Abstract
Avian influenza highlights the need for novel vaccination techniques that would allow for the rapid design and production of safe and effective vaccines. An ideal platform would be capable of inducing both protective antibodies and potent cellular immune responses. These potential advantages of DNA vaccines remain unrealized due to a lack of efficacy in large animal studies and in human trials. Questions remain regarding the potential utility of cellular immune responses against influenza virus in primates. In this study, by construct optimization and in vivo electroporation of synthetic DNA-encoded antigens, we observed the induction of cross-reactive cellular and humoral immune responses individually capable of providing protection from influenza virus infection in the rhesus macaque. These studies advance the DNA vaccine field and provide a novel, more tolerable vaccine with broad immunogenicity to avian influenza virus. This approach appears important for further investigation, including studies with humans.
Since highly pathogenic avian influenza (H5N1) virus began infecting humans in 1997 (4), there has been a strong drive to develop effective vaccines against it. Studies of patients that have succumbed to infection with this virus reveal a fundamentally different pathology than that of epidemic influenza. Human influenza is primarily an infection of the upper respiratory tract (URT), predominantly infecting epithelial cells of the trachea and bronchi (29). In contrast, H5N1 influenza virus primarily infects the lower respiratory tract (LRT) of humans, including type II pneumocytes and alveolar macrophages (28, 29). This infection leads to diffuse acute edema, alveolar damage, and the induction of severe cytokinemia resulting from alveolar macrophage infection (5, 7). Restriction to the LRT likely contributes to the inability of the virus to pass easily from person to person, which requires a high level of replication in the URT to allow for aerosolization and transmission of viral particles. The severe cytokine dysregulation seen with H5N1 influenza virus is also reminiscent of the devastating pathology associated with the 1918 influenza virus (14). Modern histopathological analyses of autopsy samples from human influenza cases from 1918 revealed significant damage to the lungs, with acute, focal bronchitis and alveolitis associated with massive pulmonary edema, hemorrhage, and rapid destruction of the respiratory epithelium (13). Furthermore, recent studies have suggested that death from the 1918 influenza was likely the result of secondary bacterial pneumonia (3). Death from infection with H5N1 is linked directly to viral infection and replication, suggesting a virus potentially more pathogenic than the 1918 influenza virus. Concerns about a potential pandemic revolve around the possibility that this virus could acquire the ability to replicate to high titers in the URT of humans, while retaining its significant pathogenicity.
Complicating efforts to develop an effective vaccine is the diversity of strains within the H5N1 subtype. Most viruses sequenced fall into one of several distinct clades, including clades 1, 2.1, and 2.2 or 2.3, 2.4, and 2.5, some of which further segregate into distinct subclades. However, there is little antibody cross-reactivity between these clades, and newer isolates of clade 1 viruses, in addition to a number of clade 2.2 and 2.3 viruses, demonstrate antigenic heterogeneity (30). The ability of consensus sequences to induce protective and cross-reactive immune responses against H5N1 has been previously reported. We have previously shown the ability of such DNA vaccines to induce cross-reactive cellular immune responses (18). In addition, Chen et al. has shown that DNA-encoded consensus H5 hemagglutinin can induce antibodies capable of inhibiting multiple subclades within H5N1 and providing various levels of protection in a murine challenge model (6).
Due to the lack of suitable challenge models, there are questions that remain unanswered in the development of next-generation influenza virus vaccines for both epidemic and pandemic influenza. Nonhuman primates (NHPs), whose physiology most closely resembles that of humans, have several limitations as models. Similar to humans infected with seasonal influenza, infected monkeys will often develop subclinical infections with few markers to assess disease severity and correlates of immunity. Infection of macaques with highly pathogenic H5N1 influenza virus has proven unsatisfactory as a model for human infection, as infection in macaques is associated with pathology in the LRT (16, 23) resulting from viral replication in type I pneumocytes rather than type II pneumocytes, as seen in human infection, and limited infection in alveolar macrophages (23, 29). These and other factors contribute to a challenge model with important differences from human infection: challenge studies report a low or transient fever, no weight loss, no cytokine dysregulation, and no fatal outcome (12, 16, 23, 25). However, these studies show that the virus can replicate and induce significant pathology in the NHP lung, which allowed us to ask important questions unanswered by challenge of species with lower phylogenies. Importantly, this includes the potential of cellular immunity to impact viral respiratory infection. While many studies using small animal models of infection suggest that cell-mediated immune responses can provide protection from influenza morbidity and mortality, there have been no prospective experiments in primates to confirm these results, and many clinicians believe antibodies are necessary and sufficient. The NHP model is very relevant for studying the induction of both cellular and humoral immune responses and comparing different routes of immunization to achieve these results.
The effect of vaccination upon H5N1 infection has been studied, using inactivated virus to induce protective humoral immune responses in NHPs. While these responses demonstrated inhibition of replication, the initial viral loads in these challenges were relatively low, and the impact of these responses on lung pathology remained unclear.
We previously reported protection from H5N1 influenza virus in a ferret infection model following immunization by electroporation of a three-plasmid synthetic consensus vaccine cocktail (SynCon) or a single vaccine formulation containing a plasmid encoding the influenza virus nucleoprotein (which induces no protective antibodies) (18). The current NHP studies were designed to extend these findings in multiple directions as follows: (i) to determine if immunization by electroporation of the consensus plasmid vaccine could induce strong immune responses in primates; (ii) to determine the optimal route of immunization to induce the correlates of immunity previously studied; and (iii) to compare the clinical impact of those correlates in vivo following viral infection.
MATERIALS AND METHODS
DNA vaccines.
The DNA vaccine combination (pComb) and the pNP-only plasmid vaccine used in this study have been described previously (18). Briefly, consensus sequences were generated by aligning multiple primary sequences (obtained from the Los Alamos National Laboratory influenza sequence database) and choosing the most common amino acid at each position to generate a sequence not necessarily found in nature (synthetic) but which retained characteristics of the component sequences chosen. Sequences were then optimized for codon usage, RNA structure, and GC content and were synthesized. Synthetic genes were then subcloned into a pVax expression vector (Invitrogen). For the NHP studies, DNA preparations were made at VGX Pharmaceuticals, Inc. (The Woodlands, TX), as previously described (11) and formulated at 10 mg/ml in water plus 1% (wt/wt) poly-l-glutamate sodium salt.
Challenge virus.
The high-pathogenicity avian influenza virus strain A/Vietnam/1203/2004 (H5N1), which was used for the monkey challenge, was obtained from the Centers for Disease Control and Prevention (Atlanta, GA). Stock virus was expanded in the allantoic cavities of 10-day-old embryonated chicken eggs at 37°C for 24 h and stored at −70°C. The 50% egg infective dose of this virus stock was 108.7/ml. All experiments with the high-pathogenicity avian influenza virus were conducted in the Bioqual, Inc., biosafety level 3+ containment facility (Rockville, MD), approved for use by the U.S. Department of Agriculture and the CDC. The low-pathogenicity avian influenza virus H5N1-PR8 reassortant strains used for the hemagglutination inhibition (HAI) assay were obtained from the CDC or from NIBSC, Hertfordshire, United Kingdom. Experiments with the low-pathogenicity avian influenza virus strains were conducted in a biosafety level 2+ containment facility approved for use by the USDA and the CDC. Seed stocks were expanded in 10-day-old embryonated chicken eggs at 35 to 37°C for 48 h and stored at −70°C.
Macaque studies.
Rhesus macaques were housed at Bioqual, Inc., in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. Animals were allowed to acclimate for at least 30 days in quarantine prior to any immunization. Four groups of five rhesus macaques were immunized at weeks 0, 4, and 8 with 1 mg/construct (at a concentration of 10 mg/ml) of pVax (intramuscularly [i.m.]); pH5HA, pNP, and pN1NA (i.m.); pH5HA, pNP, and pN1NA (intradermally [i.d.]); and pNP (i.m.). DNA was delivered into the quadriceps muscle (i.m.) or skin (i.d.), followed by in vivo electroporation. Square-wave pulses were used in all experiments and administered by using an adaptive constant current electroporator, the Cellectra device (VGX Pharmaceuticals, Inc., The Woodlands, TX). Two types of arrays were used. The electrode array used for i.m. electroporation is a circular array (1-cm diameter) of five equally spaced 21-gauge solid stainless steel needle electrodes mounted on a nonconductive material. All i.m. electroporation immunizations were performed at 0.5 A, with 3 pulses at 52 ms/pulse and 1-s intervals between pulses. The i.d. microelectrode array consisted of three 26-gauge solid stainless steel needle electrodes, 3 mm in length, placed in an isosceles triangle formation (the two long sides are 5 mm in length, and the short side is 3 mm in length) and mounted on nonconductive material. All i.d. electroporation immunizations were performed at 0.2 A, with 2 pulses at 52 ms/pulse and 1-s intervals between pulses, followed by a 3-s rest period and another 2 pulses under identical conditions (2 by 2 pulse pattern).
Blood collection.
Animals were bled every 2 weeks. Ten milliliters of blood was collected in EDTA tubes, and peripheral blood mononuclear cells were isolated by standard Ficoll-Hypaque centrifugation and resuspension in complete culture medium (RPMI 1640 with 2 mM/liter l-glutamine, 10% heat-inactivated fetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 55 μM/liter β-mercaptoethanol). Red blood cells were lysed with ammonium chloride-potassium (ACK) lysis buffer (Cambrex BioScience, East Rutherford, NJ).
HAI assay.
Sera were treated with receptor-destroying enzyme by diluting 1 part serum with 3 parts enzyme and were incubated overnight in a 37°C water bath. The enzyme was inactivated by a 30-min incubation at 56°C, followed by the addition of 6 parts phosphate-buffered saline for a final dilution of 1/10. HAI assays were performed in V-bottomed 96-well microtiter plates, using 4 hemagglutination units of virus and 1% horse red blood cells as previously described (27). Viruses used for the HAI assay are reassortant strains obtained from either the influenza branch of the CDC (Atlanta, GA) [clade 1, A/Vietnam/1203/2004 (H5N1)/PR8-IBCDC-RG; clade 2.1, A/Indonesia/05/2005 (H5N1)/PR8-IBCDC-RG2; and clade 2.3.4, Anhui/01/2005/PR8-IBCDC-RG5] or from NIBSC (United Kingdom) (clade 2.2, A/turkey/Turkey/1/2005/NIBRG-23 [reference strain]).
Microneutralization assay.
Neutralizing antibody activity was analyzed in a microneutralization assay based on the methods of the pandemic influenza virus reference laboratories of the CDC (24, 26, 27). All sera were treated with a receptor-destroying enzyme overnight, followed by heat inactivation. Low-pathogenicity H5N1 viruses (A/Vietnam/1203/2004 [SJCRH, clade 1]; A/Indonesia/5/2005 [PR8-IBCDC-RG2, clade 2.1]; A/turkey/Turkey/1/2005 [NIBRG-23, clade 2.2]; and A/Anhui/1/2005 [IBCDC-RG5, clade 2.3.4]), generated by reverse genetics, were obtained from St. Jude's Hospital (Memphis, TN), the CDC, and NIBSC.
Tracheal lavage.
Primates were sedated as previously described for the challenge and placed in dorsal recumbency, and the mouth was opened manually. The epiglottis was opened with a laryngoscope, and a syringe containing 2 ml of sterile, buffered, physiological saline used for washing was attached to a sterile tube 2 to 3 mm in diameter and inserted into the epiglottis. Two milliliters of sterile saline solution was pushed through the tubing and then collected and aliquoted into two vials.
ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were conducted as previously described (17). Briefly, ELISPOT 96-well plates (Millipore, Billerica, MA) were coated with anti-human (clone GZ-4; Mabtech, Cincinnati, OH) gamma interferon (IFN-γ) capture antibody and incubated overnight at 4°C. The following day, plates were washed with phosphate-buffered saline and blocked for 2 h with R10. Peripheral blood mononuclear cells from each group were added to each well and stimulated overnight at 37°C in the presence of R10 peptide (negative control) or specific peptide antigens (10 μg/ml) (Invitrogen, Carlsbad, CA). The peptide pools consisted of 15-mer peptides overlapping by 11 amino acids. After 24 h of stimulation, the cells were washed and incubated for 24 h at 4°C with anti-human (clone 7-B6-1; Mabtech) IFN-γ capture antibody. The plates were washed, streptavidin-alkaline phosphatase (R&D Systems, Minneapolis, MN) was added to each well, and the mixtures were incubated for 2 h at room temperature. The plates were washed, and BCIP (5-bromo-4-chloro-3-indolylphosphate)/Nitro Blue Tetrazolium chromogen (R&D Systems, Minneapolis, MN) was added. The plates were then rinsed with distilled water and dried. Spots were counted by an automated ELISPOT reader (Cellular Technology Limited, Shaker Heights, OH).
Viral challenge.
Virus (A/Vietnam/1203/2004) was diluted in L-15 tissue culture medium to a concentration of 1 × 106 50% egg infectious dose per ml. One milliliter of the virus dilution was inoculated into the trachea and 0.5 ml into each nostril. Animals were sedated with ketamine hydrochloride (10 mg/kg i.m.). For the intratracheal inoculation, the animal was placed in dorsal recumbency, and the mouth was opened manually. The epiglottis was opened with a large laryngoscope, and the small end of a sterile French rubber feeding tube (size 10, cut to 4 in. in length) was inserted into the glottis with the inoculation syringe attached. Once in place, the inoculum (1 ml) was injected into the trachea. The catheter was left in place while the inoculum syringe was removed and replaced with a syringe containing 2 ml of sterile, buffered physiological saline used for washing the catheter to ensure that all of the inoculum had been introduced into the trachea. For the i.n. inoculation, 0.5 ml inoculum was administered with a sterile luer-tip syringe introduced approximately 3 to 5 mm into each nostril. Body weights, temperature, and food intake were measured, and clinical observations were recorded.
Determination of viral titers.
For RNA isolation, tracheal lavage samples were spun down at 10,000 × g for 1 h, liquid was poured off, and 1 ml of RNA Stat-60 (IsoTex Diagnostics, Friendswood, TX) was added. Samples were then incubated at room temperature for 5 min and resuspended in 250 μl of chloroform by vortexing. The samples were spun down at 10,000 × g for 1 h, the aqueous top layer was removed, 0.5 ml of isopropanol and 10 μl of tRNA (10 μg/ml) were added, and the mixture was precipitated overnight at −20°C. Samples were spun down for 1 h, washed with cold 75% ethanol, and spun again for another hour. RNA was resuspended in 30 μl RNase-free water. For real-time PCR, 10% RNA was added to TaqMan reagents (Applied Biosystems, Foster City, CA) along with primers and probe (listed below) and amplified in a 7700 sequence detection system (Applied Biosystems). Briefly, the sample was reverse transcribed at 48°C for 30 min, held at 95°C for 10 min, and then run for 40 cycles at 95°C for 15 s and at 60°C for 1 min. The signal was compared to a standard curve of known concentrations of RNA, from 106 copies/ml down to 1 copy/ml, and multiplied by 10, giving a detection range from 20 to 107 copies/ml. All samples were run in triplicate. The primers and probe were designed to bind to a highly conserved region on the nucleoprotein gene. The primer sequences were VIETA-U, 5′-CGT CTC AAG GCA CCA AAC G-3′, and VIETA-D, 5′-GTA GAA CCT CCC AAT GCC AC-3′. The probe sequence was VIETA-P, FAM-GGA ACG CCA GAA TGC TAC TGA GAT CAG GGC-TAMRA, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine.
Histopathology.
Early deaths did not occur among the animals in the study; all animals survived to the scheduled necropsy time point at 10 days after challenge. Animals were euthanized via sodium pentobarbital overdose (>100 mg/kg, intravenously). At termination, a complete gross necropsy was conducted by testing facility personnel. Tissues were preserved in 10% neutral buffered formalin and sent to Pathology Associates International's Frederick, MD, facility for processing and histopathological evaluation. Appropriate tissues were trimmed, processed, embedded in paraffin, sectioned at approximately 5 μm, and stained with hematoxylin and eosin. The resulting glass slides were examined by a board-certified veterinary pathologist, and all microscopic pathology findings were directly entered into a Microsoft Office Excel (version 2003 SP2) spreadsheet. Histopathological evaluations were performed by the undersigned veterinary pathologist on the livers, spleens, tracheas, tonsils, hearts, and lungs from the 20 macaques. For each animal, at least 5 fields were analyzed; the analyzer was blinded to animal group. The sections were visualized, using a Zeiss Axioplan 2 microscope with a 10× objective. Digital images of the slides were captured, using a CoolSnap digital color camera (Roper Scientific, Tucson, AZ) equipped with MetaMorph software (Universal Imaging Corporation, Downington, PA).
Statistical analysis.
Statistical analysis of the data was performed using Microsoft Excel or GraphPad Prism software. Data analysis was carried out with treatment comparisons using the Wilcoxon signed-rank test or one-way analysis of variance, where statistically significant results were defined as having a P value of less than 0.05. The viral load comparison between the groups at each time point was performed using the unpaired Student's t test, where statistically significant results were defined as having a P value of less than 0.05. Correlations were established by using the Pearson test.
RESULTS
Humoral immunogenicity of plasmid-based influenza virus vaccines in nonhuman primates.
Primates were immunized three times with a previously described (18) consensus SynCon influenza pComb vaccine by i.m. or i.d. electroporation or received i.m. a pNP plasmid that induces only cellular immunity (see Materials and Methods). HAI assays were performed on clade-matched (clade 1) and divergent H5N1 viruses to compare the ability of our synthetic vaccine to induce relevant and cross-reactive antibody responses in primates, in addition to comparing the routes of immunization in inducing such responses. As detailed in Table 1, following the second immunization, 100% of macaques (5 of 5) in both the i.m. and i.d. groups developed HAI titers over 1:40 against a clade 1 virus. Each of these animals also developed HAI titers above 1:40 against a divergent clade 2.3.4 virus. In addition, 4 of 5 macaques in both the i.m. and i.d. groups had detectable levels of inhibition against a clade 2.2 virus. Inhibition of a clade 2.1 virus was detectable following the second immunization, although at a lower level.
TABLE 1.
HAI and microneutralization data for multiple clades of H5N1 influenza virus
| Assay and parameters | Mean titers (range) of antibodies to indicated clade:a
|
|||
|---|---|---|---|---|
| Clade 1 A/Vietnam | Clade 2.1 A/Indonesia | Clade 2.2 A/Turkey | Clade 2.3.4 A/Anhui | |
| HAI | ||||
| 2nd Immunization | ||||
| i.m. | 160 (80-320) | 36 (20-80) | 110 (0-320)4/5 | 80 (40-160) |
| i.d | 664 (40-1,280) | 120 (20-320) | 205 (0-320)4/5 | 592 (40-1,280) |
| 3rd Immunization | ||||
| i.m. | 288 (160-640) | 32 (0-80)3/5 | 36 (20-80) | 84 (20-160) |
| i.d. | 416 (160-640) | 64 (0-160)2/5 | 145 (20-320) | 276 (20-640) |
| Microneutralization | ||||
| 3rd Immunization | ||||
| i.m. | 144 (40-360) | 8 (0-40)1/5 | 32 (0-80)2/5 | 88 (0-160)4/5 |
| i.d. | 740 (20-2,560) | 96 (0-320)3/5 | 296 (0-1,280)3/5 | 1,172 (20-2,560) |
Superscript values represent the number of responders in the group.
Microneutralization assays were also performed on sera following the third immunization. In contrast to HAI assays, which test the ability of antibodies to inhibit virus-receptor binding, microneutralization assays test the ability of antibodies to inhibit actual infection in vitro. Both i.m. and i.d electroporation induced relatively high microneutralization titers against a clade 1 virus; i.d. immunization induced higher levels of clade-matched and cross-clade neutralization against all clades in a majority of macaques. Furthermore, i.d. immunization induced significant neutralization of clade 2.3.4 viruses in 4 of 5 macaques.
Cellular immunogenicity of plasmid-based influenza vaccines in nonhuman primates.
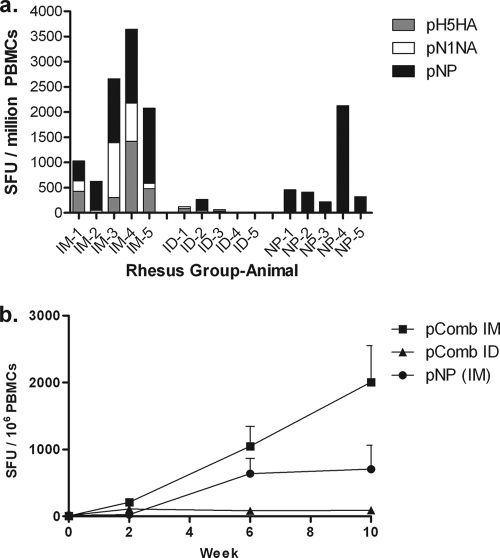
We analyzed the ability of the i.m. and i.d. routes of immunization to induce cellular immune responses as determined by IFN-γ ELISPOT assays. We included a group immunized i.m. with only pNP in order to determine the impact of cellular immune responses in the absence of antibody during viral challenge. Figure 1a shows the results of the IFN-γ ELISPOT assay 2 weeks following the third immunization: i.m. immunization with electroporation (mean count, 2,007 ± 897; range, 623 to 3,644) or with pNP alone (mean count, 707 ± 315; range, 218 to 2,128) induced strong cellular immune responses to each administered antigen in the cocktail. In contrast, i.d. immunization with electroporation induced weaker responses (mean count, 90 ± 40; range 0 to 263). The average responses over time are also shown (Fig. 1b), each time point being 2 weeks after the previous vaccination. Naïve macaques showed no antigen-specific responses at any point during the study.
FIG. 1.
Cellular immune responses. Fig. 1a shows ELISPOT assay results from individual macaques 2 weeks after the third immunization against each of the administered antigens. Figure 1b shows the mean ELISPOT assay results for each group over time (each time point being 2 weeks following the previous immunization), with each error bar showing ± 1 standard error of the mean. SFU, spot forming units; PBMC, peripheral blood mononuclear cell; IM, intramuscular; ID, intradermal.
H5N1 challenge of vaccinated macaques.
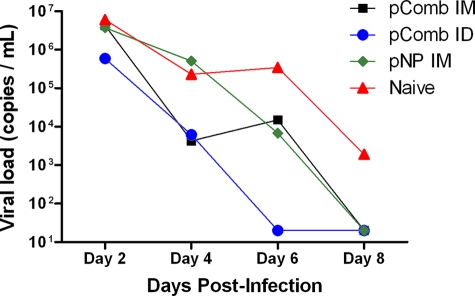
To determine if the induced immune responses were capable of impacting infection, macaques were inoculated through the i.n. and intratracheal routes with H5N1 influenza virus. The challenge virus (A/Vietnam/1203/04) was able to infect and replicate in rhesus macaques and, as expected, did not cause lethal disease. The high levels of replication in naïve animals allowed us to study the impact of vaccine-induced immune responses on viral shedding. As shown in Fig. 2, naïve macaques had high levels of replication by day 2. In a trend that continued throughout the remainder of the study, macaques immunized i.d. with pComb showed the greatest reduction in viral loads compared to vector-immunized controls, with an average reduction of 5 logs by day 6. Nevertheless, viral loads were significantly decreased in all treated groups (P < 0.003, as determined by one-way analysis of variance). Viral load reductions were also analyzed independently, with the pComb-treated group (i.m. or i.d.; P < 0.05) showing significant differences compared to the naïve group or pNP group at day 4 postchallenge. At day 6 postchallenge, all treated groups had significantly lower viral loads than the naïve animals (pComb, P < 0.01; i.d. pComb, P < 0.0001; pNP, P < 0.03), while the group treated i.d. with pComb also had significantly lower titers than the pNP group (P < 0.003). A strong correlation between decreased viral titers and prechallenge HAI was seen in the group vaccinated i.d. with pComb at day 4 postchallenge (r2 = 0.75), with a weaker correlation detected in the group vaccinated i.m. with pComb at day 4 postchallenge (r2 = 0.36).
FIG. 2.
Viral replication following challenge. Real-time PCR was performed on tracheal lavage samples every other day after challenge. Average viral loads ± standard errors of the means are shown. Viral loads were significantly decreased by pComb treatment (i.m. or i.d.; P < 0.05) compared to those for the naïve or pNP group at day 4 postchallenge. At day 6 postchallenge, all treated groups had significantly lower viral loads than the naïve animals (pComb, P < 0.01; i.d. pComb, P < 0.0001; pNP, P < 0.03), while the group treated i.d. with pComb also had significantly lower titers than the pNP group (P < 0.003). IM, intramuscular; ID, intradermal.
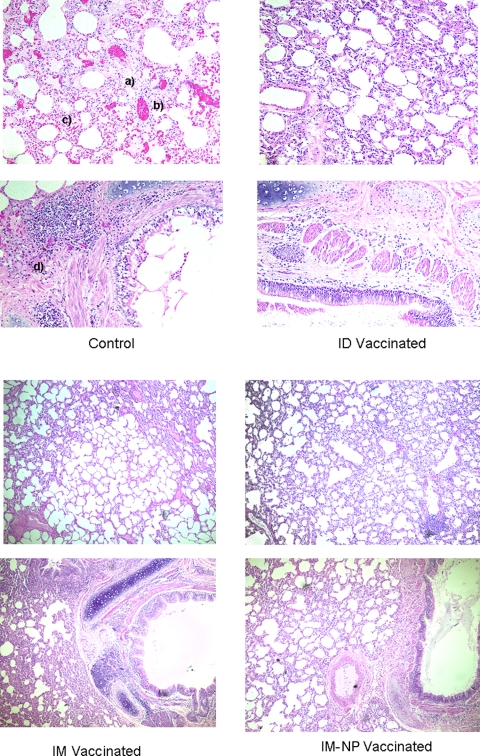
Histopathology findings.
Significant differences in lung pathology were observed between immunized and control macaques (Fig. 3). The principal histopathological changes evident in lung sections of both vaccinated and vector-treated control macaques included minimal to moderate interstitial pneumonia. Control macaques exhibited significant pathology, with peribronchobronchiolar cellular inflammation and infiltration by mixed inflammatory cells (mainly lymphocytes, eosinophils, and macrophages) that extended into adjacent alveolar septa, in addition to significant edema. Macaques immunized both i.m. and i.d. with pComb showed significant protection from inflammation, infiltration, and edema: vaccinated animals had little or no microscopic edema, peribronchobronchiolar infiltration, or inflammation. Interestingly, however, the macaques vaccinated with pNP alone showed moderate inflammation and edema but highly decreased levels of infiltration. There was no significant pathology detected in the livers, spleens, tracheas, tonsils, and hearts of any of the groups.
FIG. 3.
Histopathological analysis of lung tissue. Two panels are shown for each control, i.m., i.d., and i.m. pNP group. All slides have been stained with hematoxylin and eosin. The histopathology of lung tissue after animal sacrifice 10 days after challenge is shown. For the control panels, “a” indicates edema, “b” indicates infiltration, “c” indicates inflammation, and “d” indicates peribronchobronchiolar infiltration and inflammation. IM, intramuscular; ID, intradermal.
Clinical observations.
No vaccine-related adverse clinical signs or effects on body weight gain were observed with any of the vaccines. NHP weights remained stable throughout the study, with no monkey having a greater than ±1.5% change in body weight. Animals were sacrificed on day 10 postchallenge.
DISCUSSION
Current strategies for vaccination against influenza virus rely on the induction of serotype-specific antibodies to prevent infection. One of the initial challenges encountered in developing vaccines against H5N1 influenza virus was the inability to reliably induce seroconversion to the target antigen (H5 hemagglutinin) (20). Reasons for this include the difficulty in producing conformationally correct recombinant H5 hemagglutinin and the difficulty in growing the virus in chicken eggs, owing to its own pathogenicity. DNA vaccines avoid this problem by taking advantage of the host cellular machinery that would otherwise produce virus. However, induction of seroconversion in primates with DNA vaccines has proven difficult. Furthermore, this problem is compounded by the diversity of circulating H5N1 viruses, which have proven difficult to target with a single vaccine (8, 19). The current study combined the novel strategy of in vivo electroporation with synthetic consensus vaccines delivered by either i.m. or i.d. immunization. This technique has been used for a series of vaccines, with some entering human clinical trials (1). Nevertheless, systematic studies regarding the impact of the route of vaccination of DNA vaccines delivered by in vivo electroporation on the type of immune responses generated in NHP and on the impact these responses have on primary infection have not been performed. In our case, we showed that significant protection from severe disease can be offered by conventional i.m. immunization in addition to the easier-to-administer i.d. methodology.
The induction of protective titers of antibody against clade-matched virus in all animals, vaccinated both i.m. and i.d., following two immunizations showed that the DNA is transcribed and translated into an antigen bearing functionally relevant epitopes and that in vivo electroporation is an efficient means of delivering this vaccine. Significant cross-reactivity was also observed against clade 2.2 and 2.3.4 viruses (with all macaques having detectable levels of inhibition against each clade by the third immunization), with detectable levels of inhibition against the more difficult clade 2.1. Interestingly, each of these responses trended upward in the i.d.-administered vaccine group. These responses proved capable of reducing both viral replication and the primary causes of respiratory distress seen in influenza virus-infected individuals (edema, inflammation, and infiltration).
Extensive research with small animal models (2, 10, 15, 22), in addition to several analyses in humans (9, 21), suggests that cellular immunity could play a role in protection from the morbidity and mortality associated with influenza virus infection. Contributing to the rational behind this approach are the highly conserved nature of influenza virus nucleoprotein, conserved regions within other influenza virus antigens, and the promiscuous nature of T cells. Inducing these responses in primates has proven difficult, and thus this hypothesis remains inconclusive. In our study, in contrast to that for antibody responses, the level of induction of cellular immune responses was considerably higher in i.m.-vaccinated macaques. Both i.m.-administered pComb and i.m.-administered pNP produced responses of significant magnitude and breadth against administered antigens, while responses in the i.d.-administered pComb group were detectable but significantly lower. In the setting of infection with H5N1 influenza virus, greater protection was provided by the immune response elicited by i.d. vaccination. This may have been due to the higher level of antibody responses observed in this group or to the induction of a qualitatively different cellular immune response not detectable by the ELISPOT assays. Better protection from challenge, as shown by the decreased viral loads, was obtained by using i.d. immunization. This finding is supportive of the superiority of antibody responses over cell-mediated immunity in mediating protection against H5N1 infection in monkeys, but this issue will be addressed in depth in future studies. However, the ability of cellular immune responses alone to impact influenza virus infection with significant inhibition of viral replication and protection from virus-induced lung pathology is an important finding in this study. While the presence of antibody would be beneficial, having potent cellular immune responses present to augment incomplete or absent antibody recognition could significantly impact the toll of epidemic and pandemic influenza. Human clinical trials will be conducted to confirm these findings in the ultimate model of protection.
Acknowledgments
We thank Bradley W. Finneyfrock and Tina Long for assistance with animal care and procedures. We also thank Ruben Donis of the Influenza Branch, CDC, Atlanta, GA, for providing the H5N1-PR8 reassortant viruses A/Vietnam/1203/2004(H5N1)/PR8-IBCDC-RG, A/Indonesia/05/2005(H5N1)/PR8-IBCDC-RG2, and Anhui/01/2005/PR8-IBCDC-RG5 and Robert Newman of NIBSC, Hertfordshire, United Kingdom, for providing A/turkey/Turkey/1/2005.
This work was supported in part by NIH grants awarded to D. Weiner.
We note possible commercial conflicts associated with this work, which may include Wyeth, VGX, BMS, Virxsys, Ichor, Merck, Althea, and Aldeveron.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Bodles-Brakhop, A. M., and R. Draghia-Akli. 2008. DNA vaccination and gene therapy: optimization and delivery for cancer therapy. Expert Rev. Vaccines 71085-1101. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. M., A. M. Dilzer, D. L. Meents, and S. L. Swain. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 1772888-2898. [DOI] [PubMed] [Google Scholar]
- 3.Brundage, J. F., and G. D. Shanks. 2008. Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg. Infect. Dis. 141193-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention 1997. Isolation of avian influenza A(H5N1) viruses from humans—Hong Kong, May-December 1997. MMWR Morb. Mortal. Wkly. Rep. 461204-1207. [PubMed] [Google Scholar]
- 5.Chan, M. C., C. Y. Cheung, W. H. Chui, S. W. Tsao, J. M. Nicholls, Y. O. Chan, R. W. Chan, H. T. Long, L. L. Poon, Y. Guan, and J. S. Peiris. 2005. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, M. W., T. J. Cheng, Y. Huang, J. T. Jan, S. H. Ma, A. L. Yu, C. H. Wong, and D. D. Ho. 2008. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. USA 10513538-13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong, M. D., C. P. Simmons, T. T. Thanh, V. M. Hien, G. J. Smith, T. N. Chau, D. M. Hoang, N. V. Chau, T. H. Khanh, V. C. Dong, P. T. Qui, B. V. Cam, D. Q. Ha, Y. Guan, J. S. Peiris, N. T. Chinh, T. T. Hien, and J. Farrar. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 121203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich, H. J., M. Muller, H. M. Oh, P. A. Tambyah, C. Joukhadar, E. Montomoli, D. Fisher, G. Berezuk, S. Fritsch, A. Low-Baselli, N. Vartian, R. Bobrovsky, B. G. Pavlova, E. M. Pollabauer, O. Kistner, and P. N. Barrett. 2008. A clinical trial of a whole-virus H5N1 vaccine derived from cell culture. N. Engl. J. Med. 3582573-2584. [DOI] [PubMed] [Google Scholar]
- 9.Epstein, S. L. 2006. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 19349-53. [DOI] [PubMed] [Google Scholar]
- 10.Epstein, S. L., W. P. Kong, J. A. Misplon, C. Y. Lo, T. M. Tumpey, L. Xu, and G. J. Nabel. 2005. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 235404-5410. [DOI] [PubMed] [Google Scholar]
- 11.Hirao, L. A., L. Wu, A. S. Khan, A. Satishchandran, R. Draghia-Akli, and D. B. Weiner. 2008. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine 26440-448. [DOI] [PubMed] [Google Scholar]
- 12.Itoh, Y., H. Ozaki, H. Tsuchiya, K. Okamoto, R. Torii, Y. Sakoda, Y. Kawaoka, K. Ogasawara, and H. Kida. 2008. A vaccine prepared from a non-pathogenic H5N1 avian influenza virus strain confers protective immunity against highly pathogenic avian influenza virus infection in cynomolgus macaques. Vaccine 26562-572. [DOI] [PubMed] [Google Scholar]
- 13.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. Garcia-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445319-323. [DOI] [PubMed] [Google Scholar]
- 15.Kreijtz, J. H., R. Bodewes, G. van Amerongen, T. Kuiken, R. A. Fouchier, A. D. Osterhaus, and G. F. Rimmelzwaan. 2007. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25612-620. [DOI] [PubMed] [Google Scholar]
- 16.Kuiken, T., G. F. Rimmelzwaan, G. Van Amerongen, and A. D. Osterhaus. 2003. Pathology of human influenza A (H5N1) virus infection in cynomolgus macaques (Macaca fascicularis). Vet. Pathol. 40304-310. [DOI] [PubMed] [Google Scholar]
- 17.Kutzler, M. A., T. M. Robinson, M. A. Chattergoon, D. K. Choo, A. Y. Choo, P. Y. Choe, M. P. Ramanathan, R. Parkinson, S. Kudchodkar, Y. Tamura, M. Sidhu, V. Roopchand, J. J. Kim, G. N. Pavlakis, B. K. Felber, T. A. Waldmann, J. D. Boyer, and D. B. Weiner. 2005. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J. Immunol. 175112-123. [DOI] [PubMed] [Google Scholar]
- 18.Laddy, D. J., J. Yan, M. Kutzler, D. Kobasa, G. P. Kobinger, A. S. Khan, J. Greenhouse, N. Y. Sardesai, R. Draghia-Akli, and D. B. Weiner. 2008. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS ONE 3e2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levie, K., I. Leroux-Roels, K. Hoppenbrouwers, A. D. Kervyn, C. Vandermeulen, S. Forgus, G. Leroux-Roels, S. Pichon, and I. Kusters. 2008. An adjuvanted, low-dose, pandemic influenza A (H5N1) vaccine candidate is safe, immunogenic, and induces cross-reactive immune responses in healthy adults. J. Infect. Dis. 198642-649. [DOI] [PubMed] [Google Scholar]
- 20.Lin, J., J. Zhang, X. Dong, H. Fang, J. Chen, N. Su, Q. Gao, Z. Zhang, Y. Liu, Z. Wang, M. Yang, R. Sun, C. Li, S. Lin, M. Ji, Y. Liu, X. Wang, J. Wood, Z. Feng, Y. Wang, and W. Yin. 2006. Safety and immunogenicity of an inactivated adjuvanted whole-virion influenza A (H5N1) vaccine: a phase I randomised controlled trial. Lancet 368991-997. [DOI] [PubMed] [Google Scholar]
- 21.McElhaney, J. E., D. Xie, W. D. Hager, M. B. Barry, Y. Wang, A. Kleppinger, C. Ewen, K. P. Kane, and R. C. Bleackley. 2006. T cell responses are better correlates of vaccine protection in the elderly. J. Immunol. 1766333-6339. [DOI] [PubMed] [Google Scholar]
- 22.Powell, T. J., T. Strutt, J. Reome, J. A. Hollenbaugh, A. D. Roberts, D. L. Woodland, S. L. Swain, and R. W. Dutton. 2007. Priming with cold-adapted influenza A does not prevent infection but elicits long-lived protection against supralethal challenge with heterosubtypic virus. J. Immunol. 1781030-1038. [DOI] [PubMed] [Google Scholar]
- 23.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 756687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe, T., R. A. Abernathy, J. Hu-Primmer, W. W. Thompson, X. Lu, W. Lim, K. Fukuda, N. J. Cox, and J. M. Katz. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruat, C., C. Caillet, A. Bidaut, J. Simon, and A. D. Osterhaus. 2008. Vaccination of macaques with adjuvanted formalin-inactivated influenza A virus (H5N1) vaccines: protection against H5N1 challenge without disease enhancement. J. Virol. 822565-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephenson, I., R. G. Das, J. M. Wood, and J. M. Katz. 2007. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine 254056-4063. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson, I., J. M. Wood, K. G. Nicholson, A. Charlett, and M. C. Zambon. 2004. Detection of anti-H5 responses in human sera by HI using horse erythrocytes following MF59-adjuvanted influenza A/Duck/Singapore/97 vaccine. Virus Res. 10391-95. [DOI] [PubMed] [Google Scholar]
- 28.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2006. H5N1 virus attachment to lower respiratory tract. Science 312399. [DOI] [PubMed] [Google Scholar]
- 29.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 1711215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. September 2008, posting date. Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/avian_influenza/guidelines/recommendationvaccine.pdf.