lipid extracts for 30 min and subsequently examined liposome binding by equilibrium gradient centrifugation (Fig. 5A). The control protein β-Gal did not bind to liposomes generated with either lipid extract (Fig. 5A, lower gradient, and data not shown). In contrast, protein A associated with liposomes generated from E. coli lipid extracts, where approximately 25% partitioned into the low-density fractions (Fig. 5A, middle gradient). Only minimal protein A binding was seen with liposomes generated from brain extracts (Fig. 5A, upper gradient).
FIG. 5.
Protein A is a lipid-binding protein with affinity for specific anionic phospholipids. (A) In vitro-translated protein A or β-Gal was incubated with liposomes generated from E. coli or brain lipid extracts and subjected to equilibrium centrifugation in sucrose gradients, and fractions were analyzed by SDS-PAGE and fluorography. (B) In vitro-translated protein A was incubated with liposomes generated from equal amounts of the indicated purified lipids and analyzed as described above.
To further explore potential phospholipid selectivity, we analyzed the in vitro binding capabilities of protein A with liposomes generated from equal amounts of purified individual membrane phospholipids (Fig. 5B). We observed substantial protein A binding to the anionic phospholipids CL and PA, moderate binding to the anionic phospholipids PG and PS, and negligible binding to the anionic phospholipids PI and the neutral lipids PC and PE. The absence of significant protein A binding to PC was not due to decreased flotation, as control experiments with fluorescent PC demonstrated almost complete partitioning of this neutral lipid into upper low-density gradient fractions (data not shown). Of interest with the purified lipid experiments was protein A binding to the mitochondrion-enriched anionic phospholipids CL, PA, and PG (17, 55), and in particular the mitochondrion-specific CL, where protein A was recovered almost exclusively in the low-density fractions (Fig. 5B, top gradient, fractions 1 and 2).
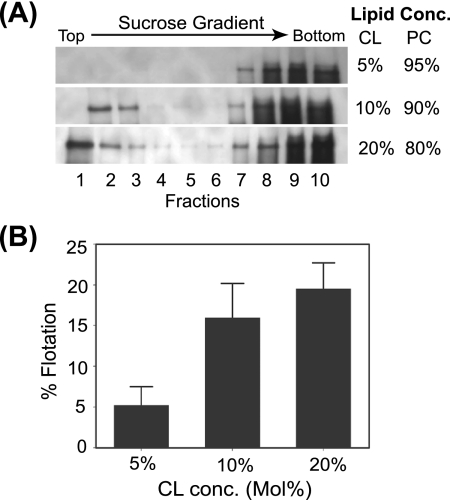
The diphosphatidyl-based lipid CL is found in small concentrations in the mitochondrial outer membrane, with the notable exception of the contact sites between mitochondrial outer and inner membranes, where it can increase to roughly 20% of total membrane phospholipids (3, 51). To further confirm the CL-binding capabilities of protein A, we generated liposomes containing increasing concentrations of CL relative to PC and assayed protein A in vitro binding by gradient flotation assays (Fig. 6). Protein A showed CL concentration-dependent flotation, where approximately 20% of protein A partitioned to low-density fractions when the total CL concentration was 20% (Fig. 6A, bottom gradient, and B). The positive correlation between CL concentration and protein A flotation (Fig. 6B) (R = 0.92) was not due to changes in liposome partitioning within sucrose gradients, as control experiments with fluorescent PC-CL liposomes demonstrated equivalent flotation characteristics regardless of CL concentration (data not shown).
FIG. 6.
Protein A binding to liposomes is correlated with CL content. (A) In vitro-translated protein A was incubated with liposomes containing the indicated CL and PC concentrations (mol%) and analyzed as described for Fig. 5. (B) Correlation of CL content and protein A-liposome binding. Percent flotation was calculated as the ratio of the amount of protein A in the upper four gradient samples (fractions 1 to 4) compared to the total signal (fractions 1 to 10).
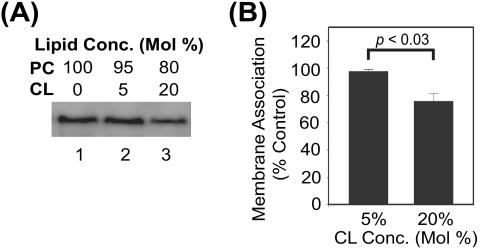
To examine the role of CL in protein A association with intact mitochondria, we initially conducted in vitro membrane association assays with mitochondria purified from Δcrd1 yeast, which lack the biosynthetic enzyme CL synthase and hence produce no CL (12). Protein A membrane association in vitro was not significantly different between mitochondria from Δcrd1 and wild-type yeast (data not shown). However, the deletion of CL synthase results in the increased accumulation of PG (12), which along with PA is a precursor in the CL biosynthetic pathway and an anionic phospholipid to which protein A was also significantly bound in flotation assays (Fig. 5B). To avoid the potential confounding effects of altered phospholipid composition in yeast strains with deletions or mutations of specific phospholipid biosynthesis genes, we subsequently conducted differential centrifugation competition experiments with mitochondria from wild-type yeast and CL-containing liposomes (Fig. 7). We initially used fluorescent PC-CL liposomes to optimize differential centrifugation conditions and minimize liposome fusion or cosedimentation with intact mitochondria, and we achieved conditions under which approximately 15% of the input lipid-associated fluorescence signal pelleted with mitochondria regardless of the CL liposome concentration (data not shown). Under these optimized conditions, liposomes containing 20% CL reduced protein A association with wild-type mitochondria by approximately 25% compared to PC-only liposomes, whereas minimal competition was seen with 5% CL liposomes (Fig. 7A and B). Similar results were obtained with mitochondria from Drosophila cells (data not shown). Taken together, these data indicated that FHV protein A is a lipid-binding protein with affinity for specific anionic phospholipids, and in particular the mitochondrion-specific phospholipid CL.
FIG. 7.
CL-containing liposomes disrupt protein A binding to wild-type mitochondria. (A) Mitochondria from YPH499 yeast (50 μg total protein) were incubated with in vitro-translated protein A and liposomes (50 μg) containing 100% PC (lane 1), 95% PC-5% CL (lane 2), or 80% PC-20% CL (lane 3) for 10 min at 25°C. Samples were separated by differential centrifugation at 1,000 × g for 5 min at 4°C, pellets were resuspended in SDS-PAGE buffer, and samples were analyzed by fluorography. (B) Quantitative analysis of liposome competition assays. Data are presented as the percentages of protein A present in the mitochondrial pellet compared to competition with 100% PC liposomes.
DISCUSSION
In this study, we investigated the interactions between FHV protein A and intracellular membranes, an initial and important step in the assembly of viral RNA replication complexes. Based on our results we conclude that protein A is a lipid-binding protein with particular affinity for specific anionic phospholipids. Furthermore, both biochemical and genetic studies indicated that protease-accessible membrane proteins, and in particular the TOM complex components Tom20, Tom70, and Tom40, were not required for protein A interactions with the mitochondrial membrane. However, we cannot exclude a role for protease-resistant and possibly membrane-embedded non-TOM complex mitochondrial outer membrane proteins in FHV protein A-membrane interactions. Indeed, although we saw almost complete association of in vitro-translated protein A with intact mitochondria, we obtained only partial association with lipid extracts or liposomes, with the exception of 100% CL liposomes. Furthermore, changes in membrane-embedded protein conformation may have accounted for the dramatic temperature dependence we observed in protein A mitochondrial binding. Nonetheless, the results with protein A in this study are consistent with those published for some endogenous mitochondrial outer membrane proteins that contain an amino-terminal signal-anchor transmembrane domain that resemble the targeting sequence of FHV protein A, suggesting a potentially similar pathway for membrane insertion (27, 57). The published study with Mcr1 used mitochondrial import assays, similar to the system described in this manuscript, including the use of protease digestion, TOM receptor component deletion strains, and Su9-DHFR competition experiments, but did not explore lipid binding (27). Further experiments with both cellular mitochondrial proteins and FHV protein A will be required to fully define this pathway.
The observation that protein A has lipid-binding capabilities is consistent with the well-described association between positive-strand RNA virus replication complexes and intracellular membranes (32, 49). Host membranes likely play multiple roles in viral RNA replication, which may include (i) serving as a scaffold for assembly of a macromolecular structure such as an RNA replication complex, (ii) shielding viral RNA replication intermediates such as double-stranded RNA from cellular innate antiviral pathways, and (iii) providing cofactors for optimal enzymatic activity of viral replicase proteins. These functions could be mediated by either protein or lipid constituents within particular organelle membranes. Thus far the predominant emphasis in the field has been placed on identifying either membrane-resident proteins or cytosolic proteins that become membrane associated upon viral RNA replication complex assembly (13, 34). The results presented in this report indicate that lipids, and in particular anionic phospholipids, may also play important roles in viral RNA replication complex assembly.
The speculative role of lipids in replication complex assembly is consistent with the observation that the replicase protein nsP1 from Semliki Forest virus, a positive-strand RNA virus that assembles its replication complexes on membranes derived from endosomes and lysosomes (52), also binds anionic phospholipids (1). FHV protein A binding to predominantly anionic phospholipids suggests that ionic forces may mediate in part the interactions with phospholipids. Indeed, for nsP1 the anionic phospholipid interaction domain was mapped to an amphipathic α-helix with a cluster of positively charged residues (1). Sequence and predicted secondary structural analyses have revealed the presence of several similar amphipathic α-helices within the FHV protein A-coding region that may be involved in anionic phospholipid interactions (K. Stapleford and D. Miller, unpublished data). However, the selectivity of protein A for certain anionic phospholipids suggests that nonionic forces also contribute to protein A-lipid interactions.
The ability of protein A to interact with several different anionic phospholipids suggests a level of “promiscuity” that may help explain the robustness of FHV RNA replication in cells derived from multiple organisms from several kingdoms (24, 30, 41) and the relative ease with which FHV RNA replication complexes can be retargeted to alternative intracellular membranes such as the ER (31). However, the ubiquitous nature of anionic phospholipids and the substantial amounts of PI and PS present in many cellular membranes, including the ER (62), indicate that net membrane charge cannot be the sole determinant of replication complex targeting. Nonetheless, protein A did not bind all anionic phospholipids, but rather showed preferential interactions with CL, PA, and PG, which are enriched in mitochondrial membranes (55), suggesting that specific anionic phospholipids can influence FHV replication complex targeting. Furthermore, we cannot exclude a role for local charge clusters within membrane microdomains or the impact of protease-resistant endogenous membrane protein interactions with anionic phospholipids in the membrane-specific targeting of FHV RNA replication complexes.
The hypothesis that anionic phospholipids play targeting or structural roles in FHV RNA replication complex assembly is particularly interesting given the results with CL, which we identified as a significant interaction partner of protein A. CL is a cellular phospholipid whose distribution is almost entirely limited to the mitochondria (17). Although the majority of CL is found in the inner mitochondrial membrane, it is also present in high localized concentrations in the outer membrane at contacts sites between inner and outer membranes (3, 44). One might speculate that FHV RNA replication complexes may initially assemble at or near these contact sites via interactions between protein A and CL. This proposed mechanism resembles the targeting of the proapoptotic promoter tBID to CL that can initiate cytochrome c-mediated apoptotic cell death (10, 25). The potential similarity between protein A-CL and tBID-CL interactions is intriguing given the recent demonstration that FHV infection induces apoptosis in cultured Drosophila cells (50), although the delayed onset of FHV-induced apoptosis until approximately 12 h postinfection suggests that a threshold of protein A-CL interactions may be required. In addition, it is possible that protein A-mediated disruption of the inner-outer membrane contact sites, an essential substructure needed to maintain mitochondrial shape (44), may lead to some of the morphological alterations that are seen in mitochondria from yeast or Drosophila cells that contain active FHV RNA replication complexes (30, 31). This is consistent with the observation that components of the endogenous mitochondrial fission and fusion machinery can localize to these contact sites (44, 53) and that disruption of the normal fission-fusion processes can lead to abnormal mitochondrial morphology (36, 43) that also resemble mitochondria in cells with active FHV RNA replication (29, 31). Furthermore, the dimeric nature of CL, which consists of four acyl chains attached to diphosphatidylglycerol, imparts a unique conical structure that favors a hexagonal HII phase that may play a role in the membrane curvatures necessary to produce the spherules associated with FHV RNA replication complexes (22, 30).
Although the focus of this study was to identify host components involved in protein A-membrane interactions, an initial step in FHV replication complex assembly, we found that protein A translated in RRLs had RNA polymerase activity when provided with an excess of exogenous virion template RNA template (data not shown). However, we did not observe the FHV RNA replication complex activity that has been described with membrane preparations from FHV-infected Drosophila cells (59, 60) and replicon-expressing yeast (31), which includes the production of single-stranded products, with in vitro-translated protein A in the presence of whole mitochondria or liposomes. The particularly difficult feat of de novo assembly of fully functional viral RNA replication complexes using a cell-free in vitro translation system has only been accomplished for a select few positive-strand RNA viruses, including poliovirus, which requires the use of uninfected mammalian cell extracts (33), the plant pathogens tomato mosaic virus, brome mosaic virus, and turnip crinkle virus, which require the use of evacuolated plant cell extracts (21), and tomato bushy stunt virus, which uses a related system that employs a yeast cell extract and purified recombinant viral proteins (39). Further studies with FHV will be required to identify the optimal conditions under which fully functional viral RNA replication complexes can be formed in vitro, and both the results presented in this report and others (59, 60) indicate that the inclusion of specific phospholipids may be a particularly important aspect of these studies.
In summary, the studies presented in this report demonstrate that FHV protein A mitochondrial association and membrane insertion is mediated by a TOM complex-independent mechanism, similar to what has been seen previously for mitochondrial outer membrane signal-anchored proteins, and provide evidence for the importance of host membrane-specific phospholipids in positive-strand RNA virus replication complex assembly. Future in vitro and in vivo studies using this established host-pathogen system will give further insight into the role of phospholipids in membrane-specific targeting as well as the biochemical mechanisms involved in positive-strand RNA virus replication complex assembly and function.
Acknowledgments
We thank Donna Gschwend for administrative assistance, F. Ulrich Hartl, Donna Gordon, and Nickolaus Pfanner for providing reagents, and all the Miller laboratory personnel for helpful comments on the research and manuscript.
This work was supported by National Institutes of Health grants R01-AI062749 and T32-GM007315.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 183164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahting, U., T. Waizenegger, W. Neupert, and D. Rapaport. 2005. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 28048-53. [DOI] [PubMed] [Google Scholar]
- 3.Ardail, D., J. P. Privat, M. Egret-Charlier, C. Levrat, F. Lerme, and P. Louisot. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 26518797-18802. [PubMed] [Google Scholar]
- 4.Becker, T., S. Pfannschmidt, B. Guiard, D. Stojanovski, D. Milenkovic, S. Kutik, N. Pfanner, C. Meisinger, and N. Wiedemann. 2008. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 283120-127. [DOI] [PubMed] [Google Scholar]
- 5.Brix, J., K. Dietmeier, and N. Pfanner. 1997. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 27220730-20735. [DOI] [PubMed] [Google Scholar]
- 6.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 7512370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckerle, L. D., C. G. Albarino, and L. A. Ball. 2003. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 31795-108. [DOI] [PubMed] [Google Scholar]
- 8.Endo, T., and D. Kohda. 2002. Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta 15923-14. [DOI] [PubMed] [Google Scholar]
- 9.Galiana-Arnoux, D., C. Dostert, A. Schneemann, J. A. Hoffmann, and J. L. Imler. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7590-597. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalvez, F., J. J. Bessoule, F. Rocchiccioli, S. Manon, and P. X. Petit. 2005. Role of cardiolipin on tBid and tBid/Bax synergistic effects on yeast mitochondria. Cell Death Differ. 12659-667. [DOI] [PubMed] [Google Scholar]
- 11.Graham, J. M. 2001. Isolation of mitochondria from tissues and cells by differential centrifugation. Curr. Protoc. Cell Biol. 33.3. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, F., M. T. Ryan, M. Schlame, M. Zhao, Z. Gu, M. Klingenberg, N. Pfanner, and M. L. Greenberg. 2000. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 27522387-22394. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, Y., E. Serviene, J. Gal, T. Panavas, and P. D. Nagy. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 807394-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, K. L., and L. A. Ball. 1999. Induction and maintenance of autonomous Flock House virus RNA1 replication. J. Virol. 737933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K. L., and L. A. Ball. 1997. Replication of Flock House virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 713323-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonczyk, M., K. B. Pathak, M. Sharma, and P. D. Nagy. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362320-330. [DOI] [PubMed] [Google Scholar]
- 17.Joshi, A. S., J. Zhou, V. M. Gohil, S. Chen, and M. L. Greenberg. 2009. Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793212-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kampmueller, K. M., and D. J. Miller. 2005. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 796827-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemper, C., S. J. Habib, G. Engl, P. Heckmeyer, K. S. Dimmer, and D. Rapaport. 2008. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 1211990-1998. [DOI] [PubMed] [Google Scholar]
- 20.Komiya, T., S. Rospert, G. Schatz, and K. Mihara. 1997. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 164267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 1011863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krimmer, T., D. Rapaport, M. T. Ryan, C. Meisinger, C. K. Kassenbrock, E. Blachly-Dyson, M. Forte, M. G. Douglas, W. Neupert, F. E. Nargang, and N. Pfanner. 2001. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, R., M. Maduro, F. Li, H. W. Li, G. Broitman-Maduro, W. X. Li, and S. W. Ding. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 4361040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutter, M., M. Fang, X. Luo, M. Nishijima, X. Xie, and X. Wang. 2000. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2754-761. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, D., and A. Schneemann. 2001. Specific packaging of nodaviral RNA2 requires the N-terminus of the capsid protein. Virology 285165-175. [DOI] [PubMed] [Google Scholar]
- 27.Meineke, B., G. Engl, C. Kemper, A. Vasiljev-Neumeyer, H. Paulitschke, and D. Rapaport. 2008. The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett. 582855-860. [DOI] [PubMed] [Google Scholar]
- 28.Meisinger, C., T. Sommer, and N. Pfanner. 2000. Purification of Saccharomyces cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 287339-342. [DOI] [PubMed] [Google Scholar]
- 29.Miller, D. J., and P. Ahlquist. 2002. Flock House virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 769856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 7511664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 7712193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 2541647-1651. [DOI] [PubMed] [Google Scholar]
- 34.Nagy, P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46217-242. [DOI] [PubMed] [Google Scholar]
- 35.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 808459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otsuga, D., B. R. Keegan, E. Brisch, J. W. Thatcher, G. J. Hermann, W. Bleazard, and J. M. Shaw. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathak, K. B., Z. Sasvari, and P. D. Nagy. 2008. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379294-305. [DOI] [PubMed] [Google Scholar]
- 38.Pfanner, N., N. Wiedemann, C. Meisinger, and T. Lithgow. 2004. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 111044-1048. [DOI] [PubMed] [Google Scholar]
- 39.Pogany, J., J. Stork, Z. Li, and P. D. Nagy. 2008. In vitro assembly of the tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. USA 10519956-19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price, B. D., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 761610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 7411724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rapaport, D. 2002. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27191-197. [DOI] [PubMed] [Google Scholar]
- 43.Rapaport, D., M. Brunner, W. Neupert, and B. Westermann. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 27320150-20155. [DOI] [PubMed] [Google Scholar]
- 44.Reichert, A. S., and W. Neupert. 2002. Contact sites between the outer and inner membrane of mitochondria-role in protein transport. Biochim. Biophys. Acta 159241-49. [DOI] [PubMed] [Google Scholar]
- 45.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 164049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlossmann, J., and W. Neupert. 1995. Assembly of the preprotein receptor MOM72/MAS70 into the protein import complex of the outer membrane of mitochondria. J. Biol. Chem. 27027116-27121. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 27644052-44063. [DOI] [PubMed] [Google Scholar]
- 48.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 666728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz, M., J. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. USA 10111263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Settles, E. W., and P. D. Friesen. 2008. Flock House virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J. Virol. 821378-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simbeni, R., L. Pon, E. Zinser, F. Paltauf, and G. Daum. 1991. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 26610047-10049. [PubMed] [Google Scholar]
- 52.Spuul, P., A. Salonen, A. Merits, E. Jokitalo, L. Kaariainen, and T. Ahola. 2007. Role of the amphipathic peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 81872-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stojanovski, D., M. Rissler, N. Pfanner, and C. Meisinger. 2006. Mitochondrial morphology and protein import: a tight connection? Biochim. Biophys. Acta 1763414-421. [DOI] [PubMed] [Google Scholar]
- 54.Stuart, R. A., and C. M. Koehler. 2007. In vitro analysis of yeast mitochondrial protein import. Curr. Protoc. Cell Biol. 1111.19. [DOI] [PubMed] [Google Scholar]
- 55.van Meer, G., D. R. Voelker, and G. W. Feigenson. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waizenegger, T., T. Stan, W. Neupert, and D. Rapaport. 2003. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J. Biol. Chem. 27842064-42071. [DOI] [PubMed] [Google Scholar]
- 57.Walther, D. M., and D. Rapaport. 2009. Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 179342-51. [DOI] [PubMed] [Google Scholar]
- 58.Wiedemann, N., V. Kozjak, A. Chacinska, B. Schonfisch, S. Rospert, M. T. Ryan, N. Pfanner, and C. Meisinger. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424565-571. [DOI] [PubMed] [Google Scholar]
- 59.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 8911136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, S. X., and P. Kaesberg. 1991. Synthesis of template-sense, single-strand Flock House virus RNA in a cell-free replication system. Virology 183392-396. [DOI] [PubMed] [Google Scholar]
- 61.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 11241-50. [DOI] [PubMed] [Google Scholar]
- 62.Zinser, E., and G. Daum. 1995. Isolation and characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 11494-536. [DOI] [PubMed] [Google Scholar]