Abstract
Primary effusion lymphoma (PEL) is a distinct type of B-cell non-Hodgkin lymphoma characterized by the presence of Kaposi's sarcoma-associated herpesvirus (KSHV/human herpesvirus 8). Despite having a genotype and gene expression signature of highly differentiated B cells, PEL does not usually express surface or cytoplasmic immunoglobulin (Ig). We show the lack of Oct-2 and OCA-B transcription factors to be responsible, at least in part, for this defect in Ig production. Like Ig genes, ORF50, the key regulator of the switch from latency to lytic reactivation, contains an octamer motif within its promoter. We therefore examined the impact of Oct-2 and OCA-B on ORF50 activation. The binding of Oct-1 to the ORF50 promoter has been shown to significantly enhance ORF50 transactivation. We found that Oct-2, on the other hand, inhibited ORF50 expression and consequently lytic reactivation by competing with Oct-1 for the octamer motif in the ORF50 promoter. Our data suggest that Oct-2 downregulation in infected cells would be favorable to KSHV in allowing for efficient viral reactivation.
Primary effusion lymphoma (PEL) is a distinct type of B-cell non-Hodgkin lymphoma that presents most frequently in body cavities as lymphomatous effusions. Kaposi's sarcoma-associated herpesvirus (KSHV) is considered to be the etiologic agent responsible for PEL as well as for Kaposi's sarcoma and plasmablastic variants of multicentric Castleman's disease (11, 13, 62). The identification of clonal immunoglobulin (Ig) gene rearrangements and somatic hypermutation of Ig genes suggests that PEL arises from post-germinal-center B cells (2, 9, 11, 19, 27, 32, 35, 43, 48, 70). Consistent with this notion is the expression of plasma cell markers such as CD138/Syndecan-1 and somatic hypermutation of the noncoding region of the BCL-6 gene (22, 23). Recently, analysis of PEL gene expression led to its classification as immunoplasmacytoid, since it showed features most similar to AIDS immunoblastic lymphoma and multiple myeloma (30, 34). Despite the clear B-cell derivation, PEL cells lack expression of many B-cell markers, including CD19, CD20, and CD22 (2, 11, 27, 32, 35, 48, 70). Moreover, they lack surface Ig (sIg) expression despite having rearranged Ig genes without crippling mutations (2, 11, 27, 32, 35, 48, 70). Given their pre-plasma-cell derivation, PEL cells may express cytoplasmic Ig (cIg) rather than sIg. In this study we found only very low levels of cIg, and therefore, we investigated the underlying cause of this deficiency.
A cis-regulatory octamer motif, ATGCAAAT, found in virtually all Ig variable-region promoters (20, 50) and heavy-chain intronic enhancers (Eμ) (5, 24, 25), as well as in some 3′ Ig(H) locus enhancers (16, 36, 41, 44, 52), plays an important role in Ig gene transcription (1, 7, 18, 24, 31, 42, 46, 75). POU family transcription factors, the ubiquitously expressed Oct-1 and B-cell-restricted Oct-2, bind this motif (15, 47, 60, 65) and recruit the coactivator OCA-B/Bob-1/OBF-1 (28, 40, 64). The B-cell specificity of Oct-2 and OCA-B suggest that these factors play a role in Ig gene expression. Recent studies support a crucial role for these factors in sustaining Ig gene expression in plasmacytoma-T-cell-lymphoma fusion hybrids (59). Furthermore, they show that Oct-1 cannot substitute for Oct-2 in this context (59). Moreover, abnormal Ig gene expression in Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin disease has been attributed to Oct-2 and OCA-B downregulation (56, 68). Therefore, we examined whether the deficiency in Oct-2 and OCA-B expression accounts for the lack of Ig transcription in PEL. Here we show that both Oct-2 and OCA-B play important roles in Ig gene transcription in PEL and that their absence explains the defective PEL Ig gene expression profile.
ORF50, regarded as the molecular switch between KSHV latency and lytic reactivation (26, 38, 39, 66, 76), contains an octamer motif within its promoter to which Oct-1 binds, facilitating ORF50 binding and thereby enhancing its autoactivation (58). With that in mind, we were interested in examining whether Oct-2 and OCA-B exert any influence on ORF50 activation. We provide evidence that Oct-2 can bind the octamer element in the ORF50 promoter, resulting in the suppression of ORF50 transactivation, lytic gene expression, and virion production. While KSHV establishes a persistent latent infection in B cells, it will switch to lytic-phase growth upon some form of cellular stress or immune suppression. Our findings indicate that the presence of Oct-2 in PEL would impede the ability of KSHV to undergo lytic reactivation under these circumstances and that this may explain the downregulation of Oct-2 in infected cells.
MATERIALS AND METHODS
Cell lines.
The PEL cell lines BC1, BC2, BC3, BC5, BCBL1, JCS1, and BC1/Tet-on/Oct-2, the Burkitt lymphoma cell lines BJAB and Namalwa, and the lymphoblastoid cell line LCL9001 were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 40 μg/ml gentamicin (Sigma-Aldrich, St. Louis, MO) at 37°C under 5% CO2. 293T cells were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 40 μg/ml gentamicin. The BC1/Tet-on/Oct-2 stable cell line was created by transfecting BC1 cells with linearized MS4A-Oct-2. Oct-2-expressing colonies were selected 24 h after transfection by addition of hygromycin (100 μg/ml; Sigma-Aldrich) to the culture medium. Oct-2 expression was induced by addition of doxycycline (2 μg/ml; Sigma-Aldrich) and was verified by immunoblotting and flow cytometry.
Plasmids.
The Ig(H) reporter plasmids VH luc Eμ and VH luc 3A 1,2 3B 4 were kindly provided by R. Roeder (63). pCGN-Oct1, pCGN-Oct2, pCGN-OCA-B, and their parent plasmid, pCGN, were generous gifts from W. Herr (4, 67). Plasmids pcDNA3/Rta and pRpluc have been described previously (17, 66). MS4A-IκBα was a generous gift from G. W. Bornkamm (21). To create MS4A-Oct-2, pCGN-Oct2 was digested with XbaI and BamHI while MS4A-IκBα was digested with SfiI to release the IκBα insert. Oct-2 was then inserted into MS4A by blunt-end ligation. The pmax-GFP plasmid was obtained from Amaxa (Gaithersburg, MD).
Antibodies.
Anti-Oct-1 (YL15) was purchased from Upstate. Anti-Oct-2 (C-20) and anti-Bob-1 (C-20) were obtained from Santa Cruz (Santa Cruz, CA). Antiactin was obtained from Sigma-Aldrich. Anti-κ light chains (A8B5) and anti-λ light chains (N10/2) were obtained from DakoCytomation (Carpinteria, CA). Anti-ORF59 was purchased from Advanced Biotechnologies Inc. (Columbia, MD). An Alexa Fluor 488-conjugated F(ab′)2 fragment of goat anti-rabbit IgG (H+L) (A11070) and an Alexa Fluor 594-conjugated F(ab′)2 fragment of goat anti-mouse IgG (H+L) were purchased from Invitrogen.
Immunohistochemistry.
Immunostaining of cytospins of cell lines was performed with a two-step polymer-based immunoperoxidase staining technique using EnVision+ System-HRP (DakoCytomation) and Liquid DAB+ Substrate-Chromogen (DakoCytomation) as a substrate. Briefly, cytospins were air dried for 1 h, fixed in absolute acetone for 10 min, and again air dried for 5 min. A monoclonal anti-λ antibody at a dilution of 1:100 or anti-κ antibody at a dilution of 1:100 in Tris-buffered saline, pH.7.4, with 0.05% Tween 20 (TTBS) (Sigma-Aldrich), was applied for 30 min. Slides were then washed three times in 0.05% TTBS and incubated with anti-mouse EnVision-HRP for 30 min. Slides were then washed (three times, for 5 min each time) in TTBS buffer, and the peroxidase reaction was developed using Liquid DAB+ for 5 min. Cytospins were then counterstained with Mayer's hematoxylin (DakoCytomation), dehydrated in alcohol, and mounted in Permount medium (Fisher Scientific, Hampton, NH). The Namalwa (λ light chain-expressing Burkitt's lymphoma) and BJAB (κ light chain-expressing Burkitt's lymphoma) cell lines were used as controls and stained in parallel. Immunohistochemical staining of clinical samples was performed on a TechMate 500 automated immunostainer (Ventana Medical Systems, Inc., Tucson, AZ) using a ChemMate ABC peroxidase secondary detection system and employing diaminobenzidine as a chromogen (Ventana Medical Systems, Inc.) to detect Ig expression. Anti-κ and anti-λ antibodies were used at a dilution of 1:10,000/ml. Prior to immunostaining, sections were retrieved for 40 min in a water bath at 95°C using Dako Target Retrieval Solution (DakoCytomation).
Immunoblot analysis.
Whole-cell lysates were prepared in radioimmunoprecipitation assay buffer supplemented with 5 μl/ml protease inhibitor cocktail III (Calbiochem, San Diego, CA), 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM Na3VO4 (Sigma-Aldrich). Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). Proteins were then separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) by semidry transfer. Blots were probed with primary antibodies overnight at 4°C and were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The Immobilon Western chemiluminescent HRP substrate (Millipore) was used for signal detection. Blots were reprobed for actin after exposure to stripping buffer (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris [pH 6.8]) for 30 min at 50°C.
Electrophoretic mobility shift assay.
Nuclear proteins were isolated as previously described (33). Briefly, the OCT1 consensus oligonucleotide (Promega, Madison, WI) was end labeled with T4 kinase and [γ-32P]ATP. Unincorporated ATP was removed using G-25 Sephadex columns (Roche, Indianapolis, IN). Four micrograms of nuclear protein was incubated with DNA binding mixture [2 μg poly(dI-dC), 0.25% NP-40, 5% glycerol, 10 mM Tris-HCl, 50 mM KCl, 1 mM dithiothreitol, and 1 mM EDTA] for 10 min at room temperature prior to the addition of the probe. The radiolabeled oligonucleotide (2 × 104 cpm) was incubated with the reaction mixture for 15 min at room temperature. A 50-fold molar excess of unlabeled oligonucleotide was added before the addition of the labeled probe for cold competition experiments. For supershift experiments, nuclear proteins were incubated with human antibodies to Oct-1, Oct-2, and OCA-B for 30 min on ice prior to the addition of the radiolabeled oligonucleotide. Samples were separated on 7% acrylamide gels and assessed by autoradiography.
Reverse transcriptase PCR (RT-PCR).
RNA was isolated with an RNeasy kit (Qiagen, Germantown, MD). One microgram of RNA was transcribed into cDNA using a reverse transcription system (Promega) and random primers (Promega) in a final volume of 20 μl according to the manufacturer's instructions.
Seminested PCR.
The VDJ joining region of Ig(H) was amplified by a previously described seminested PCR procedure (55, 71). Briefly, the primers used were as follows: Ig-FR3 (third framework region), 5′-ACACGGC(C/T)(G/C)TGTATTACTGT-3′; Ig-JH(out) (joining region), 5′-TGAGGAGACGGTGACC-3′; Ig-JH(in) (joining region), 5′-GTGACCAGGGT(A/G/C/T)CCTTGGCCCCAG-3′. The initial PCR mixture contained 50 ng of template cDNA, 20 pmol each of primers Ig-FR3 and Ig-JH(out), 200 mM deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 4.5 mM MgCl2 in a final reaction volume of 25 μl. Initial denaturation was held at 94°C for 5 min while 0.25 U of Taq polymerase (Roche) was added, followed by 25 cycles at 93°C for 40 s, 50°C for 45 s, and 72°C for 90 s. For the second reaction, 1 μl of the first reaction product was used along with primers Ig-FR3 and Ig-JH(in). The second reaction consisted of 20 cycles of 93°C for 40 s, 55°C for 45 s, and 72°C for 90 s, followed by a final primer extension at 72°C for 7 min. The final PCR product was separated by electrophoresis on a 10% polyacrylamide gel and visualized by ethidium bromide staining.
qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR) was performed with the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) with Sybr green PCR master mix (Applied Biosystems) by using 6 μl of diluted cDNA in a 20-μl final reaction mixture. The following primer sets were used: vIL6 (5′-CGTTGATGGCTGGTAGTTCAG-3′ and 5′-GCAAGTTGCCGGACGC-3′), K8.1 (5′-TTCCACACAGATTCGCACAGA-3′ and 5′-GGCACGCCACCAGACAA-3′), ORF59 (5′-CGTCGGTAGCGGCTTCA-3′ and 5′-GGCTATGCCAGCGTCGAGTA-3′), LANA (5′-GGTGATGTTCTGAGTACATAGCGG-3′ and 5′-CCGAGGACGAAATGGAAGTG-3′), and GAPDH (5′-GGAGTCAACGGATTTGGTCGTA-3′ and 5′-GGCAACAATATCCACTTTACCAGAGT-3′). For analysis of KSHV replication, the following genome-specific primers within ORF73/72 were used: 5′-CGTCGTCGATGGGAGAACC-3′ and 5′-CGAGGGCGGGTTATTGG-3′. Melting curve analysis was performed to verify the specificity of the products. Data were analyzed using the ΔCT method (except in the case of KSHV viral replication analysis). The expression of each target gene was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression by taking the difference between threshold cycle (CT) values for target genes and GAPDH (ΔCT value). These values were then calibrated to that of the control sample to give the ΔΔCT value. The relative target gene expression is given by the formula 2−ΔΔCT.
Transfections and reporter gene assays.
For luciferase reporter assays, BCBL1, BC3, and BC1/Tet-on/Oct-2 cells were plated at a density of 6 × 105/ml, and 293T cells were plated at 60% confluence 24 h before transfection. Cells were transfected with TransFectin (Bio-Rad) according to the manufacturer's instructions. The pRL-RSV vector (Promega) was used as an internal control. Cells were collected 48 h posttransfection and were lysed in passive lysis buffer (Promega). Luciferase assays were performed using the Dual Luciferase assay system (Promega) according to the manufacturer's instructions. Firefly and Renilla luciferase activities were measured with an MLX microplate luminometer (Dynex Technologies, Inc., Chantilly, VA). The ratio of firefly to Renilla luciferase activity was calculated to normalize for transfection efficiency. For studies of lytic replication, 2.5 × 106 cells were resuspended in 100 μl of Nucleofector solution (Solution V; Amaxa Biosystems, Germany) with 2 μg of DNA and were pulsed using the Amaxa Nucleofector apparatus program T-001. Immediately after nucleofection, prewarmed RPMI was added to each cuvette, and cells were then transferred to a 24-well plate.
Flow cytometry.
Forty-eight hours posttransfection, BC3 and BC1/Tet-on/Oct-2 cells were washed in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. Cells were washed in PBS containing 0.5% bovine serum albumin, washed with staining buffer (PBS containing 1% bovine serum albumin, 0.01 M HEPES, and 0.1% saponin), and stained with an anti-ORF59 or anti-Oct-2 antibody in staining buffer at room temperature for 30 min. Cells were then washed, incubated with a corresponding secondary antibody at room temperature for 30 min, washed again, and analyzed with a FACSCalibur flow cytometer (BD Pharmingen, San Diego, CA). Transfected populations were gated according to the expression of green fluorescent protein (GFP), and 4 × 104 cells were analyzed for ORF59 to measure lytic induction and for Oct-2 to confirm its expression.
ChIP.
Chromatin immunoprecipitations (ChIPs) were performed according to Upstate Biotechnology protocols. Briefly, 107 cells were resuspended in SDS lysis buffer, cross-linked with formaldehyde, sonicated to produce DNA fragments of approximately 500 bp, and immunoprecipitated (IP) with anti-Oct-1, anti-Oct-2, or a corresponding control IgG. Genomic DNA was isolated from the IP complex by phenol-chloroform extraction and was used as a template for PCR with primers specific for the ORF50 promoter (5′-GGTACCGAATGCCACAATCTGTGCCCT-3′ and 5′-CATTTTTGTGGCTGCCTGGACAGTATTC-3′).
Examination of KSHV replication.
BC1/Tet-on/Oct-2 cells were lysed 48 h posttreatment and analyzed for both episomal DNA and encapsidated genomic DNA. To measure the level of episomal DNA, the cell lysate was subjected to phenol-chloroform extraction, and DNA was precipitated with ethanol. Two microliters was used for qRT-PCR, as described above. To assess the amount of encapsidated DNA, the cell lysate was treated with DNase I (New England Biolabs, Ipswich, MA) at 37°C for 10 min to remove viral episomal and host cell DNA. DNase I was then heat inactivated at 75°C for 10 min, and proteinase K (New England Biolabs) was added for 1 h at 56°C. The enzyme was then inactivated at 95°C for 30 min. DNA was purified by phenol-chloroform extraction and ethanol precipitation, and 2 μl was used for real-time PCR.
RESULTS
PEL tumor cells express low levels of cIg.
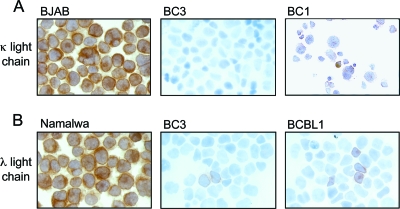
No expression of cIg light or heavy chains was identified by using standard methodology for cIg detection in plasma cells and multiple myeloma specimens (data not shown). When the sensitivity was increased to levels that detect light chains in nonplasma B cells, such as the Burkitt lymphoma cell lines Namalwa and BJAB, we found that only a small percentage of PEL cells were weakly positive for either λ or κ light chains (Fig. 1; Table 1). The level of cIg expression detected in PEL cell lines is summarized in Table 1. Little to no light-chain expression was found by immunohistochemistry and RT-PCR in all PEL cell lines. In a previous study, we found that only 4 out of 26 PEL clinical samples expressed low levels of λ light-chain-restricted Ig despite having clonal Ig gene rearrangements in 84% of cases (12). Our data, taken together with previous reports demonstrating the absence of crippling mutations in Ig genes, indicate that the defect in Ig expression in PEL lies at the transcriptional level.
FIG. 1.
PEL cells express low levels of cIg. Cytospins of PEL cell lines were analyzed for expression of κ (A) and λ (B) light chains by immunohistochemistry. BC1 showed very faint κ expression. BC3 was negative for κ light chains but showed rare λ-positive cells. A few BCBL1 cells were positive for λ light chains. BJAB and Namalwa cells were used as positive controls for κ and λ light chain expression, respectively. Experiments were performed at least three independent times.
TABLE 1.
Immunohistochemical analysis of λ and κ light chains in PEL
| Cell linea | % of cellsb with the following light chain:
|
|
|---|---|---|
| κ | λ | |
| BC1 | 10 | − |
| BC2 | − | − |
| BC3 | − | <10 |
| BC5 | − | − |
| BCBL1 | − | 10 |
| BCP1 | − | 10 |
| JCS1 | − | − |
| BJAB | + | − |
| Namalwa | − | + |
PEL cell lines are BC1, BC2, BC3, BC5, BCBL1, BCP1, and JCS1. Burkitt lymphoma cell lines, used as positive controls, are BJAB and Namalwa.
−, no light chain detected; +, light chain detected.
PEL cells lack expression of Oct-2 and OCA-B.
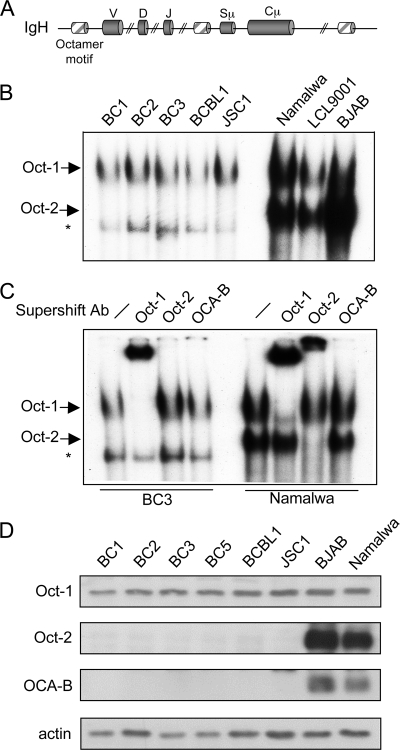
The octamer motif ATTTGCAT is the only conserved regulatory element in virtually all heavy- and light-chain variable (V) promoters and Eμ and in several Ig(H) locus 3′ enhancers (Fig. 2A), and it is essential for efficient Ig promoter activity both in vivo and in vitro (1, 7, 18, 24, 31, 42, 46, 75). We examined the ability of POU transcription factors Oct-1 and Oct-2 and the coactivator OCA-B to bind an octamer motif in PEL cells by an electrophoretic mobility shift assay. While two complexes were observed in the B-cell lymphoma control cell lines Namalwa, LCL9001, and BJAB, only the upper complex was seen in PEL cell lines (Fig. 2B). Supershift analysis revealed that Oct-1 was present in the upper complex (Fig. 2C, second and seventh lanes) and Oct-2 was part of the lower complex (Fig. 2C, third and eighth lanes). OCA-B is likely to be present in both complexes; however, we were unable to detect a shift in either complex with an anti-OCA-B antibody (Fig. 2C, fourth and ninth lanes). In all PEL cell lines, a band was observed whose makeup was not determined, but it is unlikely to contain Oct-1 or Oct-2, since it migrated further than the lower complex in Namalwa cells and was not supershifted with either the anti-Oct-1 or the anti-Oct-2 antibody. Immunoblot analysis of Oct-1, Oct-2, and OCA-B showed normal expression of Oct-1, while levels of Oct-2 and OCA-B were undetectable (Fig. 2D), confirming a previous report (3).
FIG. 2.
PEL cells lack expression of Oct-2 and OCA-B. (A) Schematic representation of octamer motifs found in the Ig(H) locus. Sμ is the switch μ region and Cμ is the constant μ region of Ig(H). (B) Electrophoretic mobility shift assay analysis of the PEL cell lines BC1, BC2, BC3, BCBL1, and JSC1 and the control B-cell lymphoma cell lines Namalwa, LCL9001, and BJAB using an oligonucleotide probe specific for octamer binding proteins. (C) Supershift analysis revealing the makeup of each protein complex. Antibodies to Oct-1, Oct-2, and OCA-B were used. The Oct-1- and Oct-2-containing complexes are labeled; the asterisk indicates a nonspecific complex. (D) Immunoblot analysis of Oct-1, Oct-2, and OCA-B in PEL cells. The Burkitt lymphoma cell lines BJAB and Namalwa were used as positive controls. Data are representative of triplicate experiments.
IgVH gene expression increases upon the introduction of Oct-2 and OCA-B.
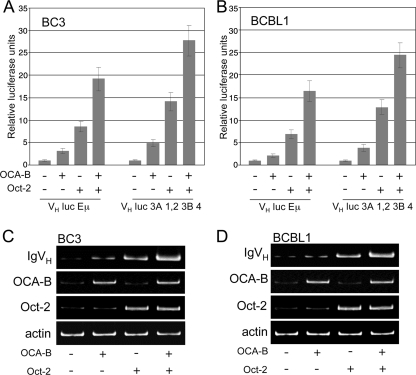
To determine whether the defect in Ig transcription was due to the lack of Oct-2 and OCA-B, we examined whether restoring their expression could induce IgVH gene transcription. BC3 and BCBL1 cells were transfected with Oct-2 and OCA-B in addition to either the VH luc Eμ reporter plasmid, containing Ig(H) promoter and Eμ elements, or the VH luc 3A 1,2 3B 4 reporter plasmid, containing Ig(H) promoter and 3′ enhancer elements (described by Stevens et al., 2000 [63]). The addition of OCA-B increased reporter activity in both cell lines, and transfection of Oct-2 caused an even greater increase. Moreover, cotransfection of OCA-B and Oct-2 synergistically enhanced the expression of both reporters (Fig. 3A and B). We then examined whether addition of these factors could induce endogenous Ig expression. By seminested PCR we detected low levels of IgVH transcript that corresponded to the low levels of Oct-2 and OCA-B transcripts (Fig. 3C and D, first lanes). As in our reporter gene assays, transfection of either Oct-2 or, to a lesser extent, OCA-B resulted in elevated IgVH transcription, while cotransfection further enhanced IgVH gene expression. This increase, however, was not detectable at the protein level by immunohistochemistry or flow cytometry (data not shown).
FIG. 3.
IgVH gene expression increases upon ectopic expression of Oct-2 and OCA-B. (A and B) IgVH luciferase reporter activity in BC3 (A) and BCBL1 (B) cells transfected with Oct-2 and/or OCA-B. Both the VH luc Eμ and the VH luc 3A 1,2 3B 4 reporter construct, described in Materials and Methods, were used. Data, representative of three independent experiments, are shown as means of triplicate values ± standard errors of the means. (C and D) Expression of endogenous IgVH transcripts in BC3 (C) and BCBL1 (D) cells, transfected as indicated. IgVH expression was determined by seminested PCR. Expression of Oct-2 and OCA-B was verified by RT-PCR. Data are representative of three independent experiments.
Oct-2 represses ORF50 transactivation.
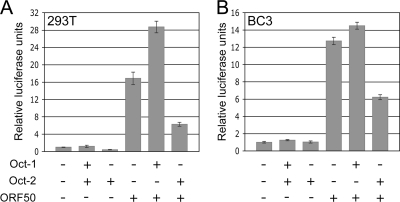
The binding of Oct-1 to an octamer element in the ORF50 promoter has been reported to enhance its transactivation (58). Since Oct-2 also associates with octamer motifs, we were interested in whether it could affect ORF50 activation. In contrast to Oct-1, which enhanced ORF50 promoter activity when cotransfected with ORF50, Oct-2 inhibited ORF50 reporter activation in both 293T and BC3 cells (Fig. 4A and B). The increase in ORF50 reporter activity after cotransfection of ORF50 with Oct-1, compared to transfection of ORF50 alone, was much larger in 293T cells than in BC3 cells, most likely because the latter express Oct-1 constitutively. Ectopic expression of OCA-B alone or in conjunction with ORF50, Oct-1, or Oct-2 had no additive effect on reporter activity (data not shown).
FIG. 4.
Oct-2 represses ORF50 transactivation. Shown is pRpORF50 luciferase reporter activity in 293T (A) and BC3 cells (B) transfected as indicated. Experiments were performed independently in triplicate, and representative data are shown as means ± standard errors of the means.
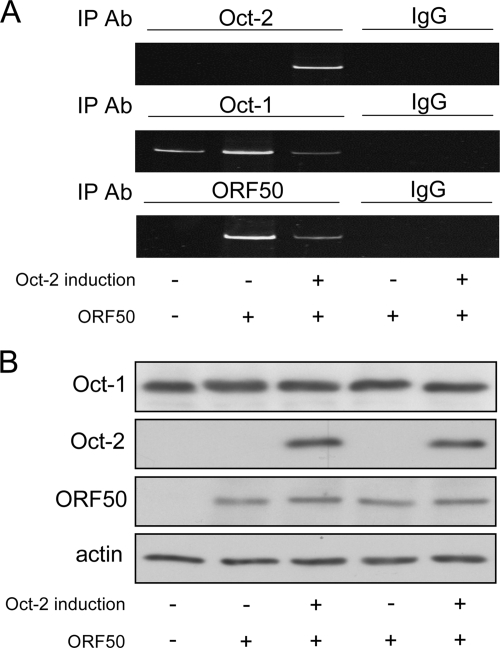
Next we examined the interactions of Oct-1 and Oct-2 with the endogenous ORF50 promoter by ChIP analysis. For this purpose, we used a stable Oct-2-inducible cell line (BC1/Tet-on/Oct-2). In uninduced cells, endogenous Oct-1 associated with the ORF50 promoter (Fig. 5A, first lane); transfection of ORF50 further enhanced its binding (Fig. 5A, second lane). Induction of Oct-2 resulted in its association with the ORF50 promoter and a decrease in the binding of Oct-1 and of ORF50 (Fig. 5A, third lane). Our data suggest that Oct-2 can compete with Oct-1 for the octamer site within the ORF50 promoter and can consequently affect the ability of ORF50 to bind its own promoter.
FIG. 5.
Oct-2 represses the binding of Oct-1 and ORF50 to the ORF50 promoter. (A) ChIP assay results show the binding of Oct-1, Oct-2, and ORF50 to the endogenous ORF50 promoter in BC1/Tet-on/Oct-2 cells under specified conditions. A control IgG corresponding to each IP antibody was used to verify specificity. Data shown are representative of three independent experiments. (B) Expression of Oct-1, Oct-2, and ORF50 under the various conditions was confirmed by Western blotting.
Oct-2 inhibits lytic reactivation.
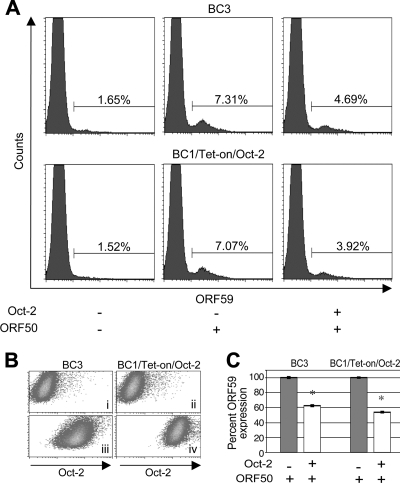
Given that Oct-2 inhibits ORF50 transactivation, we examined the impact of Oct-2 on KSHV lytic replication. For this purpose, BC3 and BC1/Tet-on/Oct-2 cells were transfected with ORF50, and the level of ORF59, an early-lytic-phase protein, was measured by flow cytometry. Cells were cotransfected with a plasmid encoding GFP to indicate transfected cells. BC3 cells were also transfected with Oct-2, whereas BC1/Tet-on/Oct-2 cells were induced to express Oct-2 with doxycycline. Forty-eight hours after transfection or induction of Oct-2, cells were fixed, permeabilized, and stained for ORF59 or Oct-2. In both cell lines, expression of Oct-2 was confirmed (Fig. 6B). GFP-positive cells were gated, and ORF59 expression was measured (Fig. 6A). On average, 40 to 45% of cells expressed GFP (data not shown). Among BC3 and BC1/Tet-on/Oct-2 cells, 1.65% and 1.52%, respectively, were positive for ORF59, in accordance with previous reports of less than 2% of PEL cells undergoing lytic replication (Fig. 6A, left). Upon ORF50 transfection, the percentage of ORF59-positive cells increased to 7.31% for BC3 and 7.07% for BC1/Tet-on/Oct-2 cells (Fig. 6A, center). Upon expression of Oct-2, the proportion of ORF59-positive cells decreased to 4.69% for BC3 and 3.92% for BC1/Tet-on/Oct-2 cells (Fig. 6A, right), corresponding to decreases in ORF59 expression of 37% and 46% in BC3 and BC1/Tet-on/Oct-2 cells, respectively (Fig. 6C). The somewhat larger decrease in ORF59 expression in BC1/Tet-on/Oct-2 cells can be explained by the higher expression of Oct-2 in these cells.
FIG. 6.
Oct-2 represses lytic reactivation. (A) BC3 and BC1/Tet-on/Oct-2 cells were analyzed for ORF59 expression by flow cytometry under untreated conditions (left), upon ORF50 transfection (middle), and upon coexpression of ORF50 and Oct-2 (right). Cells were cotransfected with GFP, and GFP-positive cells were gated to demarcate the transfected population. (B) Expression of Oct-2 in GFP-positive untransfected BC3 cells (i), uninduced BC1/Tet-on/Oct-2 cells (ii), Oct-2-transfected BC3 cells (iii), and (iv) induced BC1/Tet-on/Oct-2 cells. (C) Graph showing reduction in ORF59 expression in ORF50-transfected cells upon Oct-2 expression. All experiments were performed three independent times. P values of ≤0.01, according to Student's t test, were considered significant (*).
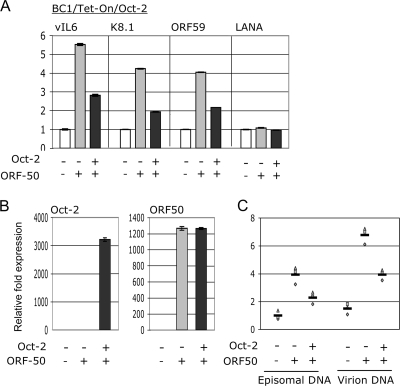
We further examined the ability of Oct-2 to inhibit lytic reactivation by looking at the transcription of lytically expressed genes. BC1/Tet-on/Oct-2 cells were transfected with ORF50, and the expression of vIL6, K8.1, and ORF59 was assessed 48 h posttransfection by real-time PCR. The expression of LANA, a latent transcript, was also assessed. Oct-2 induction reduced the level of each of the lytic transcripts by roughly 50% (Fig. 7A), while LANA expression remained unchanged.
FIG. 7.
Oct-2 represses lytic replication. (A) qRT-PCR analysis of vIL6, K8.1, ORF59, and LANA in BC1/Tet-on/Oct-2 cells under the specified conditions. (B) qRT-PCR analysis of Oct-2 and ORF50 expression. (C) Analysis of KSHV episomal and encapsidated genomic DNA by real-time PCR. For panels A and B, data are reported as means of triplicate values ± standard errors of the means, and representative data from three independent experiments are shown. In panel C, data from three independent experiments are graphed.
Finally, we analyzed the effect of Oct-2 expression on KSHV virion production in PEL. BC1/Tet-on/Oct-2 cells were transfected with ORF50. After 48 h they were lysed, and episomal and virion DNA was harvested. Expression of Oct-2 and ORF50 was confirmed by qRT-PCR (Fig. 7B). Unlike other PEL cell lines, BC1 cells are defective in their ability to release encapsidated KSHV genomes (45); therefore, upon lytic reactivation, virions accumulate within BC1 cells. Nevertheless, the protected viral DNA in virions can be distinguished from unencapsidated (episomal) DNA by digestion of the lysates with DNase prior to capsid digestion. The level of DNA was assessed by real-time PCR using KSHV genome-specific primers within ORF73/72. Transfection of ORF50 enhanced the level of episomal and virion DNA, while induction of Oct-2 opposed this ORF50-mediated increase in both forms of KSHV genomic DNA (Fig. 7C).
DISCUSSION
PEL cells are defective in their ability to produce Ig. Our results indicate that this is due, in large part, to a lack of Oct-2 and OCA-B expression. Ectopic expression of both proteins led to increased Ig heavy-chain reporter activity and increased endogenous Ig heavy-chain transcription. These results are supported by previous reports that these transcription factors are necessary for proper Ig gene expression to occur (1, 7, 18, 75).
Although Oct-2 and OCA-B play a large role in the production of Ig, other factors are also involved in Ig gene regulation. The control of Ig expression is rather complex and involves a diversity of interactions between B-cell-specific factors and regulatory elements (reviewed by Singh, 1994 [61]; Reya and Grosschedl, 1998 [57]; and Henderson and Calame, 1998 [29]). Another of these tissue-restricted factors involved in Ig transcription is PU.1, which is also absent in PEL cells (3). PU.1 is regulated through an octamer motif in its promoter, and its downregulation results from a lack of Oct-2 (3). Oct-2 and OCA-B deficiency in PEL can, therefore, impact Ig expression both directly, by not binding octamer elements within Ig genes, and indirectly, through their inability to regulate PU.1 expression.
Although Ig transcripts were detectable by seminested PCR upon addition of Oct-2 and OCA-B, the level of expression was low compared to that in B-cell lines, such as Namalwa and BJAB, and expression was undetectable at the protein level. Ushmorov et al. found that treatment of HRS cells with the demethylating agent 5-aza-2-deoxycytidine potentiated the effects of Oct-2 and OCA-B on Ig expression, although expression still did not reach levels comparable to those of normal Ig-producing B cells (69). DNA methylation may similarly contribute to the Ig gene expression profile in PEL; however, as in HRS cells, the full complement of tissue-specific factors is likely to be necessary for normal Ig transcription to be fully restored. Studies are currently under way to examine the impact of DNA methylation on Ig expression.
ORF50 orchestrates the switch from viral latency to lytic reactivation by activating a number of lytic gene promoters, including its own. It is both necessary and sufficient to induce the complete lytic cycle (26, 38, 39, 66, 76). The binding of Oct-1 to the ORF50 promoter enhances ORF50 autoactivation (58). Our data show that the presence of ORF50 protein facilitates Oct-1 promoter binding. In addition, we found that induction of Oct-2 reduced Oct-1 and ORF50 binding, suggesting that it competes with Oct-1 for the octamer element within the ORF50 promoter and that this competition reduces the ability of ORF50 to associate with the promoter. Furthermore, we show that Oct-2 repressed ORF50-mediated activation of several lytic genes, resulting in a reduction in lytic replication and virion production.
Oct-2 expression consistently resulted in a roughly 40 to 45% decrease in ORF50-mediated lytic reactivation by the various methods used. This is impressive, considering that ORF50 expression is not solely regulated by the octamer element within its promoter; other cellular transcription factors and regulatory elements in the ORF50 and lytic gene promoters have also been shown to be involved. For example, the CAAT/enhancer binding protein alpha (C/EBPα) can stimulate ORF50, ORF57, and K8 expression (72, 73), and the recombination signal binding protein (RBP-Jk) promotes the activation of ORF50, ORF57, ORF6, K-bZIP, and vGPCR (10, 74). The presence of these factors may account for the level of lytic reactivation that persists despite the inhibitory influence of Oct-2.
The herpes simplex virus (HSV) VP16 protein is essential for lytic replication (6, 8, 51, 53). Like the KSHV ORF50 protein, VP16 forms a complex with Oct-1 on octamer elements within immediate-early (IE) promoters, leading to efficient lytic gene activation (49, 54). HSV is unable to undergo lytic reactivation in neuronal cells due to the presence of Oct-2 (37). Oct-2 binds octamer motifs in HSV IE gene promoters; however, unlike Oct-1, it is unable to associate with VP16. This inability to form a complex with VP16 has been attributed to a 1-amino-acid difference between Oct-1 and Oct-2 (14). Our data suggest that, in analogy to the observations for HSV, the negative influence of Oct-2 on ORF50 transactivation stems from its inability to form a complex with the ORF50 protein on the ORF50 promoter. Future studies will examine more closely the structural differences between Oct-1 and Oct-2 that are responsible for the discrepancy in binding ORF50 protein. The consistent absence of Oct-2 in PEL leads us to speculate that this deficiency is selected for in order for ORF50 autoactivation to take place unimpeded and for lytic reactivation to follow.
Acknowledgments
This study has been partially supported by a grant from the NIH (CA68939) and an Irma T. Hirschl and Monique Weill-Caulier Career Scientist Award to E.C. and by a Cancer Research Institute Tumor Immunology Predoctoral Training Grant to D.L.D.B.
Footnotes
Published ahead of print on 18 February 2009.
REFERENCES
- 1.Annweiler, A., S. Zwilling, R. A. Hipskind, and T. Wirth. 1993. Analysis of transcriptional stimulation by recombinant Oct proteins in a cell-free system. J. Biol. Chem. 2682525-2534. [PubMed] [Google Scholar]
- 2.Ansari, M. Q., D. B. Dawson, R. Nador, C. Rutherford, N. R. Schneider, M. J. Latimer, L. Picker, D. M. Knowles, and R. W. McKenna. 1996. Primary body cavity-based AIDS-related lymphomas. Am. J. Clin. Pathol. 105221-229. [DOI] [PubMed] [Google Scholar]
- 3.Arguello, M., M. Sgarbanti, E. Hernandez, Y. Mamane, S. Sharma, M. Servant, R. Lin, and J. Hiscott. 2003. Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene 22964-973. [DOI] [PubMed] [Google Scholar]
- 4.Babb, R., M. A. Cleary, and W. Herr. 1997. OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol. Cell. Biol. 177295-7305. (Erratum, 18:2430, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji, J., L. Olson, and W. Schaffner. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33729-740. [DOI] [PubMed] [Google Scholar]
- 6.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergman, Y., D. Rice, R. Grosschedl, and D. Baltimore. 1984. Two regulatory elements for immunoglobulin kappa light chain gene expression. Proc. Natl. Acad. Sci. USA 817041-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 9.Carbone, A., A. Gloghini, E. Vaccher, V. Zagonel, C. Pastore, P. Dalla Palma, F. Branz, G. Saglio, R. Volpe, U. Tirelli, and G. Gaidano. 1996. Kaposi's sarcoma-associated herpesvirus DNA sequences in AIDS-related and AIDS-unrelated lymphomatous effusions. Br. J. Haematol. 94533-543. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, K. D., W. Bu, D. Palmeri, S. Spadavecchia, S. J. Lynch, S. A. Marras, S. Tyagi, and D. M. Lukac. 2006. Kaposi's sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J. Virol. 809697-9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 3321186-1191. [DOI] [PubMed] [Google Scholar]
- 12.Chadburn, A., E. Hyjek, S. Mathew, E. Cesarman, J. Said, and D. M. Knowles. 2004. KSHV-positive solid lymphomas represent an extra-cavitary variant of primary effusion lymphoma. Am. J. Surg. Pathol. 281401-1416. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 14.Cleary, M. A., S. Stern, M. Tanaka, and W. Herr. 1993. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 772-83. [DOI] [PubMed] [Google Scholar]
- 15.Clerc, R. G., L. M. Corcoran, J. H. LeBowitz, D. Baltimore, and P. A. Sharp. 1988. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 21570-1581. [DOI] [PubMed] [Google Scholar]
- 16.Dariavach, P., G. T. Williams, K. Campbell, S. Pettersson, and M. S. Neuberger. 1991. The mouse IgH 3′-enhancer. Eur. J. Immunol. 211499-1504. [DOI] [PubMed] [Google Scholar]
- 17.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 813043-3048. [DOI] [PubMed] [Google Scholar]
- 18.Dreyfus, M., N. Doyen, and F. Rougeon. 1987. The conserved decanucleotide from the immunoglobulin heavy chain promoter induces a very high transcriptional activity in B-cells when introduced into an heterologous promoter. EMBO J. 61685-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fais, F., G. Gaidano, D. Capello, A. Gloghini, F. Ghiotto, S. Roncella, A. Carbone, N. Chiorazzi, and M. Ferrarini. 1999. Immunoglobulin V region gene use and structure suggest antigen selection in AIDS-related primary effusion lymphomas. Leukemia 131093-1099. [DOI] [PubMed] [Google Scholar]
- 20.Falkner, F. G., and H. G. Zachau. 1984. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature 31071-74. [DOI] [PubMed] [Google Scholar]
- 21.Feuillard, J., M. Schuhmacher, S. Kohanna, M. Asso-Bonnet, F. Ledeur, R. Joubert-Caron, P. Bissieres, A. Polack, G. W. Bornkamm, and M. Raphael. 2000. Inducible loss of NF-κB activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood 952068-2075. [PubMed] [Google Scholar]
- 22.Gaidano, G., D. Capello, A. M. Cilia, A. Gloghini, T. Perin, S. Quattrone, A. Migliazza, F. Lo Coco, G. Saglio, V. Ascoli, and A. Carbone. 1999. Genetic characterization of HHV-8/KSHV-positive primary effusion lymphoma reveals frequent mutations of BCL6: implications for disease pathogenesis and histogenesis. Genes Chromosomes Cancer 2416-23. [DOI] [PubMed] [Google Scholar]
- 23.Gaidano, G., A. Gloghini, V. Gattei, M. F. Rossi, A. M. Cilia, C. Godeas, M. Degan, T. Perin, V. Canzonieri, D. Aldinucci, G. Saglio, A. Carbone, and A. Pinto. 1997. Association of Kaposi's sarcoma-associated herpesvirus-positive primary effusion lymphoma with expression of the CD138/syndecan-1 antigen. Blood 904894-4900. [PubMed] [Google Scholar]
- 24.Gerster, T., P. Matthias, M. Thali, J. Jiricny, and W. Schaffner. 1987. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 61323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillies, S. D., S. L. Morrison, V. T. Oi, and S. Tonegawa. 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33717-728. [DOI] [PubMed] [Google Scholar]
- 26.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 746207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green, I., E. Espiritu, M. Ladanyi, R. Chaponda, R. Wieczorek, L. Gallo, and H. Feiner. 1995. Primary lymphomatous effusions in AIDS: a morphological, immunophenotypic, and molecular study. Mod. Pathol. 839-45. [PubMed] [Google Scholar]
- 28.Gstaiger, M., L. Knoepfel, O. Georgiev, W. Schaffner, and C. M. Hovens. 1995. A B-cell coactivator of octamer-binding transcription factors. Nature 373360-362. [DOI] [PubMed] [Google Scholar]
- 29.Henderson, A., and K. Calame. 1998. Transcriptional regulation during B cell development. Annu. Rev. Immunol. 16163-200. [DOI] [PubMed] [Google Scholar]
- 30.Jenner, R. G., K. Maillard, N. Cattini, R. A. Weiss, C. Boshoff, R. Wooster, and P. Kellam. 2003. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 10010399-10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenuwein, T., and R. Grosschedl. 1991. Complex pattern of immunoglobulin mu gene expression in normal and transgenic mice: nonoverlapping regulatory sequences govern distinct tissue specificities. Genes Dev. 5932-943. [DOI] [PubMed] [Google Scholar]
- 32.Karcher, D. S., and S. Alkan. 1997. Human herpesvirus-8-associated body cavity-based lymphoma in human immunodeficiency virus-infected patients: a unique B-cell neoplasm. Hum. Pathol. 28801-808. [DOI] [PubMed] [Google Scholar]
- 33.Keller, S. A., E. J. Schattner, and E. Cesarman. 2000. Inhibition of NF-κB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood 962537-2542. [PubMed] [Google Scholar]
- 34.Klein, U., A. Gloghini, G. Gaidano, A. Chadburn, E. Cesarman, R. Dalla-Favera, and A. Carbone. 2003. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood 1014115-4121. [DOI] [PubMed] [Google Scholar]
- 35.Knowles, D. M., G. Inghirami, A. Ubriaco, and R. Dalla-Favera. 1989. Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenetic role of the Epstein-Barr virus. Blood 73792-799. [PubMed] [Google Scholar]
- 36.Lieberson, R., S. L. Giannini, B. K. Birshtein, and L. A. Eckhardt. 1991. An enhancer at the 3′ end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res. 19933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lillycrop, K. A., C. L. Dent., S. C. Wheatley, M. N. Beech, N. N. Ninkina, J. N. Wood, and D. S. Latchman. 1991. The octamer-binding protein Oct-2 represses HSV immediate-early genes in cell lines derived from latently infectable sensory neurons. Neuron 7381-390. [DOI] [PubMed] [Google Scholar]
- 38.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 739348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252304-312. [DOI] [PubMed] [Google Scholar]
- 40.Luo, Y., and R. G. Roeder. 1995. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 154115-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madisen, L., and M. Groudine. 1994. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. 82212-2226. [DOI] [PubMed] [Google Scholar]
- 42.Mason, J. O., G. T. Williams, and M. S. Neuberger. 1985. Transcription cell type specificity is conferred by an immunoglobulin VH gene promoter that includes a functional consensus sequence. Cell 41479-487. [DOI] [PubMed] [Google Scholar]
- 43.Matolcsy, A., R. G. Nador, E. Cesarman, and D. M. Knowles. 1998. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am. J. Pathol. 1531609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthias, P., and D. Baltimore. 1993. The immunoglobulin heavy chain locus contains another B-cell-specific 3′ enhancer close to the alpha constant region. Mol. Cell. Biol. 131547-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizushima-Sugano, J., and R. G. Roeder. 1986. Cell-type-specific transcription of an immunoglobulin kappa light chain gene in vitro. Proc. Natl. Acad. Sci. USA 838511-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller, M. M., S. Ruppert, W. Schaffner, and P. Matthias. 1988. A cloned octamer transcription factor stimulates transcription from lymphoid-specific promoters in non-B cells. Nature 336544-551. [DOI] [PubMed] [Google Scholar]
- 48.Nador, R. G., E. Cesarman, A. Chadburn, D. B. Dawson, M. Q. Ansari, J. Sald, and D. M. Knowles. 1996. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpesvirus. Blood 88645-656. [PubMed] [Google Scholar]
- 49.O'Hare, P., C. R. Goding, and A. Haigh. 1988. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 74231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parslow, T. G., D. L. Blair, W. J. Murphy, and D. K. Granner. 1984. Structure of the 5′ ends of immunoglobulin genes: a novel conserved sequence. Proc. Natl. Acad. Sci. USA 812650-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 825870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersson, S., G. P. Cook, M. Bruggemann, G. T. Williams, and M. S. Neuberger. 1990. A second B cell-specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature 344165-168. [DOI] [PubMed] [Google Scholar]
- 53.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24555-565. [DOI] [PubMed] [Google Scholar]
- 54.Preston, C. M., M. C. Frame, and M. E. Campbell. 1988. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell 52425-434. [DOI] [PubMed] [Google Scholar]
- 55.Ramasamy, I., M. Brisco, and A. Morley. 1992. Improved PCR method for detecting monoclonal immunoglobulin heavy chain rearrangement in B cell neoplasms. J. Clin. Pathol. 45770-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Re, D., M. Muschen, T. Ahmadi, C. Wickenhauser, A. Staratschek-Jox, U. Holtick, V. Diehl, and J. Wolf. 2001. Oct-2 and Bob-1 deficiency in Hodgkin and Reed Sternberg cells. Cancer Res. 612080-2084. [PubMed] [Google Scholar]
- 57.Reya, T., and R. Grosschedl. 1998. Transcriptional regulation of B-cell differentiation. Curr. Opin. Immunol. 10158-165. [DOI] [PubMed] [Google Scholar]
- 58.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 756894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salas, M., and L. A. Eckhardt. 2003. Critical role for the Oct-2/OCA-B partnership in Ig-secreting cells. J. Immunol. 1716589-6598. [DOI] [PubMed] [Google Scholar]
- 60.Scheidereit, C., J. A. Cromlish, T. Gerster, K. Kawakami, C. G. Balmaceda, R. A. Currie, and R. G. Roeder. 1988. A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature 336551-557. [DOI] [PubMed] [Google Scholar]
- 61.Singh, H. 1994. Genetic analysis of transcription factors implicated in B lymphocyte development. Immunol. Res. 13280-290. [DOI] [PubMed] [Google Scholar]
- 62.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 861276-1280. [PubMed] [Google Scholar]
- 63.Stevens, S., J. Ong, U. Kim, L. A. Eckhardt, and R. G. Roeder. 2000. Role of OCA-B in 3′-IgH enhancer function. J. Immunol. 1645306-5312. [DOI] [PubMed] [Google Scholar]
- 64.Strubin, M., J. W. Newell, and P. Matthias. 1995. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80497-506. [DOI] [PubMed] [Google Scholar]
- 65.Sturm, R. A., G. Das, and W. Herr. 1988. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 21582-1599. [DOI] [PubMed] [Google Scholar]
- 66.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9510866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60375-386. [DOI] [PubMed] [Google Scholar]
- 68.Theil, J., H. Laumen, T. Marafioti, M. Hummel, G. Lenz, T. Wirth, and H. Stein. 2001. Defective octamer-dependent transcription is responsible for silenced immunoglobulin transcription in Reed-Sternberg cells. Blood 973191-3196. [DOI] [PubMed] [Google Scholar]
- 69.Ushmorov, A., O. Ritz, M. Hummel, F. Leithauser, P. Moller, H. Stein, and T. Wirth. 2004. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood 1043326-3334. [DOI] [PubMed] [Google Scholar]
- 70.Walts, A. E., I. P. Shintaku, and J. W. Said. 1990. Diagnosis of malignant lymphoma in effusions from patients with AIDS by gene rearrangement. Am. J. Clin. Pathol. 94170-175. [DOI] [PubMed] [Google Scholar]
- 71.Wan, J. H., K. J. Trainor, M. J. Brisco, and A. A. Morley. 1990. Monoclonality in B cell lymphoma detected in paraffin wax embedded sections using the polymerase chain reaction. J. Clin. Pathol. 43888-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 779590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jκ in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 35919-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wirth, T., L. Staudt, and D. Baltimore. 1987. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature 329174-178. [DOI] [PubMed] [Google Scholar]
- 76.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 793479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]