Abstract
Dengue virus relies on a conformational change in its envelope protein, E, to fuse the viral lipid membrane with the endosomal membrane and thereby deliver the viral genome into the cytosol. We have determined the crystal structure of a soluble fragment E (sE) of dengue virus type 1 (DEN-1). The protein is in the postfusion conformation even though it was not exposed to a lipid membrane or detergent. At the domain I-domain III interface, 4 polar residues form a tight cluster that is absent in other flaviviral postfusion structures. Two of these residues, His-282 and His-317, are conserved in flaviviruses and are part of the “pH sensor” that triggers the fusogenic conformational change in E, at the reduced pH of the endosome. In the fusion loop, Phe-108 adopts a distinct conformation, forming additional trimer contacts and filling the bowl-shaped concavity observed at the tip of the DEN-2 sE trimer.
Dengue virus, a member of the flavivirus family, imposes one of the largest social and economic burdens of any mosquito-borne viral pathogen (6, 11). Three structural proteins (C, M, and E) and a lipid bilayer package the positive-strand RNA genomes of flaviviruses (13). The core nucleocapsid protein, C, binds directly to genomic RNA, while the major envelope glycoprotein, E, and the membrane protein, M, form the outer protein shell (9). C-terminal α-helical hairpins anchor E and M in the lipid membrane. E binds a receptor on the host cell surface during infection. Receptor binding directs the virion to the endocytic pathway. E responds to the reduced pH of the endosome with a large conformational rearrangement (17). This rearrangement delivers the energy required to bend the host cell membrane toward the viral membrane, inducing the two membranes to fuse (17). The fusogenic conformational rearrangement is a critical step in viral entry, as it delivers the viral genome into the cytoplasm. Crystal structures of the E protein ectodomains from dengue virus type 2 (DEN-2) and from tick-borne encephalitis (TBE) virus have been determined both before and after their fusogenic conformational rearrangements (3, 16, 17, 22, 26). The structures of DEN-3 virus E and of West Nile virus E in the prefusion conformation have also been determined (8, 18, 19). These structures provide us with a detailed molecular picture of the fusion mechanism of flaviviruses (15). First, E inserts a hydrophobic anchor, the so-called fusion loop, into the outer bilayer leaflet of the host cell membrane. Second, E folds back on itself, directing its C-terminal transmembrane anchor toward the fusion loop. This fold-back forces the host cell membrane (held by the fusion loop) and the viral membrane (held by the C-terminal transmembrane anchor) against each other, resulting in fusion of the two membranes. Here we report the crystal structure of a soluble fragment of the E protein (sE) from DEN-1 containing residues 1 to 400, that is, all but the last 50 residues of the ectodomain (Fig. 1). The protein is in the postfusion conformation even though it was never exposed to a lipid membrane or detergent.
FIG. 1.
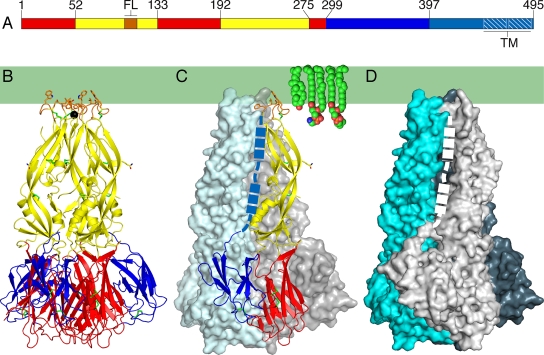
Structure of the DEN-1 sE trimer. (A) Domain I is in red, domain II is in yellow, and domain III is in blue. The fusion loop (FL) is in orange. A 53-residue “stem” (cyan) links the ectodomain to a two-helix C-terminal transmembrane anchor (TM; white hatching). (B to D) DEN-1 sE trimer, colored as in panel A, shown in a ribbon diagram (B) and as surface representations (C and D). The overall fold resembles that of DEN-2 sE (17). The hydrophobic region of the outer leaflet of the fused viral-endosomal membrane is represented by a green rectangle, with representative lipids shown as space fillers. The presumed location of the stem region is shown by a dashed line (a thicker line indicates segments that are α-helical in the mature virus) (25). (B and C) Shown in an all-atom representation are the six disulfide bonds (green), hydrophobic residues in the fusion loop (orange), and Asn67 (yellow), which is glycosylated. A chloride ion (black sphere) binds on the threefold axis near the exposed fusion loops.
MATERIALS AND METHODS
Expression, purification, and crystallization.
DEN-1 sE was expressed in Drosophila melanogaster S2 cells from the pMTBip/V5-His vector (Invitrogen) in frame with the BiP chaperone protein signal sequence. Cells were cotransfected with a hygromycin resistance marker (Invitrogen) and cultured for 6 weeks in 0.5 μg/ml hygromycin to obtain a population of expressor cells. Expression was induced with 0.5 mM CuSO4. Recombinant sE was purified from the culture medium 5 to 7 days after induction. After buffer exchange into 50 mM MES (morpholineethanesulfonic acid; pH 6.1), sE was purified by cation-exchange and size exclusion chromatographies. Crystals were grown at 16°C by hanging-drop vapor diffusion by mixing 0.4 μl of sE (at 13 g/liter in 50 mM MES, pH 6.1, 0.15 M NaCl) with 0.3 μl of the reservoir solution (12.5% polyethylene glycol 550 monomethyl ether [MME] 0.1 M MES, pH 6.5, 10 mM ZnSO4). Crystals grew after 4 weeks as small hexagonal plates. Crystals were transferred into cryoprotectant solution (30% polyethylene glycol 550 MME, 0.1 M MES, pH 6.5, 10 mM ZnSO4) and frozen in liquid nitrogen. Crystallographic data were collected at the Argonne and Brookhaven National Laboratories (see Table 1 for data collection statistics).
TABLE 1.
Data collection and refinement statistics
| Data collecteda | Value(s) |
|---|---|
| Resolution (Å) | 50.0-3.5 (3.63-3.50) |
| Rmerge | 0.223 (0.783) |
| I/σI | 13.25 (1.64) |
| % Completeness | 98.7 (92.0) |
| Redundancy | 12.1 (7.1) |
| Refinement | |
| Resolution (Å) | 25-3.5 |
| No. of reflections (working set) | 6,475 |
| No. of reflections (test set) | 311 |
| Rwork, Rfree | 0.2084, 0.2967 |
| No. of protein atoms | 2,905 |
| Avg B factors (residual after TLS refinementb) | |
| Protein (Å2) | 63 |
| Chloride ion, water (Å2) | 19 |
| RMS deviations (geometry) | |
| Bond length (Å) | 0.0179 |
| Bond angle (°) | 1.69 |
Rmerge, Σh,k,lΣi|Ii(hkl) − 〈I(hkl)〉|/Σh,k,lΣiIi(hkl), where I is an intensity that is observed i times; I/σI, signal-to-noise ratio (average observed intensity divided by average standard deviation of the observed intensity); Rwork, Σh,k,l||Fobs| − |Fcalc||/Σh,k,l|Fobs|, where h, k, l cover the “working set” of observed structure factor amplitude (Fobs) reflections used in refinement (all reflections minus the test set) and Fcalc is the calculated structure factor amplitude; Rfree, Σh,k,l||Fobs| − |Fcalc||/Σh,k,l|Fobs|, where h, k, l cover the “test set,” 5% of randomly selected Fobs reflections sequestered before refinement and not used in refinement; RMS, root mean square. Values for the highest-resolution shell (3.63 to 3.50 Å) are shown in parentheses.
See the Protein Data Bank entry for TLS tensor refinement parameters.
Structure determination.
The structure of DEN-1 sE was determined by molecular replacement using a monomer of the postfusion DEN-2 sE structure (Protein Data Bank code 1OK8 [17]) as the search model in the program PHASER 2.1 (14). The three domains in the single molecule in the asymmetric unit were first refined as rigid bodies with CNS software (4). Coordinates were then refined by simulated annealing with torsion angle dynamics with CNS and rebuilt with the software program O (7) in iterative cycles, using 2Fo − Fc and Fo − Fc Fourier maps and density-modified maps. A sharpening B factor of −80 Å2 was applied to the structure factors to obtain the most-informative maps. Later refinement cycles included restrained refinement of B factors for individual atoms and energy minimization against a maximum-likelihood target. In the final cycles, rigid-body motions of the protein molecules in the crystal were taken into account with REFMAC5 (24) in terms of “TLS” tensors for translation (T), libration (L) and correlations of libration and translation (S). The final model contains residues 1 to 144, 158 to 245, and 258 to 397; one water molecule (on a threefold crystallographic axis and visible only with B-factor sharpening); and one chloride ion (see Table 1 for refinement statistics). Ramachandran angles are good for data at a 3.5-Å resolution, with favored, additional, generous, and disallowed values of 73.0%, 26.4%, 0.6%, and 0.0%, respectively. The pseudoatomic model used to generate the entire outer protein shell of the DEN-1 virion is available upon request.
Protein Data Bank accession number.
The atomic coordinates and structure factors of the DEN-1 sE structure are available from the Protein Data Bank (entry 3G7T.pdb).
RESULTS
DEN-1 sE is in the trimeric, postfusion conformation.
The DEN-2 and TBE sE postfusion trimers are poorly soluble in the absence of detergent due to exposure of the fusion loop to the solvent upon rearrangement to the postfusion state (1, 23). Hence, because we purified DEN-1 sE without exposing it to either lipid or detergent, we expected the structure of DEN-1 sE to be in the prefusion conformation, despite the acidic pH (6.5) of the crystallization environment. Unexpectedly, DEN-1 sE crystallized in the same trimeric conformation as postfusion DEN-2 sE (17) (Fig. 1). The overall root mean square distance between atoms in the two superimposed structures is 1.38 Å (Fig. 2A). Thus, unlike DEN-2 sE and TBE virus sE, DEN-1 sE can undergo the transition to the postfusion state in the absence of lipid or detergent. Chemical cross-linking studies show, however, that our DEN-1 sE material is mostly in the monomeric, prefusion conformation in solution, even following exposure to the crystallization buffer (data not shown). We note that a longer construct of the TBE virus E ectodomain, containing helix 1 from the stem region, can form trimers in the absence of target membranes (2).
FIG. 2.
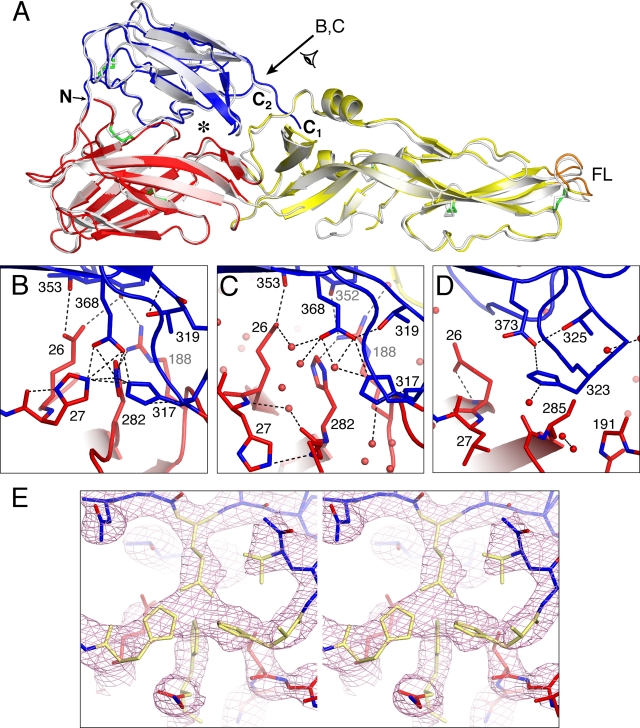
Comparison of the DEN-1 sE and DEN-2 sE structures. (A) Structure of a monomer of DEN-2 sE in the postfusion conformation (in gray) (17) superimposed on a DEN-1 sE monomer (colored as in Fig. 1), using domain I as the reference. The C terminus of DEN-1 sE (labeled C1) extends 10 Å closer to the fusion loop (FL) than the C terminus of DEN-2 sE (C2). The cluster of polar residues at the interface between domains I and III is marked with an asterisk. The direction of the view labeled B,C is shown with an arrow. N, N terminus. (B) Polar cluster at the domain I-domain III interface of DEN-1. His-27, His-282, His-317, and Glu-368 form a tight cluster at the interface between domains I and III. His-317 is required for “sensing” by E with the reduced pH of the endosome (5). In DEN-2 (C) and TBE virus (D), the homologous residues do not form any interdomain contacts. However, His-317/-323 and Glu-368/-373 still form an intradomain salt bridge. (E) Stereoscopic view of the electron density near the polar cluster at the domain I-domain III interface of DEN-1 sE. A 2Fo-Fc Fourier omit map calculated at a 25- to 3.5-Å resolution is shown in pink contoured at 1.5 σ. The polar cluster residues and the side chain atoms of Thr-319 were omitted from the coordinate set for map calculation (omitted atoms are shown in yellow). The map unambiguously shows the positions of His-27, His-317, Glu-368, and His-282 at the left, right, top, and bottom of the cluster, respectively. The map was “sharpened” by −80 Å2 (see Materials and Methods). The view is rotated ∼30° around the horizontal axis relative to the view in B.
The conformational change that drives membrane fusion consists mainly of large-scale movements of the three domains of E relative to each other, and the core architecture of each domain is conserved during the transition (17). The proteins most similar to DEN-1 E are DEN-3 and DEN-2 E, with sequence identities of 77.5% and 69.3%, respectively. As expected at these levels of sequence identity, the three individual domains of DEN-1 sE adopt the same overall three-dimensional folds as the corresponding domains in the prefusion structures of DEN-3 sE (18) and other flaviviral E proteins (8, 17, 22, 26). Domain I organizes the structure, which consists mostly of β-strands. Two long insertions in domain I form the elongated domain II, which bears the fusion loop at its tip. Domain III is an immunoglobulin C-like module (20) and is thought to contain the receptor binding site (12). The root mean square distances of atoms in domains I, II, and III from the corresponding atoms of postfusion DEN-2 sE (the most closely related postfusion structure) are 0.69 Å, 0.87 Å, and 1.00 Å, respectively. The root mean square distances between atoms in domains II and III of postfusion DEN-1 sE and prefusion DEN-3 sE (the prefusion structure with the most closely related amino acid sequence) are comparable, at 1.56 Å and 1.60 Å, respectively. The root mean square distance between domain I atoms in the two structures is much higher (5.39 Å) due to rearrangements in the secondary structure of domain I that accompany the fusogenic conformational change (17). The relative orientations of the three domains with respect to each other are essentially the same in the DEN-1 and DEN-2 sE trimers.
Tight cluster of polar residues at the domain I-domain III interface.
While the overall flavivirus architecture is conserved in the DEN-1 sE structure, there are significant differences between the postfusion structures of DEN-1 and DEN-2 sE at the domain I-domain III interface, where four polar residues—His-27, His-282, His-317, and Glu-368—form a tight cluster that nucleates the interdomain interface in DEN-1 sE (Fig. 2B). Each residue in the cluster is within hydrogen bonding distance of each of the other three residues in the cluster. Since two of the residues are in domain I and the other two are in domain III, the cluster stabilizes the domain I-domain III interface in the postfusion trimer. His-282, His-317, and Glu-368 are conserved in flaviviruses, including dengue, TBE, yellow fever, West Nile, and Japanese encephalitis viruses. His-27 is conserved in dengue virus but not in other flaviviruses. Despite this high degree of sequence conservation, however, the cluster is not observed in the DEN-2 or TBE virus postfusion E structures (Fig. 2C and D). The presence of the cluster is likely due to the different environments used to express and crystallize the DEN-1 trimer. Specifically, unlike postfusion DEN-2 sE and TBE virus sE, postfusion DEN-1 sE was crystallized at a pH that is typical of early endosomes, pH 6.5. His-317 is a critical part of the “pH sensor” that triggers the fusogenic conformational change in response to the reduced pH of the endosome. Indeed, protonation of His-317 at low pH destabilizes the prefusion domain I-domain III interface and stabilizes the postfusion E trimer (5). The basis for this trimer stabilization is unclear from previous studies. Our structure provides a possible explanation, at least for DEN-1, since His-317 in domain III forms contacts with both His-27 and His-282 in domain I, thereby stabilizing the postfusion domain I-domain III interface (Fig. 2B). The presence of an interdomain contact between His-317 and His-282 in the postfusion trimer may also explain the reduced thermal stability and less efficient trimer formation of a TBE virus E mutant in which the TBE virus His-282 homolog, His-287, and one other histidine have been mutated (5).
Because the polar cluster contains only one negative charge, on Glu-368, protonation of all three histidine side chains in the cluster to their positively charged forms would be electrostatically unfavorable. Based on our structure, His-282 is unlikely to be protonated because of its proximity to the positively charged side chain of Arg-188. His-27 is likely to be protonated because of contacts with both the main-chain carbonyl oxygen and the negatively charged Glu-368 side chain. For His-317, our structure is consistent with either the protonated state or the unprotonated state. The proposed pH-sensing role of His-317 suggests a protonated state at endosomal pH (5). We note that the pH of the crystallization solvent of DEN-1 sE, pH 6.5, is close to the pKa of the histidine side chain.
Distinct shape of the fusion loop membrane anchor.
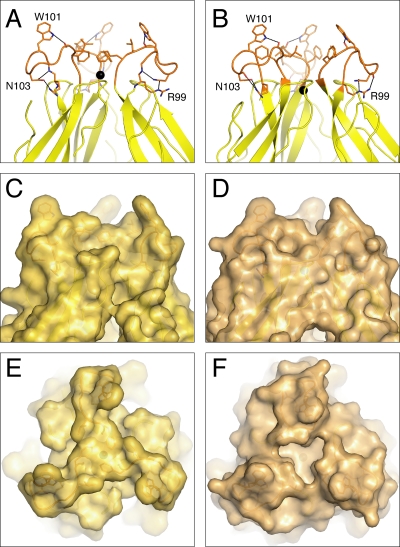
In the first stage of the fusogenic conformational change, the fusion loop becomes exposed on the viral surface. Trimer contacts then begin to form throughout the E protein, and the fusion loop inserts into the outer lipid leaflet of the host cell endosomal membrane (17). Despite its limited penetration into the endosomal membrane, the fusion loop is anchored sufficiently firmly in the membrane to withstand the physical forces created during the second half of the fusogenic conformational change, when contacts created as E trimerizes bend the viral and endosomal membranes toward one another to the point of membrane fusion (17). The backbone conformations of the fusion loops in the DEN-1 and DEN-2 sE postfusion structures are similar. As in DEN-2 sE, there can be no lipid micelle covering the fusion loop in DEN-1 sE, as this region is involved in crystal contacts with residues in domain III of a symmetry-related molecule. However, in DEN-1 sE, the side chain of Phe-108 adopts a different conformation, forming additional trimer contacts and filling the bowl-shaped concavity observed in DEN-2 sE at the tip of the trimer. As a result, the apex of the DEN-1 sE trimer has the flat shape of a mesa rather than of a concave bowl (Fig. 3). This difference is likely due to the different environments used to crystallize the DEN-1 and DEN-2 sE trimers. Indeed, unlike other postfusion sE structures, DEN-1 sE was crystallized in the absence of detergent. However, long-range effects of neighboring unconserved regions could also contribute to the difference. If the conformation of the DEN-1 sE fusion loop is maintained during membrane insertion, its bulkier shape may displace a larger number of lipid molecules upon insertion into the membrane. This could result in a more pronounced nipple-like protrusion with positive membrane curvature upon fusion loop insertion into the membrane, possibly enhancing fusion activity (10).
FIG. 3.
Structure of the fusion loop region of the sE trimer. Closeup of the aromatic anchor formed by Trp-101 and Phe-108 in the fusion loops (orange) of DEN-1 sE (A) and DEN-2 sE (B). The chloride ion (black) is shifted by 1.9 Å along the threefold axis in DEN-1 sE relative to its position in DEN-2 sE. The three clustered fusion loops form an apex that is mesa shaped in DEN-1 sE (C and E) and bowl shaped in DEN-2 sE (D and F). The view in panels E and F is down the threefold axis and rotated 90° relative to the views of the other panels.
Mapping sequence conservation to the viral surface.
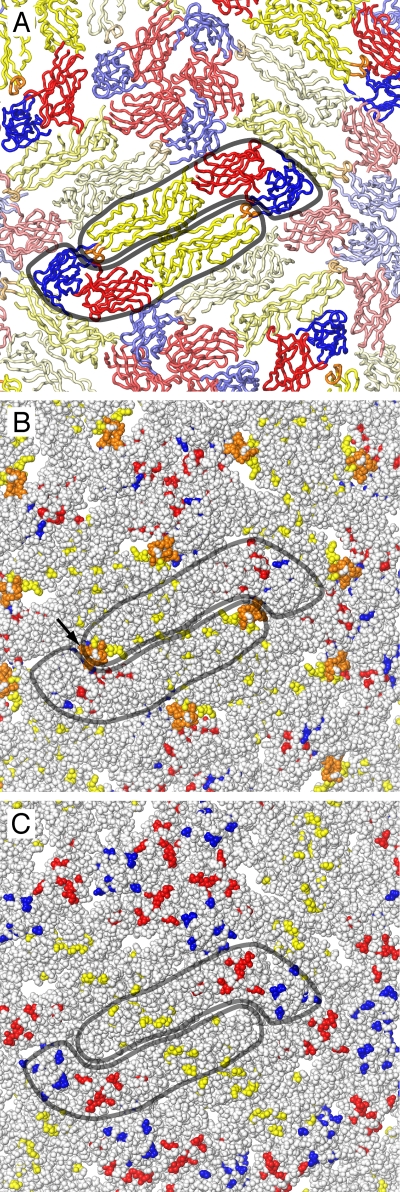
A structure of the mature DEN-2 virion has been determined by electron cryomicroscopy (cryoEM) image reconstruction at a 9.5-Å resolution (25), allowing a highly accurate fitting of the atomic coordinates of the prefusion DEN-2 sE crystal structure into the cryoEM structure (25). By superimposing the three individual domains of our DEN-1 sE structure onto the cryoEM-fitted DEN-2 sE atomic coordinates (Protein Data Bank entry 1THD.pdb) and applying icosahedral symmetry, we generated a pseudoatomic model of the entire outer protein shell of the DEN-1 virion (Fig. 4). As expected from the high degree of structural similarity of the individual domains of DEN-1 sE to homologous domains from other flaviviral sE structures (8, 16, 18, 20, 22, 26), the 180 subunits of E fit well into the in the cryoEM electron density. The pseudoatomic model shows which residues are exposed on the viral surface. Of the 75 residues that are conserved in West Nile, TBE, Japanese encephalitis, yellow fever, and dengue viruses, few are exposed on the viral surface (Fig. 4B). The largest surface-exposed cluster consists of 9 residues in the region of the fusion loop. Conserved clusters such as these may be good targets for broad-spectrum therapeutic antibodies or other antiviral therapeutics. Of the 36 DEN-1 residues that are not conserved in any of the flaviviruses listed above, 7 cluster into a ridge on the central surface of domain II, 11 are distributed across the surface of domain I, and 8 are exposed on the surface of domain III (Fig. 4C). One hundred twenty-three residues are conserved in all four dengue types but not in other flaviviruses. About two-thirds of these dengue-specific residues are exposed on the viral surface. These exposed dengue virus-specific residues determine the receptor specificity, vector preference, host range, and tropism of dengue virus.
FIG. 4.
Distribution of conserved and variable residues on the surface of mature DEN-1 virus. (A) Backbone trace of the atomic model of the DEN-1 outer protein shell based on the 9.5-Å-resolution electron microscopy reconstruction of DEN-2 (25). Two E subunits are outlined in gray. (B) Seventy-five residues are strictly conserved in the West Nile, TBE, Japanese encephalitis, yellow fever, and dengue viruses. Few of these residues are visible on the viral surface (shown as space fillers, with the same coloring scheme as in previous figures). The largest surface-exposed cluster (labeled with a black arrow) consists of 9 residues in the region of the fusion loop. (C) Same view as in panel B, with residues shown in color only if they are unique to DEN-1. Of the 36 such DEN-1-specific residues, 7 cluster in a ridge on the central surface of domain II (yellow), 11 are distributed across the surface of domain I (red), and 8 are exposed on the surface of domain III (blue).
DISCUSSION
DEN-1 sE was crystallized in the postfusion conformation. This was unexpected because the protein's solvent was not acidified in the presence of lipid membranes, a treatment that is required to convert prefusion DEN-2 sE and TBE virus sE to the postfusion conformation (17, 23). Comparison of the postfusion structures of sE from DEN-1 and DEN-2 shows that the differences are, as anticipated, relatively subtle. The presence of a polar cluster at the domain I-domain III interface suggests that the DEN-1 sE trimer may be more stable than the DEN-2 sE trimer. This could result in altered fusion properties for DEN-1, such as a higher pH threshold. Confirmation of this hypothesis will require additional careful analysis of the ability of live virus or subviral particles to fuse with target membranes at different pHs.
Surprisingly, the most-notable differences between the postfusion DEN-1 E and DEN-2 E structures are in regions with conserved amino acid sequences, namely, in the polar cluster between domains I and III and in the fusion loop. These differences are likely due to the different environments used to crystallize the DEN-1 and DEN-2 E trimers. Long-range effects of neighboring unconserved regions may, however, also contribute to the differences. Indeed, while the fusion loop itself is strictly conserved, it is immediately flanked by less conserved surfaces (formed by residues 70 to 82 and 242 to 248). These types of structural variations may modulate the efficacy of antibody neutralization from one dengue serotype to another, providing a possible explanation for the tendency of antibodies against the fusion loop to enhance infection by noncognate serotypes (21). On a more general level, the observation that conserved sequences can adopt different structures in different dengue virus types illustrates the complexity of the E proteins as antigens and the need for structural comparisons to complement genetic sequence analysis.
Acknowledgments
This work was supported by a Burroughs Wellcome Fund Investigators in Pathogenesis of Infectious Disease Program grant to Y.M. and by NIH grant R41-AI069664-01 to M.L.
We thank Rajashankar Kanagalaghatta and other staff at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source (APS), supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. We thank staff at the X25 and X29A beamlines of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. Use of the APS (under contract no. DE-AC02-06CH11357) and NSLS is supported by the Offices of Biological and of Basic Energy Sciences of the U.S. Department of Energy.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 735605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 5.Fritz, R., K. Stiasny, and F. X. Heinz. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead, S. B. 2007. Dengue. Lancet 3701644-1652. [DOI] [PubMed] [Google Scholar]
- 7.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 8.Kanai, R., K. Kar, K. Anthony, L. H. Gould, M. Ledizet, E. Fikrig, W. A. Marasco, R. A. Koski, and Y. Modis. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 8011000-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzmin, P. I., J. Zimmerberg, Y. A. Chizmadzhev, and F. S. Cohen. 2001. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA 987235-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyle, J. L., and E. Harris. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 6271-92. [DOI] [PubMed] [Google Scholar]
- 12.Lee, J. W., J. J. Chu, and M. L. Ng. 2006. Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor alphaVbeta3 integrin. J. Biol. Chem. 2811352-1360. [DOI] [PubMed] [Google Scholar]
- 13.Lindenbach, B. D., H.-J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., p.1102-1152. Lippincott Williams and Wilkins, Philadelphia, PA.
- 14.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modis, Y. 2006. Class II fusion proteins. In S. Pöhlmann and G. Simmons (ed.), Virus entry into cells. Landes Bioscience, Georgetown, TX.
- 16.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 1006986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427313-319. [DOI] [PubMed] [Google Scholar]
- 18.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 791223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 8011467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nybakken, G. E., T. Oliphant, S. Johnson, S. Burke, M. S. Diamond, and D. H. Fremont. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliphant, T., G. E. Nybakken, M. Engle, Q. Xu, C. A. Nelson, S. Sukupolvi-Petty, A. Marri, B. E. Lachmi, U. Olshevsky, D. H. Fremont, T. C. Pierson, and M. S. Diamond. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 8012149-12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375291-298. [DOI] [PubMed] [Google Scholar]
- 23.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 763784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374300-321. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 121607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]