Abstract
Human cytomegalovirus (HCMV) is a common cause of morbidity and mortality in immunocompromised and immunosuppressed individuals. During infection, HCMV is known to employ host transcription factors to facilitate viral gene expression. To further understand the previously observed delay in viral replication and protein expression in p53 knockout cells, we conducted microarray analyses of p53+/+ and p53−/− immortalized fibroblast cell lines. At a multiplicity of infection (MOI) of 1 at 24 h postinfection (p.i.), the expression of 22 viral genes was affected by the absence of p53. Eleven of these 22 genes (group 1) were examined by real-time reverse transcriptase, or quantitative, PCR (q-PCR). Additionally, five genes previously determined to have p53 bound to their nearest p53-responsive elements (group 2) and three control genes without p53 binding sites in their upstream sequences (group 3) were also examined. At an MOI of 1, >3-fold regulation was found for five group 1 genes. The expression of group 2 and 3 genes was not changed. At an MOI of 5, all genes from group 1 and four of five genes from group 2 were found to be regulated. The expression of control genes from group 3 remained unchanged. A q-PCR time course of four genes revealed that p53 influences viral gene expression most at immediate-early and early times p.i., suggesting a mechanism for the reduced and delayed production of virions in p53−/− cells.
Human cytomegalovirus (HCMV) is a common infectious agent, causing significant morbidity and mortality in immunosuppressed and immunocompromised individuals, such as AIDS patients and transplant recipients (38). In addition, HCMV is a major cause of birth defects, with a 30 to 40% risk of intrauterine transmission. Adverse effects are most likely if the infection occurs within the first half of gestation (50). One percent of newborns are infected yearly, and 5 to 10% of these exhibit neurological defects at birth. These defects include deafness, mental retardation, blindness, microencephaly, and cerebral calcification (2, 12, 18). HCMV has also recently been linked to the development of certain kinds of cancers, including malignant gliomas, prostate carcinomas, and colorectal cancers (13, 20).
As a member of the betaherpesvirus family, HCMV has a linear, double-stranded DNA genome of approximately 230 kb, containing 198 known open reading frames (ORFs) (gene image map for human herpesvirus 5 [http://www.oralgen.lanl.gov/oralgen/bacteria/hhv5/]). These ORFs are temporally expressed in three main stages (19, 47, 48). Immediate-early (IE) gene expression begins soon after the viral genome is deposited into the nucleus of the host cell. There, IE gene products drive the expression of early (E) genes, which primarily code for proteins responsible for replicating the viral genome. Once viral genome replication has progressed sufficiently, late (L) gene expression commences. L genes encode components of the capsid, the tegument, and the envelope of the virions.
To ensure precise timing of gene expression and a favorable environment in which to replicate, transcriptional cross talk between the virus and its host cell has evolved. In this cross talk, the virus utilizes cellular transcription factors such as AP-1, USF, E2F, TFIID, and Elk-1 to enhance viral gene expression (10, 24, 25, 46, 54). The viral IE transcription factors IE1 and IE2 indirectly alter host gene expression through protein-protein interactions, leading to multiple outcomes including the transition of cells from G0 to S phase, inhibition of cellular DNA replication, and prevention of apoptosis (1, 37, 55, 58).
As a key cell cycle regulator, the p53 protein is known to regulate the transcription of a variety of genes, including those involved in cell cycling, DNA repair, and programmed cell death (reviewed in reference 40). Besides transcriptional activation, p53 is an important player in the DNA repair machinery of the cell, working primarily via protein-protein interactions. It is stabilized in a normal cell following stress conditions such as UV irradiation or gamma-irradiation, hypoxia, starvation, extreme heat, or viral infection (reviewed in reference 34). This stabilization leads to the activation of the p53 protein and to downstream effects on the expression of cellular proteins, leading to cell cycle arrest or apoptosis. p53 can activate genes encoding p21/Waf1/Cip1, 14-3-3α, GADD45, and PCNA to arrest the cell cycle and allow DNA damage repair, or the apoptosis-promoting Bax, Noxa, PERP, PIG3, and Fas genes can be upregulated by p53 stabilization (26, 44). p53 is also known to function as a transcriptional inhibitor, and other groups have documented the repression of genes after p53 induction (31, 32). In addition, the C-terminal nonspecific DNA binding domain can modulate the binding of p53 to its specific binding site (BS) by protein modifications such as phosphorylation, methylation, or sumoylation, thus altering gene expression by conferring DNA structure specificity (11).
p53 contains four main functional domains (from the N to the C terminus): a protein-protein interaction domain, a specific DNA binding domain, a homotetramerization domain, and a nonspecific DNA binding domain. The protein-protein interaction domain is located on the N terminus of the protein. This domain interacts with proteins such as MDM2, TFIID, and the adenovirus protein E1B. It also serves as a transcriptional transactivation domain (9, 29, 57). The binding of MDM2 and E1B to this domain suppresses the transactivating function of p53, whereas the binding of the TATA box binding protein TFIID facilitates its function.
The specific DNA interaction domain is crucial to the transactivating ability of p53. Transcriptional activation or repression occurs when p53 binds to its responsive element (RE). The RE consists of the sequence RRRCWWGYYY (where R is a purine, Y is a pyrimidine, and W stands for A or T), followed by another RE located 0 to 13 nucleotides from the first (16). General features of strong REs include the conservation of the internal C and G residues and the presence of no more than three nonconsensus bases within the sequence. The RE configuration containing an AT sequence between the C and G residues at the middle of each half-site exhibits the strongest p53 binding (39, 53). Usually, transcriptional activation/inhibition of genes takes place within a few thousand nucleotides of the RE (reviewed in reference 28).
The third domain consists of a C-terminal helix-plus-basic-region motif used for the homotetramerization of the p53 protein, since p53 is most active in transcription as a tetramer (51). Besides the homotetramerization domain, the C terminus also contains the fourth functional domain, the nonspecific DNA binding domain, which is considered responsible for the DNA damage-sensing abilities of p53 (33). The nuclear import and export signals are also located in the C terminus.
Several studies involving p53 and HCMV have shown that p53 levels are rapidly stabilized following infection with HCMV in several cell types, including fibroblasts, vascular smooth muscle cells, human umbilical vein endothelial cells, and astrocytes (23, 27, 30, 36, 49). Although p53 levels are elevated upon viral infection, the cellular downstream targets of p53 are not activated (4, 23). However, p53 appears to play a functional role in the viral life cycle of HCMV. Huang and colleagues have shown that p53 is involved in the regulation of the viral protein UL94 (56). In addition, we have found that cells infected with HCMV in the absence of p53 produce fewer infectious viral particles (7). Examination of viral protein production (by Western blotting) and trafficking (by immunofluorescence) revealed delays in these processes in cells lacking p53. A lag in the formation of replication centers, potentially delaying viral encapsidation, was also observed (7).
Our earlier studies also showed that p53 was sequestered into the viral replication compartments and that its specific DNA binding domain was required for the sequestration of the protein into these centers (17, 41). When the HCMV genome was examined for p53 REs, 21 potential sites were found (41). Chromatin immunoprecipitation (ChIP) analyses revealed that p53 was temporally bound to some of these REs within the viral genome. Several of these REs were located in or near genes known to be essential for viral reproduction in tissue culture (15).
In this study, microarray analyses revealed that the expression levels of 22 viral genes were affected by the absence of p53. We have confirmed this observation for the 11 most highly affected genes by real time reverse transcriptase, or quantitative, PCR (q-PCR). In addition, q-PCR analyses at a multiplicity of infection (MOI) of 5 uncovered p53-dependent regulation of four of six genes shown to have p53 bound to their corresponding p53 BSs located in the vicinity of their start codons (41). q-PCR time courses for four viral genes regulated at a low MOI in microarray analyses indicated that, at both low and high MOIs, p53 exerted its greatest influence on gene expression at 24 h postinfection (hpi). To determine whether direct interaction of p53 with the defined BSs was affecting gene expression during infection, luciferase reporters were constructed using UL65, UL75, and UL122 upstream sequences. Varying effects were seen depending on the time postinfection (p.i.) and the particular site tested. Both UL75 and UL122 upstream sequences showed that the presence of intact p53 BSs, as well as cellular effects due to the p53 knockout environment, influenced promoter activity. These influences are most probably due to a combination of direct and indirect interactions of p53 with the virus and its genome. We propose that the presence of p53 is important for both IE and E viral gene expression and that it enhances the fitness, survival, and reproduction of the virus within the host. We also provide an explanation for the delay in viral replication and reproduction in 53−/− cells.
MATERIALS AND METHODS
Cells and cell culture.
Primary human foreskin fibroblasts (HFF; a kind gift from Steven Spector, University of California—San Diego) were grown in minimal essential medium with Earle's salts (MEM; Gibco BRL) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals), 200 U/ml penicillin, 200 μg/ml streptomycin, 2 mM glutamine, and 1.5 μg/ml amphotericin B (all from Gibco BRL). The telomerase-immortalized human lung fibroblasts (THF; a kind gift from John Sedivy, Brown University) were grown in Dulbecco's MEM-F-12 (Gibco BRL) supplemented with 10% FBS, 200 U/ml penicillin, 200 μg/ml streptomycin, and 2 mM glutamine. THF-derived p53−/− telomerase-immortalized human fibroblasts (also a kind gift from John Sedivy) were grown in Dulbecco's MEM-F-12 supplemented with 7.5% FBS, 200 U/ml penicillin, 200 μg/ml streptomycin, and 2 mM glutamine (5). All cells were maintained at 37°C under a humidified atmosphere containing 5% CO2.
Virus infection.
Cells were synchronized in G0 by serum starvation for 60 h (7), trypsinized, and replated onto 100-mm-diameter dishes at a density of 1 × 106 cells per plate for microarray and q-PCR analyses. Cells were then infected with the HCMV strain Towne 3 h after reseeding at an MOI of 1 for the microarray studies and at MOIs of 1 and 5 for q-PCR experiments. Virus-containing media were replaced by fresh media at 3 hpi; the cells were harvested by trypsinization; and the pellets were stored at −80°C until use.
Microarray analysis.
For microarray analysis, HFF, THF, and p53−/− fibroblasts were infected as described above and harvested at 12 and 24 hpi. RNA was extracted from the harvested cells with the SV Total RNA isolation kit (Promega) and was concentrated in a SpeedVac concentrator. RNA was amplified with the Superscript RNA amplification kit (Invitrogen), and the amplified RNA was labeled with Cy5 using the ULS labeling kit for CombiMatrix arrays (Kreatech Biotechnology).
The labeled RNAs obtained at 12 and 24 hpi from the infected HFF, THF, and p53−/− cells were hybridized to six custom-designed CombiMatrix microarray slides using protocols provided by CombiMatrix. Each of the four identical arrays on a slide consisted of 2,240 features (305 control features, 1,265 cellular gene features, and 670 viral gene features). Every gene on the array was represented by at least three features.
Three arrays on a slide were hybridized with three different Cy5-labeled RNA samples, gathered as biological replicates from three independent infection experiments. The fourth array served as a control and was hybridized with an equimolar mixture of the three biological replicate RNA samples used for the other three arrays on each slide. The data from these control arrays were consistent with the measurements of the three biological replicates but were not used for further calculations.
The fluorescence of each feature on the hybridized slides was read on an Axon GenePix 4200A microarray scanner (Axon, Sunnyvale, CA), and raw fluorescence values were obtained using the CombiMatrix microarray imager. The slides were then stripped using the protocol provided by CombiMatrix and were reprobed with the same RNAs used in the first round of hybridization. This stripping procedure was repeated twice to provide technical replicates, reducing protocol handling and device variability.
Data analysis.
The raw data were analyzed as follows. First, a mean value was calculated for each feature on the array from the three technical replicates. The variance stabilization (22) was calculated separately for the 12-hpi and 24-hpi values. The variance-stabilized values were used for six crosswise comparisons of the three cell types at the two time points (HFF versus THF, HFF versus p53−/− cells, and THF versus p53−/− cells, at 12 hpi and 24 hpi). The differences between the logarithmic values of corresponding features on different slides were calculated, and the t value of a two-tailed Student t test with a 95% confidence interval was used to determine the P value of every feature. The t value was generated from the three biological replicate values. For each individual gene, the average regulation of all features of the gene was calculated. The expression of a gene was considered significantly different in a particular comparison based on two criteria: at least 50% of the gene's features had to be significantly different in the t test (P < 0.05), and the gene had to have a >2-fold difference between the cell types, averaged over all the features of the gene.
q-PCR.
RNA was extracted from cells with the SV Total RNA isolation kit (Promega) and was reverse transcribed with Superscript II (Invitrogen) or ImProm-II reverse transcriptase (Promega). RNA samples were used for reverse transcription only if no contaminating genomic DNA could be detected by using oligonucleotide primers for IE1 in a 40-cycle PCR. q-PCR was performed on an ABI Prism 7900 system (Applied Biosystems) by using the Power SYBR green PCR master mix (Applied Biosystems) in 15-μl reaction mixtures for 40 PCR cycles. Calculations were based on absolute starting quantities, using reactions specific for glucose-6-phosphate dehydrogenase (G6PD) as normalization controls (45). Each measurement consisted of two biological replicates, gathered from independent infection experiments. In addition, each biological replicate consisted of three technical replicates, and their averaged values were used for downstream calculations.
Molecular cloning.
All plasmids used for q-PCR were generated with the pCR8 TOPO cloning kit (Invitrogen). Taq polymerase was used to amplify the insert fragments used as standards in the q-PCR experiments from the cDNA of HCMV-infected HFF. (cDNA was generated as described in the preceding section.) The sequences of the oligonucleotides used to amplify the insert fragments are available upon request.
For luciferase assays, plasmids carrying firefly luciferase under the control of either 2 kb of UL75 upstream sequence, 2 kb of UL122 upstream sequence, or 1 kb of UL65 upstream sequence were generated by amplifying the corresponding sequences from viral genomic DNA using Phusion polymerase (New England Biolabs) (primer information is available upon request). The constructs were designed with 600 bp 5′ to the p53 BS located closest to the start codon of these genes. For the UL65 upstream sequence, the distance between the p53 BS and the start codon was 400 bp; for the UL75 and UL122 upstream sequences, this distance was 1,400 bp. Amplicons were cloned into ptubFLuc (a kind gift from Gustavo Arrizabalaga) via the NsiI and KpnI sites present in both the oligonucleotide primers and 5′ of the luciferase ORF in ptubFLuc. The p53 BS in each of these plasmids was mutated by site-specific PCR mutagenesis using QuikChange (Stratagene) (primer information is available upon request). Site-specific mutations in these plasmids were checked by sequencing. As a normalization control in luciferase assays, a plasmid carrying Renilla luciferase under the control of the simian virus 40 (SV40) enhancer element was constructed. This plasmid was created by ligating the BglII/blunt-ended-HindIII 400-bp fragment of pGL4.13 (Promega) into the BglII/blunt-ended-PstI sites of pRL-CMV (Promega). This procedure exchanged the CMV enhancer for the SV40 enhancer element in this vector. Blunt ends were generated using T4 polymerase (Fermentas).
Luciferase assays.
A total of 2 × 106 THF and p53−/− cells were transiently transfected with 10 μg of one of the firefly luciferase plasmids mentioned above and 2 μg of the Renilla luciferase vector by electroporation at 330 V, 2,500 μF, and 75 Ω in a BTX ECM630 electrocell manipulator. Cells were seeded in 60-mm-diameter plates after electroporation and were infected with HCMV at an MOI of 5 at 48 h posttransfection. Cells were harvested at 24 or 72 hpi by trypsinization. Luciferase measurements were performed using the Dual Luciferase reporter assay system (Promega). Graphs show the mean ratio between the firefly luciferase and Renilla luciferase readings from two independent experiments.
Microarray data accession number.
The microarray data, including the list of genes on the arrays and the sequences of the spotted oligonucleotides, have been deposited at the GEO database under accession number GPL7474.
RESULTS
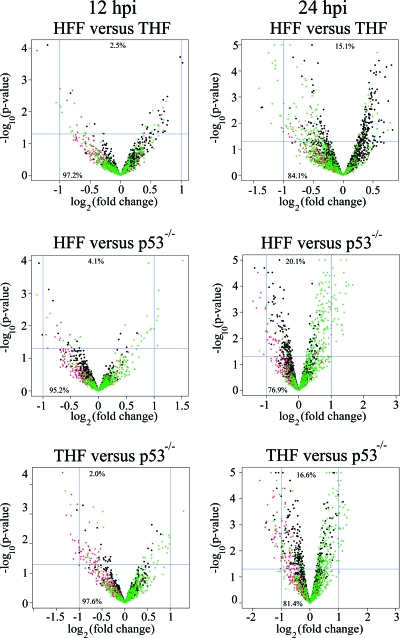
To determine the influence of p53 on the expression of viral genes, HFF, THF, and p53−/− cells were infected with the HCMV strain Towne at an MOI of 1. Cells were harvested at 12 and 24 hpi. RNA was then isolated, amplified, labeled, and hybridized to microarrays. Detailed descriptions of the hybridization procedure, the array features, and the processing of data are provided in Materials and Methods. After quantification and variance stabilization, we cross-compared the normalized fluorescence of HFF versus THF, HFF versus p53−/− cells, and THF versus p53−/− cells at 12 hpi and 24 hpi. An array feature was considered significantly different when the difference observed in the comparison of two cell types was at least twofold and the P value of the t test was below 0.05. The cross-comparisons of the analysis on the feature level are depicted in Fig. 1 as volcano plots.
FIG. 1.
Distributions of microarray features of cross-comparisons of HHF, THF, and p53−/− cells by the log2 of the n-fold regulation (on the abscissa) and the negative log10 of the P value generated from Student's t test (on the ordinate), depicted as volcano plots. Each feature is represented by a dot, with black dots representing cellular genes, red dots representing controls, and green dots representing viral genes. Significant features are represented by dots above the horizontal line at 1.3 (indicating a P value of <0.05) and left or right, respectively, of the vertical line at −1 or the vertical line at 1 (indicating a >2-fold change in one or the other cell type). The relative numbers of features in quadrants are given as percentages.
In the 12-hpi comparisons, more than 99% of all features failed at least one significance criterion. Four of 2,240 features in the HFF-versus-THF comparison (corresponding to two cellular genes, one viral gene, and one manufacturer's control gene) fulfilled both criteria of a significant change at 12 hpi. In addition, 8 of 2,240 features in the THF-versus-p53−/− cell comparison (corresponding to two cellular and four viral genes) and 16 of 2,240 features in the HFF-versus-p53−/− cell comparison (corresponding to two cellular, two viral, and nine manufacturer's control genes) were also considered significant. It should be noted that each gene has multiple features; therefore, the total number of significant features does not necessarily correlate with the sum of the corresponding genes. In the 24-hpi comparisons, 18 of 2,240 features in the HFF-versus-THF comparison (corresponding to 1 cellular, 10 viral, and 2 manufacturer's control genes), 69 of 2,240 features in the THF-versus-p53−/− cell comparison (corresponding to 3 cellular, 25 viral, and 5 manufacturer's control genes), and 68 of 2,240 features in the HFF-versus-p53−/− cell comparison (corresponding to 12 cellular, 26 viral, and 19 manufacturer's control genes) met both significance criteria. In the 24-hpi comparisons, more than 97% of all features failed at least one significance criterion.
A gene was determined to be significantly different in a particular comparison if the averaged difference over all of this gene's features was >2-fold and at least 50% of its features (with every gene being represented by at least three features on the array) had P values of <0.05. (For example, gene X had five features on the array. The changes determined for these features were 2.5-fold, 2.6-fold, 2.3-fold, 1.9-fold, and 2.8-fold, which averaged to 2.42-fold, meeting the first significance criterion. However, the t tests for each of gene X's five features resulted in two features with P values of <0.05 and three with P values of >0.05, i.e., only 40% of gene X's features were considered to be significantly different. Therefore, gene X would not be regarded as significantly changed. If three rather than two of gene X's features had had P values of <0.05, gene X would have been considered significantly regulated.) These conditions were chosen to minimize the number of false-positive results. Because our experiments concentrated on viral genes and their expression, cellular genes were not further analyzed in this study.
At 12 hpi, by application of the significance criteria defined above, no genes showed a significant difference in any of the comparisons. At 24 hpi, we found 19 genes to be significantly different in the THF-versus-p53−/− cell comparison and 14 genes to be significantly different in the HFF-versus-p53−/− cell comparison. Eleven genes were shared between HFF and THF compared to p53−/− cells (UL30, UL54, UL63, UL65, UL69, UL75, UL110, UL122, IRL4, IRS1, and TRS1) (Table 1). Eight genes were unique in the THF-versus-p53−/− cell comparison (UL48, UL68, UL84, UL100, UL106, UL109, UL111, and RL5A), and three genes were unique in the HFF-versus-p53−/− cell comparison (UL14, UL16, and US2). The 22 separate genes found in these comparisons cover ∼10% of all 198 known ORFs in the HCMV genome. When HFF and THF were compared, only US2 showed a significant difference. The expression of US2, a glycoprotein responsible for proteasomal degradation of major histocompatibility complex proteins, might therefore be dependent on the cell line (52). The average ratio of the expression level in the HFF-versus-p53−/− cell comparison to the expression level in the THF-versus-p53−/− cell comparison for the 11 shared genes was 1.2, indicating that, with the exception of US2, these two cell lines behaved similarly with respect to p53-influenced viral gene expression.
TABLE 1.
Significantly different genes in cross-comparisons in the microarray analyses
| Comparison and gene at 24 hpia | Avg fold regulation ± SD | No. of significant features/total no. of features | Kineticsb | Protein function |
|---|---|---|---|---|
| p53−/− cells vs THF | ||||
| UL30 | −2.37 ± 0.09 | 2/3 | N/A | N/A |
| UL48 | −2.93 ± 0.67 | 3/4 | L | Tegument protein |
| UL54 | −2.82 ± 0.34 | 3/3 | E | DNA polymerase subunit |
| UL63 | −5.79 ± 0.83 | 3/3 | E | N/A |
| UL65 | −7.11 ± 0.47 | 3/3 | L | N/A |
| UL68 | −2.26 ± 0.41 | 2/3 | L | N/A |
| UL69 | −2.36 ± 0.39 | 3/3 | E-L | Transcriptional regulator |
| UL75 | −3.31 ± 0.73 | 2/3 | E-L | Envelope glycoprotein |
| UL84 | −2.48 ± 0.19 | 3/4 | E-L | Transcriptional regulator |
| UL100 | −2.32 ± 0.28 | 3/3 | E-L | Envelope glycoprotein |
| UL106 | −2.07 ± 0.09 | 2/3 | L | N/A |
| UL109 | −2.45 ± 0.25 | 3/4 | IE-L | N/A |
| UL110 | −3.46 ± 0.43 | 3/3 | L | N/A |
| UL111 | −2.30 ± 0.17 | 2/3 | IE-L | N/A |
| UL122 | −2.27 ± 0.23 | 4/4 | IE | Transcriptional regulator IE2 |
| IRL4 | −2.68 ± 0.09 | 4/6 | E | N/A |
| IRS1 | −2.32 ± 0.15 | 2/4 | IE | Transcriptional regulator TRS1 |
| RL5A | −2.60 ± 0.18 | 6/6 | N/A | Probably a glycoprotein |
| TRS1 | −3.12 ± 0.21 | 4/4 | IE | Transcriptional regulator TRS1 |
| p53−/− cells vs HFF | ||||
| UL14 | 2.09 ± 0.15 | 3/5 | L | N/A |
| UL16 | 2.16 ± 0.17 | 3/3 | E | Glycoprotein, immunoevasin |
| UL30 | −2.12 ± 0.07 | 2/3 | N/A | N/A |
| UL54 | −2.25 ± 0.15 | 3/3 | E | DNA polymerase subunit |
| UL63 | −5.10 ± 1.09 | 3/3 | E | N/A |
| UL65 | −6.86 ± 0.72 | 3/3 | L | N/A |
| UL69 | −2.12 ± 0.19 | 3/3 | E-L | Transcriptional regulator |
| UL75 | −2.71 ± 0.60 | 2/3 | E-L | Envelope glycoprotein |
| UL110 | −2.17 ± 0.11 | 3/3 | L | N/A |
| UL122 | −2.08 ± 0.10 | 4/4 | E | Transcriptional regulator IE2 |
| US2 | 4.26 ± 0.35 | 4/4 | E | Glycoprotein |
| IRL4 | −2.37 ± 0.14 | 4/6 | E | N/A |
| IRS1 | −2.32 ± 0.15 | 2/4 | IE | Transcriptional regulator TRS1 |
| TRS1 | −2.13 ± 0.12 | 4/4 | IE | Transcriptional regulator TRS1 |
| HFF vs THF, US2 | 2.43 ± 0.34 | 4/4 | E | Glycoprotein |
Genes that are shared between HFF and THF cells in comparisons with p53−/− cells are shown in boldface. No significant differences in gene expression between different cell types were found at 12 hpi.
Expression kinetics of each gene as determined in reference 8. N/A, information not available.
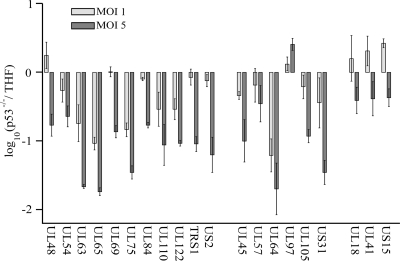
To confirm the microarray data, q-PCR was performed on a sample of 11 genes fulfilling the significance criteria of the microarray analysis (UL48, UL54, UL63, UL65, UL69, UL75, UL84, UL110, UL122, TRS1, and US2, defined as group 1 genes). These genes either showed the largest changes in the microarray analysis or were known to act at the IE stage of infection. Several other genes were also analyzed. First, UL64, located between the ORFs of UL63 and UL65, the genes with the largest downregulation in the microarray studies, was chosen for q-PCR analysis. Although no regulation in the microarray studies was observed for UL64 and no p53 binding was detected for this site in our earlier ChIP experiments, this ORF harbors a unique cluster of three overlapping p53 BSs that could influence its transcription (41). Second, five genes (UL45, UL57, UL97, UL105, and US31, defined together with UL64 as group 2 genes) previously shown to have p53 bound in their upstream sequences or in the ORF itself were chosen for q-PCR experiments (41). These genes showed an average 1.3-fold downregulation in p53−/− cells in the microarray study with an MOI of 1. Last, three genes (UL18, UL41, and US15, defined as group 3 genes) that failed the significance criteria in the microarray studies and were not associated with p53 BSs were analyzed for control purposes. The distance from the closest p53 BS to the start codon of the gene was 7.6 kb, 4.5 kb, and 18.3 kb for UL18, UL41, and US15, respectively. All q-PCR experiments were performed on THF and p53−/− cells, since THF are the parental cell line for p53−/− cells, and THF and HFF are known to have similar infection kinetics (7).
In q-PCR experiments (Fig. 2) performed at an MOI of 1, the largest changes were found in UL64, UL65, UL63, U75, UL110, and UL122: 13.9-fold, 11.0-fold, 7.0-fold, 6.5-fold, 4.0-fold, and 3.6-fold, respectively (Fig. 2). With the exception of UL64, these genes all belong to group 1. Taking into account the large standard deviations of some measurements shown in Fig. 2, differences of <3-fold in q-PCR experiments were regarded as no change. All group 3 genes analyzed as controls (UL18, UL41, and US15) were considered unchanged in p53−/− cells compared to THF at an MOI of 1. The transcription of the five group 2 genes (UL45, UL57, UL97, UL105, and US31) was also unchanged at an MOI of 1 in q-PCR experiments. This finding was consistent with our microarray results. The changes in the expression of the other six group 1 genes (UL48, UL54, UL69, UL84, TRS1, and US2), although considered significantly different in the microarray studies, fell below the threshold of threefold, and thus their expression was considered not significantly different in p53−/− cells versus THF (the average change in the expression of these genes was 1.5-fold in the q-PCR experiments and 2.6-fold in the microarray studies). However, the group 1 genes with the highest changes in the microarray studies showed similar results in q-PCR analysis.
FIG. 2.
q-PCR experiments conducted at 24 hpi on 20 viral genes at MOIs of 1 (light shaded bars) and 5 (dark shaded bars) in THF and p53−/− cells. Each bar represents the average from two independent experiments, and error bars indicate the minimum and maximum values of the single measurements. The expression of the first 11 genes (UL48 to US2) was shown to be significantly different in the microarray experiments, while the 6 genes in the middle group (UL45 to US31) were chosen to verify the results of the ChIP experiments performed by our group previously (41). The three genes in the last group (UL18 to US15) served as control genes, with no p53 BS within 3 kb of the start codons. The ordinate is scaled in a log10 manner, with negative values indicating downregulation in p53−/− cells.
The same experiment was repeated with RNA samples gathered from cells infected at an MOI of 5 (Fig. 2). Again, UL65, UL63, UL64, UL75, UL110, and UL122 were among the genes with the largest downregulation in p53−/− cells compared to THF: 55.7-fold, 45.7-fold, 35.3-fold, 29.2-fold, 14.1-fold, and 10.7-fold changes, respectively. UL97 (a group 2 gene) was the only gene that showed an increase in expression in p53−/− cells at MOIs of 1 and 5, although the change was only 2.6-fold at an MOI of 5 and was therefore disregarded due to our threshold of threefold for q-PCR experiments. The group 2 genes, implicated in our earlier ChIP experiments, showed 5- to 10-fold downregulation at an MOI of 5 in p53−/− cells relative to THF (41). The group 3 control genes were downregulated an average of only 2.6-fold at an MOI of 5. For all genes analyzed by q-PCR at 24 hpi, the number of p53−/− transcripts was on average 5.4-fold higher than the number of THF transcripts in infections with higher MOIs. As a further control, the transcript levels (numbers of gene transcripts per G6PD transcript) of all genes analyzed were determined (Table 2). For all viral genes analyzed in infected THF, the transcript levels were an average of 4.8-fold higher in cells infected at an MOI of 5 than in those infected at an MOI of 1. This follows logically from the fivefold increase in the MOI. By far the lowest expression level was measured for UL63, with 0.004 mRNA copy per G6PD copy. These q-PCR experiments suggested that the presence of p53 was crucial for transcriptional control of several HCMV genes during the IE and E stages of infection.
TABLE 2.
Transcript levels of genesa relative to G6PD transcript levels in THF at MOIs of 1 and 5 at 24 hpi
| Gene | No. of gene transcripts per G6PD transcript at an MOI of:
|
|
|---|---|---|
| 1 | 5 | |
| UL18 | 0.261 | 1 |
| UL41 | 3746 | 9,881 |
| UL45 | 73 | 136 |
| UL48 | 1288 | 3180 |
| UL54 | 20 | 47 |
| UL57 | 5 | 29 |
| UL63 | 0.004 | 0.029 |
| UL64 | 17,459 | 99,144 |
| UL65 | 186 | 1,638 |
| UL69 | 94 | 203 |
| UL75 | 93 | 135 |
| UL84 | 263 | 962 |
| UL97 | 0.050 | 2 |
| UL105 | 347 | 437 |
| UL110 | 347 | 445 |
| UL122 | 3 | 45 |
| TRS1 | 813 | 2143 |
| US2 | 47 | 155 |
| US15 | 56 | 310 |
| US31 | 71 | 270 |
Determined by q-PCR.
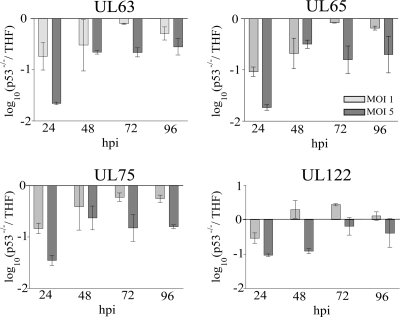
Our microarray results showed delayed expression of HCMV genes in p53−/− cells. We wondered if, as infection progressed to later time points, this reduced expression would be overcome. This would point to a major role for p53 at the IE and E phases of viral gene expression but a somewhat minor role at later stages. To assess this question and to quantify the transcriptional differences across an entire time course (24 to 96 hpi), the expression patterns of four viral genes (UL63, UL65, UL75, and UL122) in p53−/− cells and THF at MOIs of 1 and 5 were analyzed (Fig. 3). The differences between the two cell types for all genes analyzed were highest at 24 hpi at both MOIs. At 72 and 96 hpi, essentially no difference in the expression level (less than a threefold change, as defined above) of any viral gene was detected at an MOI of 1. At an MOI of 5, the difference in gene expression for UL63, UL65, and UL75 dropped to ∼5-fold beginning at 48 hpi. UL122 showed no difference between cell types beginning at 72 hpi. This confirmed that the presence of p53 influenced viral gene expression more significantly at the IE and E stages of infection. These results also suggest that as an infection progresses, viral gene expression becomes less dependent on the presence of p53.
FIG. 3.
Expression kinetics of UL63, UL65, UL75, and UL122 as determined by q-PCR over a time course of 96 hpi at MOIs of 1 (light shaded bars) and 5 (dark shaded bars). Each bar represents the average from two independent experiments, and error bars indicate the minimum and maximum values of the single measurements. The ordinate is scaled in a log10 manner, with negative values indicating downregulation in p53−/− cells.
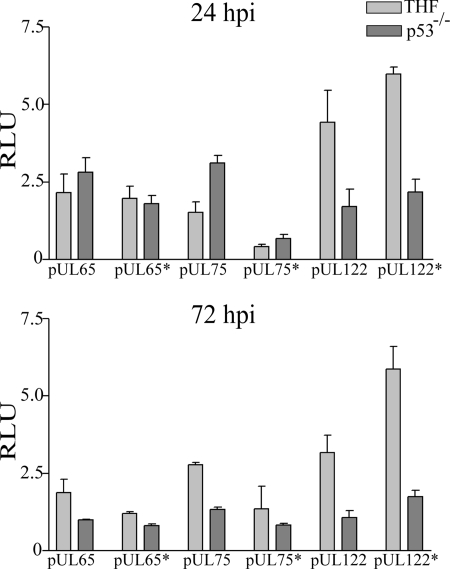
In an attempt to determine if p53 was directly involved in the transcriptional activation of viral genes or if secondary effects of the absence of p53 in the p53−/− cells were responsible for our observations, we performed luciferase reporter assays. The expression of firefly luciferase was placed under the control of three different sequences. The first sequence was 1 kb of UL65 upstream sequence, harboring the unique p53 BS with three overlapping half-sites, described previously as the UL64 BS (41). This sequence was not bound in our previous ChIP assays but was adjacent to the two genes most highly affected in microarray and q-PCR analyses. The second sequence was 2 kb of UL75 upstream sequence, harboring a p53 BS with one mismatch in each half-site. This sequence was not identified previously in the HCMV sequence analysis, due to more stringent inclusion criteria, and therefore was not tested in ChIP assays (41). Last, 2 kb of UL122 upstream sequence, harboring one predicted perfect p53 BS, was examined. p53 was bound to this site (previously referred to as the UL122/123 site) at 48 hpi in our prior ChIP experiments (41). Another set of plasmids was constructed in which the p53 BSs of the constructs described above were mutated and destroyed (see Materials and Methods for details). After electroporation of THF and p53−/− cells with either wild-type or p53 BS-deficient viral upstream sequences, cells were infected with HCMV, and luciferase measurements were performed at either 24 or 72 hpi. Constitutively expressed Renilla luciferase activity under the control of an SV40 enhancer was used as a normalization control. Only background firefly luciferase activity was observed in mock-treated cells at 24 or 72 hpi, a finding consistent with earlier observations by others using UL94 upstream sequences (21).
The first sequence analyzed was the UL65 upstream sequence, which showed differences of <1.5-fold between cell types or sequences with wild-type versus mutated p53 BSs. UL65 was therefore regarded as unchanged at both 24 and 72 hpi (Fig. 4). Although we had not seen binding of p53 to this site in the ChIP assays (41), this finding was rather surprising. UL63 and UL65 were among the genes with the highest changes in the microarray and q-PCR studies. The luciferase activity of the UL75 upstream sequence was, to a small degree, dependent on the integrity of the p53 BS, with 3.7-fold and 2-fold differences in expression between the wild-type and mutant constructs in THF at 24 hpi and 72 hpi, respectively. However, the differences observed were not due to p53 binding, since the largest differences (4.6-fold) were seen in p53−/− cells at 24 hpi. The changes seen here appear to be due to indirect effects of p53. The third sequence we analyzed was the 2-kb UL122 upstream sequence. The intact p53 BS negatively influenced the promoter activity of the UL122 gene in THF at 72 hpi, as evidenced by the 1.9-fold increase in activity after mutation of the site. This was not seen in p53−/− cells. Luciferase activity was 3-fold to 4.6-fold lower in p53−/− cells than in THF with both the wild-type and the mutated construct. This could point to binding of activating transcription factors other than p53 to this site. Overall, although the three p53 BSs we analyzed behaved differently, the observed direct influence of p53 on viral gene expression was only minor. Therefore, the major changes in transcriptional activation observed in these experiments were caused by secondary effects due to the p53 knockout environment. Our data showed that while regulation of viral gene expression was influenced by the presence of p53, intact p53 BSs in promoters of viral genes are of only minor significance to the regulation of genes observed in microarray and q-PCR analyses.
FIG. 4.
Luciferase assays using upstream sequences of UL65, UL75, or UL122 to drive the expression of firefly luciferase. Asterisks in construct designations indicate the presence of a mutated p53 BS in these sequences. Renilla luciferase expression was used as a normalization control, yielding relative light units (RLU) as the dimension on the ordinate. Each bar represents the average from two independent experiments, and error bars represent the minimum and maximum values of the single measurements.
DISCUSSION
Based on the delay in virus replication in p53−/− cells compared to p53+/+ cells, we evaluated the ability of p53 to influence viral gene expression during an infection with HCMV (7, 41). Microarray analyses were conducted with custom-designed arrays containing all known 198 HCMV ORFs at 12 and 24 hpi. At 12 hpi, no difference was detected between p53−/− cells, their parental cell line (THF), and unrelated primary fibroblasts (HFF). At 24 hpi, 19 (THF versus p53−/− cells) and 14 (HFF versus p53−/− cells) genes were found to be significantly downregulated in p53−/− cells, with 11 genes shared between the two comparisons. Our choice of an MOI of 1 and very strict criteria to identify significance in gene expression in microarray analyses may have limited the number of genes found to be regulated by the presence of p53 in this study. The data from our high-MOI q-PCR experiments indicate that additional genes are transcriptionally influenced by the presence of p53; however, our goal was to find the most obvious changes by the use of a low MOI.
The genes found to be significantly changed were classified based on their gene expression kinetics as defined by Chambers and colleagues (8). These genes included 4 IE genes (UL122, IRS1, TRS1, and UL110, corresponding to 50% of known IE genes), 10 E and E-L genes (UL16, UL54, UL63, UL69, UL75, UL84, UL100, UL106, IRL4, and US2, corresponding to 10% of known genes with E and E-L kinetics), and 6 genes with L kinetics (UL14, UL48, UL65, UL68, UL109, and UL111, corresponding to 13% of known L genes). Chambers and colleagues did not provide a timetable for the expression of the genes analyzed, but ganciclovir was used to distinguish L from IE and E genes at 72 hpi (8). Considering that replication centers are clearly visible as early as 24 hpi in HFF and THF, this does not preclude the (perhaps unstable) expression of L genes at 24 hpi (7). No information is available on the gene expression kinetics of UL30 and RL5A. Interestingly, all IE genes identified were described as transcriptional activators or enhancers. No IE apoptosis inhibitors, such as UL36 and UL37, were affected by the presence of p53 (1). This may explain the delay in viral gene expression at the IE and E stages of infection seen previously in p53−/− cells (7).
One of the most striking differences we had observed in earlier studies was the delay in viral DNA accumulation in p53−/− cells (7). q-PCR experiments revealed that the transcript levels of UL54, the catalytic subunit of the viral DNA polymerase, were downregulated twofold at an MOI of 1 (which was considered insignificant) and fourfold at an MOI of 5 in p53−/− cells. This was a moderate downregulation compared to that of other genes, such as UL63 (46-fold) or UL65 (55-fold), at an MOI of 5. However, it could help to account for the delay in viral replication when coupled with our previous data showing that other replication proteins, such as UL44 and UL57, were downregulated at the protein level in p53−/− cells (7; also unpublished data). Starisky and Hayward showed that the gene products of UL84, UL122, and UL69 (a regulator at the pre- or posttranscriptional level) are crucial for viral replication (43). In high-MOI infections, these three genes were also downregulated in p53−/− cells, as determined by q-PCR. The decreases in the levels of UL54, UL69, UL84, and UL122 transcripts could likely impair and delay viral replication in p53−/− cells (7). In addition, the transcriptional expression of five envelope glycoproteins was found to be decreased in p53−/− cells in our microarray study: UL16 (an immunoevasin), US2, UL75 (envelope glycoprotein H), UL100, and RL5A (which belongs to the RL11 glycoprotein family) (14). The damaged appearance of virions shed from p53−/− cells, observed in our unpublished electron microscopy studies, and the poor infectivity of these shed virions may be explained by the lack of glycoprotein transcripts. Indeed, previous protein studies have shown delays in tegument, capsid, and glycoprotein expression in p53−/− cells (7 and our unpublished results). Since the functions of only 12 of the 22 genes revealed in our microarray studies are known, many more processes vital to viral reproduction, such as protein particle trafficking within the cell (42), remodeling of the nuclear membrane (6), processing of replicated DNA (3), or immune response modifications (reviewed in reference 35), could, in an in vivo situation, be impaired by the observed changes in gene expression.
To relate the low-MOI microarray experiments more directly to our earlier experiments performed at a high MOI, we performed q-PCR analyses at both high and low MOIs. We found that transcription was significantly changed in a larger number of genes when infections were performed at an MOI of 5. In fact, almost all group 1 and group 2 genes were affected by the absence of p53 protein at a high MOI. It is possible that a decrease in the levels of IE transcripts in p53−/− cells results in a disproportionate reduction in the level of E transcription from the more numerous input viral genomes present at a higher MOI due to transcriptional feedback mechanisms. In contrast to these group 1 and 2 genes, the group 3 control genes showed similar expression patterns at MOIs of 1 and 5. At both MOIs, the expression of these three genes was considered unregulated.
A plausible mechanism for the transcriptional activity of p53 in the viral genome is direct binding to the viral DNA. Fourteen of the 21 p53 BSs in the viral genome were loaded with p53 protein at various times p.i. in our earlier ChIP experiments (41). To test several of these sites, luciferase reporter constructs were engineered from upstream sequences of three viral genes, containing both wild-type and mutant p53 BSs. Reporter assays were then conducted in the presence and absence of p53 to investigate if the transcript changes seen in q-PCR and microarray analyses were due to intact p53 BSs and/or to secondary effects of the absence of p53. We tested the upstream sequences of UL65, UL75, and UL122. A unique cluster of three overlapping sites exists in the UL64 ORF that may affect the transcription of UL63, UL64 itself, and UL65. Although we had not observed binding of p53 to this site in our ChIP experiments, at a high MOI these three genes showed the largest downregulation in our q-PCR experiments. Further evaluation of p53 BSs in the viral genome revealed 10 additional sites, with one mismatch in each RE. These were in addition to our original group, each of which contained one perfect RE and one with as many as three mismatches. The BS in the UL75 upstream sequence represents 1 of the 10 newly identified potential BSs. The BS in the UL122 upstream sequence represents 1 of the 21 sites previously identified. This site was shown previously to bind p53 during at least one time point during infection (41).
Transcription from the UL65 upstream sequence with its unique BS was not affected at all by the presence of p53 or its BS in these assays. In spite of the significant regulation of UL63 and UL65 in both microarray and q-PCR analyses, this result was consistent with our ChIP data, which did not show binding of this site at any time point analyzed. It should be noted that the UL65 upstream sequence consisted of only 1 kb, whereas the UL75 and UL122 sequences were 2 kb. Thus, the binding of other viral or cellular transcription factors further upstream may be necessary to explain the effects on transcription observed in q-PCR and microarray analyses. It is also possible that a larger genomic context of the p53 BS may be required in order to observe direct regulation of this site by p53. Regardless, in this context, the transcriptional effects on UL63 and UL65 appear to be due primarily to indirect effects of the absence of p53.
Luciferase assays using the UL75 upstream sequence revealed that the integrity of the p53 BS was of minor importance for transcriptional activity. However, p53 binding did not account for these changes. In addition, these assays indicated a potential time-dependent effect of the presence of p53 on the UL75 late promoter.
In luciferase assays using the UL122 upstream sequence, the downregulation observed in microarray and q-PCR experiments was reflected in decreased transcriptional activity in the p53 knockout environment in both wild-type and mutant BS configurations. It was surprising that mutation of the p53 BS upregulated the transcription of the UL122 upstream sequence. This could indicate the presence of an activating transcription factor site overlapping the p53 BS. Although the effects observed in these reporter assays were small, it should be noted that the two- to fourfold changes measured were consistent with the findings of previous studies of the effects of p53 on UL94 gene expression (56).
In future experiments, we will focus on generating recombinant viruses with mutated p53 BSs, since we are fully aware that the genomic context of these upstream sequences could be extremely important. We will also analyze the expression of the UL63 and UL65 proteins, among others, since mRNA expression and protein expression do not necessarily correlate.
In summary, this study demonstrates that p53 is a crucial player in driving viral gene expression at IE and E times p.i., and it presents an explanation for the delayed and reduced production of virions in p53−/− cells. It also contributes to further defining the role that p53 plays in the viral life cycle and expands on our earlier findings that an intact p53 DNA binding domain is essential for its sequestration and that p53 is directly bound to the viral genome at multiple times p.i. Contrary to our expectation that p53 would be implicated directly in viral transcription control, the current results indicate that the binding of p53 to the viral genome may be a mechanism the virus uses to sequester p53 in the viral replication centers, thus preventing p53 from activating its normal cellular targets.
Acknowledgments
This study was supported by NIH grants RO1-AI51463 and P20-RR015587 (COBRE program) to E.A.F.
We thank Matt Settles (Washington State University) for friendly assistance with programming and data processing and Dan Streblow (Oregon Health and Science University) for help with the initial design of experiments.
Footnotes
Published ahead of print on 18 February 2009.
REFERENCES
- 1.Andoniou, C. E., and M. A. Degli-Esposti. 2006. Insights into the mechanisms of CMV-mediated interference with cellular apoptosis. Immunol. Cell Biol. 8499-106. [DOI] [PubMed] [Google Scholar]
- 2.Boppana, S. B., K. B. Fowler, W. J. Britt, S. Stagno, and R. F. Pass. 1999. Symptomatic congenital cytomegalovirus infection in infants born to mothers with preexisting immunity to cytomegalovirus. Pediatrics 10455-60. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., K. Wagner, A. Binz, B. Sodeik, and M. Messerle. 2008. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J. Virol. 822065-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224150-160. [DOI] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 2821497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Camozzi, D., S. Pignatelli, C. Valvo, G. Lattanzi, C. Capanni, P. Dal Monte, and M. P. Landini. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89731-740. [DOI] [PubMed] [Google Scholar]
- 7.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 808390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 735757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 134107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., and M. F. Stinski. 2000. Activation of transcription of the human cytomegalovirus early UL4 promoter by the Ets transcription factor binding element. J. Virol. 749845-9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuikov, S., J. K. Kurash, J. R. Wilson, B. Xiao, N. Justin, G. S. Ivanov, K. McKinney, P. Tempst, C. Prives, S. J. Gamblin, N. A. Barlev, and D. Reinberg. 2004. Regulation of p53 activity through lysine methylation. Nature 432353-360. [DOI] [PubMed] [Google Scholar]
- 12.Cinque, P., R. Marenzi, and D. Ceresa. 1997. Cytomegalovirus infections of the nervous system. Intervirology 4085-97. [DOI] [PubMed] [Google Scholar]
- 13.Cobbs, C. S., L. Harkins, M. Samanta, G. Y. Gillespie, S. Bharara, P. H. King, L. B. Nabors, C. G. Cobbs, and W. J. Britt. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 623347-3350. [PubMed] [Google Scholar]
- 14.Davison, A. J., P. Akter, C. Cunningham, A. Dolan, C. Addison, D. J. Dargan, A. F. Hassan-Walker, V. C. Emery, P. D. Griffiths, and G. W. Wilkinson. 2003. Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J. Gen. Virol. 84657-663. [DOI] [PubMed] [Google Scholar]
- 15.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 145-49. [DOI] [PubMed] [Google Scholar]
- 17.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 722033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fowler, K. B., F. P. McCollister, A. J. Dahle, S. Boppana, W. J. Britt, and R. F. Pass. 1997. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J. Pediatr. 130624-630. [DOI] [PubMed] [Google Scholar]
- 19.Geballe, A. P., F. S. Leach, and E. S. Mocarski. 1986. Regulation of cytomegalovirus late gene expression: gamma genes are controlled by posttranscriptional events. J. Virol. 57864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harkins, L., A. L. Volk, M. Samanta, I. Mikolaenko, W. J. Britt, K. I. Bland, and C. S. Cobbs. 2002. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 3601557-1563. [DOI] [PubMed] [Google Scholar]
- 21.Huang, L., C. L. Malone, and M. F. Stinski. 1994. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J. Virol. 682108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, W., A. von Heydebreck, H. Sultmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1)S96-S104. [DOI] [PubMed] [Google Scholar]
- 23.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 696697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. M., Y. Hong, K. T. Jeang, and S. Kim. 2000. Transactivation activity of the human cytomegalovirus IE2 protein occurs at steps subsequent to TATA box-binding protein recruitment. J. Gen. Virol. 8137-46. [DOI] [PubMed] [Google Scholar]
- 25.Klucher, K. M., and D. H. Spector. 1990. The human cytomegalovirus 2.7-kilobase RNA promoter contains a functional binding site for the adenovirus major late transcription factor. J. Virol. 644189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 101054-1072. [DOI] [PubMed] [Google Scholar]
- 27.Kovacs, A., M. L. Weber, L. J. Burns, H. S. Jacob, and G. M. Vercellotti. 1996. Cytoplasmic sequestration of p53 in cytomegalovirus-infected human endothelial cells. Am. J. Pathol. 1491531-1539. [PMC free article] [PubMed] [Google Scholar]
- 28.Laptenko, O., and C. Prives. 2006. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 13951-961. [DOI] [PubMed] [Google Scholar]
- 29.Liu, X., C. W. Miller, P. H. Koeffler, and A. J. Berk. 1993. The p53 activation domain binds the TATA box-binding polypeptide in holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol. Cell. Biol. 133291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lokensgard, J. R., M. C. Cheeran, G. Gekker, S. Hu, C. C. Chao, and P. K. Peterson. 1999. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J. Hum. Virol. 291-101. [PubMed] [Google Scholar]
- 31.Maeda, Y., S. D. Seidel, G. Wei, X. Liu, and F. M. Sladek. 2002. Repression of hepatocyte nuclear factor 4α by tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol. Endocrinol. 16402-410. [DOI] [PubMed] [Google Scholar]
- 32.Maiyar, A. C., P. T. Phu, A. J. Huang, and G. L. Firestone. 1997. Repression of glucocorticoid receptor transactivation and DNA binding of a glucocorticoid response element within the serum/glucocorticoid-inducible protein kinase (sgk) gene promoter by the p53 tumor suppressor protein. Mol. Endocrinol. 11312-329. [DOI] [PubMed] [Google Scholar]
- 33.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 187621-7636. [DOI] [PubMed] [Google Scholar]
- 34.Meulmeester, E., and A. G. Jochemsen. 2008. p53: a guide to apoptosis. Curr. Cancer Drug Targets 887-97. [DOI] [PubMed] [Google Scholar]
- 35.Michelson, S. 2004. Consequences of human cytomegalovirus mimicry. Hum. Immunol. 65465-475. [DOI] [PubMed] [Google Scholar]
- 36.Muganda, P., O. Mendoza, J. Hernandez, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 688028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 803872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puchhammer-Stöckl, E., and I. Gorzer. 2006. Cytomegalovirus and Epstein-Barr virus subtypes—the search for clinical significance. J. Clin. Virol. 36239-248. [DOI] [PubMed] [Google Scholar]
- 39.Resnick, M. A., D. Tomso, A. Inga, D. Menendez, and D. Bell. 2005. Functional diversity in the gene network controlled by the master regulator p53 in humans. Cell Cycle 41026-1029. [DOI] [PubMed] [Google Scholar]
- 40.Riley, T., E. Sontag, P. Chen, and A. Levine. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9402-412. [DOI] [PubMed] [Google Scholar]
- 41.Rosenke, K., M. A. Samuel, E. T. McDowell, M. A. Toerne, and E. A. Fortunato. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 34819-34. [DOI] [PubMed] [Google Scholar]
- 42.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 792754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 707398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sax, J. K., and W. S. El-Deiry. 2003. p53-induced gene expression analysis. Methods Mol. Biol. 23465-71. [DOI] [PubMed] [Google Scholar]
- 45.Shlapobersky, M., R. Sanders, C. Clark, and D. H. Spector. 2006. Repression of HMGA2 gene expression by human cytomegalovirus involves the IE2 86-kilodalton protein and is necessary for efficient viral replication and inhibition of cyclin A transcription. J. Virol. 809951-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 992836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spaete, R. R., and E. S. Mocarski. 1985. Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans-activated by viral functions in permissive human fibroblasts. J. Virol. 56135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spector, D. H. 1996. Activation and regulation of human cytomegalovirus early genes. Intervirology 39361-377. [DOI] [PubMed] [Google Scholar]
- 49.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265391-394. [DOI] [PubMed] [Google Scholar]
- 50.Stagno, S., R. F. Pass, G. Cloud, W. J. Britt, R. E. Henderson, P. D. Walton, D. A. Veren, F. Page, and C. A. Alford. 1986. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA 2561904-1908. [PubMed] [Google Scholar]
- 51.Stürzbecher, H. W., R. Brain, C. Addison, K. Rudge, M. Remm, M. Grimaldi, E. Keenan, and J. R. Jenkins. 1992. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene 71513-1523. [PubMed] [Google Scholar]
- 52.Terauchi, M., H. Koi, C. Hayano, N. Toyama-Sorimachi, H. Karasuyama, Y. Yamanashi, T. Aso, and M. Shirakata. 2003. Placental extravillous cytotrophoblasts persistently express class I major histocompatibility complex molecules after human cytomegalovirus infection. J. Virol. 778187-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomso, D. J., A. Inga, D. Menendez, G. S. Pittman, M. R. Campbell, F. Storici, D. A. Bell, and M. A. Resnick. 2005. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc. Natl. Acad. Sci. USA 1026431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wade, E. J., K. M. Klucher, and D. H. Spector. 1992. An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J. Virol. 662407-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiebusch, L., J. Asmar, R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus immediate-early protein 2 (IE2)-mediated activation of cyclin E is cell-cycle-independent and forces S-phase entry in IE2-arrested cells. J. Gen. Virol. 8451-60. [DOI] [PubMed] [Google Scholar]
- 56.Wing, B. A., R. A. Johnson, and E. S. Huang. 1998. Identification of positive and negative regulatory regions involved in regulating expression of the human cytomegalovirus UL94 late promoter: role of IE2-86 and cellular p53 in mediating negative regulatory function. J. Virol. 721814-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 35782-85. [DOI] [PubMed] [Google Scholar]
- 58.Yu, Y., and J. C. Alwine. 2002. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 763731-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]