Abstract
Epstein-Barr virus (EBV) infection is mediated by several viral envelope glycoproteins. We have assessed gp110's functions during the virus life cycle using a mutant that lacks BALF4 (ΔBALF4). Exposure of various cell lines and primary cell samples of epithelial or lymphoid lineages to the ΔBALF4 mutant failed to establish stable infections. The ΔBALF4 virus, however, did not differ from wild-type EBV in its ability to bind and become internalized into primary B cells, in which it elicited a potent T-cell-specific immune reaction against virion constituents. These findings show that ΔBALF4 viruses can reach the endosome-lysosome compartment and dovetail nicely with the previously identified contribution of gp110 to virus-cell fusion. Other essential steps of the virus life cycle were unaffected in the viral mutant; DNA lytic replication and viral titers were not altered in the absence of gp110, and ΔBALF4 viruses complemented in trans transformed infected B cells with an efficiency indistinguishable from that observed with wild-type viruses. All of the steps of virus maturation could be observed in lytically induced 293/ΔBALF4 cells. Induction of lymphoblastoid cells generated with transiently complemented ΔBALF4 virus led to the production of rare mature virions. We therefore infer that gp110 is not required for virus maturation and egress in 293 cells or in B cells. The ΔBALF4 virus's phenotypic traits, an inability to infect human cells coupled with potent antigenicity, potentially qualify this mutant as a live vaccine. It will provide a useful tool for the detailed study of EBV-cell interactions in a physiological context.
Herpesvirus cell entry is a multistep process that includes virus binding and fusion with its target cell as well as transport of the viral DNA to the cell nucleus. It is mediated by the sequential interaction of viral glycoproteins with their cognate receptors in the various organelles and compartments of the target cells as recently reviewed (5, 20, 34, 35, 38). Herpesviruses undergo fusion with different types of cell membranes. Herpes simplex virus 1 (HSV-1) classically fuses with the plasma membrane of its target cell, but an alternative infection route through the endosomal compartment may involve fusion with the endosomal-lysosomal membranes (6). The Epstein-Barr herpesvirus (EBV) invades primary B cells after attaching to CD21, which sets off internalization and transport to the endosome within vesicles (30). B-cell lines, such as Burkitt's lymphoma cell line Raji, however, differ from their primary cell correspondents in that both cell membrane binding and fusion take place at the plasma membrane surface (28, 36). How EBV infects primary epithelial cells is less well documented, but fusion with the plasma cell membrane is the currently favored model (20, 31). The extensive differences in the biochemical composition of plasma and endosomal membranes renders it likely that the process of fusion of these two membrane types with virions is mediated by distinct cellular or even viral interaction partners. Herpesvirus fusion with its target cells appears to be particularly complex and has been shown to require cooperation of several glycoproteins; the minimal set of glycoproteins required for EBV B-cell fusion in in vitro cell-cell fusion assays has been narrowed down to gp42, gH, gL, and gp110 (16). Moreover, increased incorporation of gp110 in the viral membrane enhances the efficiency of infection by one to two orders of magnitude (31).
We wished to improve our understanding of gp110's functions during the EBV life cycle in the context of a complete viral genome. We have therefore constructed an EBV BALF4-knockout genome (ΔBALF4) mutant using a bacterial artificial chromosome recombinant virus system (7) whose phenotypic traits we report here. In particular, we focused our attention on the ability of this mutant to infect a panel of human cells that belong to different lineages.
MATERIALS AND METHODS
Cell culture conditions.
The 293 cell line was generated by transfection of the adenovirus type 5 E1a and E1b genes into human embryonic epithelial kidney cells (14). This cell line shows neuroendocrine differentiation (37). Raji is a human Burkitt's lymphoma cell line (33). SVK-CR2 cells are SVK epithelial cells stably transfected with an expression vector containing the CR2 cDNA under the control of a retroviral promoter (27). Peripheral blood mononuclear cells from peripheral blood buffy coats were purified on a Ficoll cushion as described previously (10). CD19-positive B cells were isolated using M-450 CD19 (Pan B) Dynabeads (Dynal) and were detached from Dynabeads using Detachabead (Dynal). Cells were grown in RPMI 1640 medium containing 10% fetal calf serum. Primary sphenoidal sinus epithelial cells were obtained and cultured as described previously (11).
Recombinant DNA plasmids.
The 5,610-bp XbaI/SalI fragment from p91.D2, a plasmid that carries the EBV B95.8 HindIII D fragment (EBV B95.8 coordinates 146916 to 166486; accession number NC_007605) was subcloned onto pUC19 cleaved with XbaI and SalI to produce plasmid p2354. This plasmid contains the BALF4 gene (EBV B95.8 coordinates 156752 to 159322). A 1,694-bp fragment encompassing the BALF4 gene 5′ region (EBV B95.8 coordinates 157589 to 159283) was excised from p2354 by restriction with NheI and BglII and Klenow polymerase treatment and exchanged against the kanamycin resistance gene cassette cleaved from the pCP15 plasmid by digestion with SacI and HindIII, followed by treatment with Klenow. The resulting targeting vector, p2529, was further linearized with BamHI and EcoRV to release a 5,321-bp fragment that was used for homologous recombination in recA+ Escherichia coli BJ5183 containing p2089. The latter plasmid comprises the wild-type EBV (EBV-wt) genome, the F-factor origin of replication, the chloramphenicol and hygromycin resistance genes, as well as the gene encoding green fluorescence protein (GFP) (7). Double selection with chloramphenicol and kanamycin allowed the emergence of bacterial clones that carry the mutated viral genome. DNA from one of these bacterial clones was further transformed into a recA context (E. coli DH10B) to allow viral DNA amplification (21). Plasmid DNA from antibiotic-resistant DH10B clones was then prepared and analyzed with the BamHI, EcoRI, EcoRV, and XhoI restriction enzymes to identify clones with the correct recombination pattern. Recombinant DNA from EBV-wt was digested in parallel as a reference. 293 cells were transfected with the properly recombined mutant viral DNA using Lipofectamine (Invitrogen) as described previously (21). Selection of stable 293 cell clones carrying the EBV recombinant plasmid was performed by adding hygromycin to the culture medium (100 μg/ml). Cell clones surviving selection were first assessed for GFP fluorescence, and then the positive clones were further expanded. The construction of the BALF4 expression plasmid p2670 has previously been described (31).
The original BZLF1-tet-NGFR-GFP plasmid was previously described (2). We have exchanged the PvuII fragment containing an estrogen receptor-BZLF1 fusion against a PvuII fragment carrying the native BZLF1 gene (pB262). This plasmid contains three genes (those for BZLF1, GFP, and NGF receptor [NGFR]) under the control of a bidirectional doxycyclin-inducible promoter. NGFR and GFP are separated by an internal ribosome entry site sequence. The pCMV-NGFR plasmid contains the NGFR gene cloned under the control of a cytomegalovirus (CMV) promoter.
Generation of virus supernatants and infections.
293/EBV-wt cells (that carry p2089) and 293/ΔBALF4 cells were lytically induced in six-well cluster plates by transfection of the p509 plasmid (0.5 μg each per well) that carries BZLF1 (17). Complementation of the mutant phenotype was performed by transfection of the BALF4 expression plasmid p2670 (0.5 μg each per well). Plasmids were mixed with RPMI 1640 medium in a final volume of 100 μl/well. Metafectene (Biontex; 3 μl/μg DNA) was diluted into 100 μl RPMI medium and mixed with the diluted DNA. A total of 200 μl of the mixture was added to the cells after a 20-min incubation at room temperature. The transfection mixture was left overnight on the cells and replaced by 2 ml of standard medium on the following day. Four days after transfection, supernatants were harvested, filtered (0.45 μm pore size), and kept at 4°C until further use.
Primary B cells were infected at a multiplicity of infection (MOI) of 10 and seeded into U-bottomed 96-well microtiter plates at a cell density of 104 cells per well (two plates per supernatant). In each well, γ-irradiated human embryonic lung fibroblasts served as a feeder layer.
Lytic cycle inductions in LCLs.
Lymphoblastoid cell lines (LCLs) immortalized by EBV-wt or ΔBALF4 virus were transfected by electroporation (250 V, 960 μF, 4-mm cuvettes; Bio-Rad gene pulser) with pB262 that contains the BZLF1 gene under the control of a doxycyclin-inducible promoter and pCMV-NGFR. Transfected cells were immunostained with a mouse antibody specific to NGFR (a kind gift from E. Kremmer, Munich, Germany), followed by staining with a secondary anti-mouse antibody coupled to Dynabeads (Dynal). Purified cells were incubated with doxycyclin (0.5 μg/ml final concentration) for 3 days.
Determination of viral titers and virus binding assay.
Virus titers expressed as genome equivalents per milliliter were determined by quantitative real-time PCR analysis (qPCR) using primers and labeled probe specific for the BALF5 gene, which encodes the viral DNA polymerase as previously described (10); 25 μl of filtered supernatants was treated with 10 U/ml of DNase I (Roche) for 1 h at 37°C, followed by inactivation of the enzyme at 70°C for 10 min. Viral DNA was then freed from virus particles by treatment with proteinase K (50 μg/ml for 1 h at 50°C), which in turn was inactivated by heat treatment (20 min at 75°C). Lysates were used either undiluted or at appropriate dilutions before being assayed for qPCR. For virus binding assays, 2.5 × 105 primary human B cells were incubated with filtered virus supernatants on ice at an MOI of 25 in a final volume of 1.5 ml. After 3 h, cells were washed three times with 1.5 ml of phosphate-buffered saline (PBS) and resuspended in 250 μl of standard medium. Twenty-five microliters of this cell suspension was treated the same way as virus supernatants. However, after proteinase K treatment, cellular RNA was removed by treating lysates with DNase-free RNase A (20 μg/ml for 1 h at 37°C). The resulting lysate was used directly for the determination of EBV copy number using qPCR.
Rescue of mutant EBV into E. coli.
Circular DNA molecules were extracted from 293/ΔBALF4 cells using a denaturation-renaturation method (7, 15). The integrity of the viral genomes transformed in E. coli clones was further assessed by restriction enzyme analysis.
Immunostaining and Western blot analysis.
Cells were washed three times in PBS, dried on glass slides, and treated with pure acetone for 20 min at room temperature. Slides were incubated for 30 min with a purified mouse monoclonal antibody directed against EBV glycoprotein gp110 (anti-VCA-gp125, clone MAB8184; Chemicon) or gp350 (hybridoma 72A1; ATCC). Immunostaining with antibodies specific to the EBV immediate early antigen (EA) BZLF1 (clone BZ.1), EBNA2 (clone pE2), or the EA BMRF1 (anti-EA-D MAB818; Chemicon) was performed with cells fixed with 4% paraformaldehyde for 20 min at room temperature and permeabilized with 0.1% Triton X-100 for 2 min at room temperature. Slides were then washed three times in PBS and incubated for 30 min with a secondary goat anti-mouse antibody conjugated with the Cy3 fluorochrome (Dianova). After several washes in PBS, slides were embedded with 90% glycerol and immunofluorescence evaluated by using a fluorescence microscope coupled to a charge-coupled-device camera (Leica). Western blots for gp110 have been described (31).
T-cell antigen recognition assays.
T-cell antigen recognition assays were used to identify virus internalization in the endosome-lysosome compartment as previously described (1, 10). Mini-LCLs were obtained by transformation of primary B cells with the mini-EBV, a recombinant plasmid that contains the EBV latent genes but not all lytic genes and therefore cannot enter the replicative cycle (22, 29). Mini-LCL cells (1 × 105/well) were incubated with serial dilutions of viral supernatant and then fixed by treatment with 1% paraformaldehyde for 5 min at room temperature followed by extensive washings. T cells (1 × 105) were then incubated with the infected B cells and placed in culture for 24 h. T-cell cytokines released in the culture medium following antigen-specific B-cell/T-cell interactions were quantified with an enzyme-linked immunosorbent assay system (R&D Systems). The CD4+ T-cell clones specific for gp350 (clone 1D6) and gp110 (clone A9) have been isolated from EBV-seropositive donor 001 and cultivated as described previously (1). These T-cell clones are HLA-DRB1*1301 and DRB1*0801 restricted, respectively.
Electron microscopy.
Cells were washed in PBS and fixed with 2.5% glutaraldehyde in PBS for 20 min at 4°C. Samples were postfixed in 2% osmium tetroxide in cacodylate buffer for 1 h at 4°C, stained with 0.5% uranyl acetate for 16 h at 4°C, washed twice in distilled water, dehydrated in ethanol, and embedded in Epon 812. Thin sections were examined using a Zeiss electron microscope.
Gardella gel analysis.
EBV-infected cells were loaded on an 0.8% agarose gel where they were lysed using sodium dodecyl sulfate as previously described (7, 21). The full-length viral genomes were then separated by electrophoresis and submitted to Southern blot analysis. Viral DNAs were blotted onto a Hybond-XL membrane (Amersham) and hybridized with a 32P-labeled DNA fragment specific to BamHI-W EBV (B95.8 coordinates 12001 to 15072).
RESULTS
Introduction of a BALF4 deletion in the EBV genome.
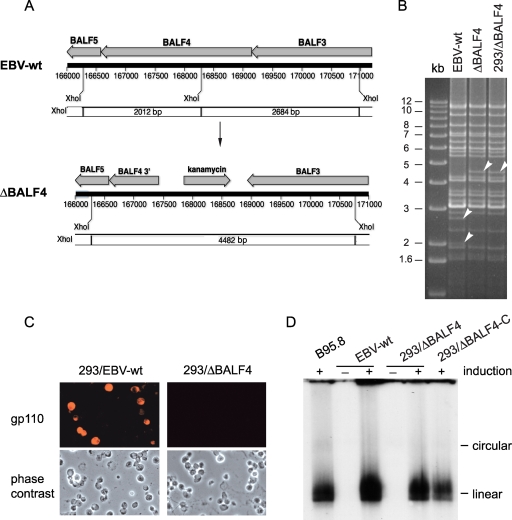
We have used a genetic approach to determine the functions of gp110 at various stages of EBV infection. To this end, we have generated an EBV genome devoid of the BALF4 gene using a linear DNA fragment that comprises the kanamycin resistance gene flanked by sequences homologous to the BALF4 gene region as a targeting vector (Fig. 1A). E. coli cells stably carrying the EBV-wt genome cloned onto a bacterial artificial chromosome vector were transformed with this targeting vector, and successfully recombined plasmids were selected by growing the bacterial cells in the presence of kanamycin. This resulted in a deletion of the first two-thirds of the BALF4 gene. DNA preparations thereof were subsequently submitted to restriction analysis with several enzymes that allowed distinction between wild-type and properly recombined genomes (Fig. 1B and data not shown). ΔBALF4 mutant DNA was then introduced into 293 cells, and stably established clones were retrieved by hygromycin selection. Twenty of these clones were tested for their permissivity to EBV lytic replication by measuring the percentage of cells expressing the late lytic protein gp350 following transfection of the BZLF1 transactivator. Several clones were found to express gp350 in approximately 20% of transfected cells, and one of these was selected for further analysis (here referred to as 293/ΔBALF4 cells). To ensure that the structure of the mutant genome in 293/ΔBALF4 cells had not been altered by the successive manipulations, we transferred the viral DNA back into E. coli cells where it was analyzed by restriction analysis. The genome was found to be structurally intact (Fig. 1B). Immunostaining and Western blot analysis of induced 293/ΔBALF4 cells with a gp110-specific mouse monoclonal antibody were negative, as expected (Fig. 1C and data not shown). In contrast, 293/EBV-wt cells produced gp110 upon induction of EBV lytic replication (Fig. 1C).
FIG. 1.
Generation and characterization of a ΔBALF4 recombinant EBV genome. (A) EBV-wt and ΔBALF4 genome maps illustrating the organization of the BALF4 gene locus. Exchange of the BALF4 gene against the kanamycin resistance cassette alters the XhoI restriction pattern; the 2,012- and 2,684-bp XhoI fragments that carry the BALF4 gene in the wild-type genome are swapped against a unique 4,482-bp fragment that carries the kanamycin cassette in the mutant. (B) The mutated genome was assayed by restriction enzyme analysis at various stages of the construction process after initial recombination with the targeting vector in E. coli and after stable transfection in 293 cells. Wild-type genomes were digested in parallel to serve as a reference. DNA fragments that differ in the wild-type and mutated genomes are indicated by arrows. The observed restriction patterns were compared with the blueprints depicted in panel A. (C) BALF4 gene expression was assayed by immunostaining in induced 293/EBV-wt and 293/ΔBALF4 cells. (D) Gardella gel analysis coupled to Southern blot analysis of lytically induced cells using the BamHI-W segment as a probe. Linear genome production was monitored for 293/EBV-wt cells, 293/ΔBALF4 cells, and 293/ΔBALF4-C cells.
The BALF4 gene is not required for DNA lytic replication and lytic protein production.
We first assessed lytic gene expression and DNA replication in induced 293/ΔBALF4 cells. The production pattern of EA-D and late antigen gp350 (products of the BMRF1 and BLLF1 genes, respectively) in induced 293/ΔBALF4 cells was very similar to that observed in their complemented counterpart (referred to as 293/ΔBALF4-C cells) or in 293/EBV-wt cells as monitored by immunostaining (EA-positive cells, 15% ± 5.7%, 17% ± 4.2%, and 13% ± 0.71%; gp350-positive cells, 9.7% ± 0.42%, 12% ± 3.7%, and 13% ± 4.9%, respectively). Therefore, the absence of gp110 does not prevent 293/ΔBALF4 cells from entering the lytic cycle. This finding is in line with earlier observations that reported late EBV gene expression in lytically induced B cells carrying a BALF4-knockout EBV genome (19, 26). We further monitored linear unit-length virion DNA synthesis in induced 293/ΔBALF4 cells by performing a Gardella gel analysis (12), followed by Southern blotting with an EBV-specific BamHI-W probe. This assay did not reveal any substantial difference between induced 293/ΔBALF4 cells, 293/ΔBALF4-C cells, or 293/EBV-wt cells (Fig. 1D). Thus, BALF4 does not appear to serve any function during DNA lytic replication.
293/ΔBALF4 cells produce properly assembled virus particles.
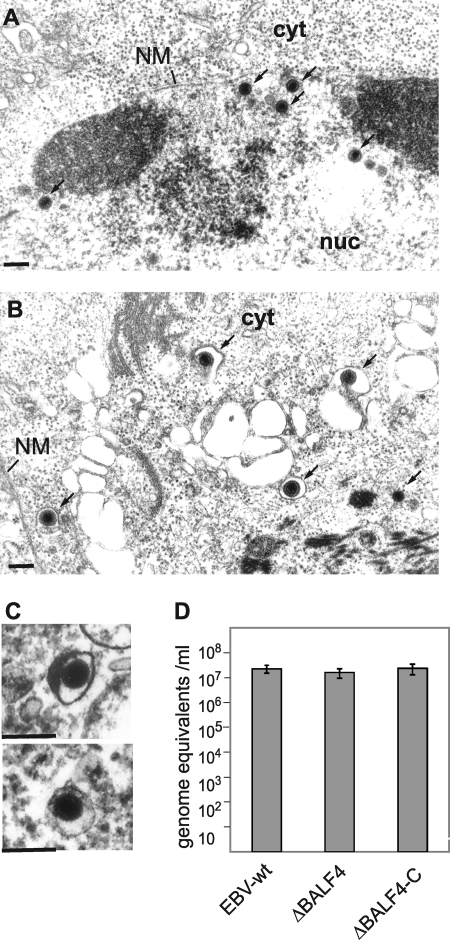
We then examined ultra-thin sections of lytically induced 293/ΔBALF4 cells by electron microscopy. Intact virus capsids at all stages of maturation were readily visible in the nuclei of cells (17% A capsids, 41% B capsids, and 42% C capsids; percentages calculated from 300 capsids observed in 15 induced cells) (Fig. 2A). Induced wild-type cells taken as a positive control showed the following capsid distribution: 20% A capsids, 49% B capsids, and 31% C capsids (percentages calculated from 300 capsids observed in 13 induced cells). Fully assembled virions containing packaged viral DNA could be further identified in the cytoplasm of both mutant and wild-type cells (Fig. 2B). From these observations, we inferred that 293/ΔBALF4 cells were very likely to deliver intact viral particles in the extracellular milieu. To further test this hypothesis, we first examined pelleted supernatants from induced 293/ΔBALF4 cells using electron microscopy (Fig. 2C). Here again, mature viral particles that displayed normal morphological features were easily detected. Second, we determined the concentration of DNase I-resistant EBV DNA by qPCR in supernatants from lytically induced cells. This analysis unequivocally showed similar amounts of capsid-packaged EBV DNA in supernatants from induced 293/ΔBALF4 cells, 293/ΔBALF4-C cells, and 293/EBV-wt cells (Fig. 2D).
FIG. 2.
Induced 293/ΔBALF4 cells efficiently assemble and release viral particles. (A and B) Lytically induced 293/ΔBALF4 cells were analyzed by electron microscopy. DNA-filled nucleocapsids (A) and cytoplasmic virions (B) are indicated by arrows. NM, nuclear membrane; cyt, cytoplasm; nuc, nucleus. Bars, 0.15 μm. (C) Electron micrographs of pelleted supernatants from induced 293/ΔBALF4 cells. (D) Viral titers (genome equivalents) in supernatants from lytically induced 293/EBV-wt cells, 293/ΔBALF4 cells, and 293/ΔBALF4-C cells were determined by qPCR using primers specific to the BALF5 gene.
ΔBALF4 viruses are no longer infectious.
The previous set of data suggested normal maturation of ΔBALF4 viruses in 293 cells. We further assessed ΔBALF4 infectious properties by exposing various cell lines (SVK-CR2, Raji, 293) and primary cells (B cells and epithelial cells) previously characterized as sensitive to EBV infection to ΔBALF4 viruses, to their complemented counterparts, or to the wild-type virus (Table 1). Evidence for infection was sought by exposing target cells to UV fluorescence, with the exception of primary B cells that do not produce substantial amounts of GFP upon infection and were therefore stained for EBNA2 expression 2 days after infection. These assays failed to identify GFP- or EBNA2-positive cells after infection with ΔBALF4 viruses. Positive controls (ΔBALF4-C virus, EBV-wt), however, showed normal infection rates (Table 1).
TABLE 1.
gp110 is absolutely essential for infection of human primary cells or cell lines of lymphoid and epithelial origin
| Cells and target | Mean efficiency of infection ± SD witha:
|
||
|---|---|---|---|
| EBV-wt | ΔBALF4 mutant | ΔBALF4-C mutant | |
| Raji (104 GFP+ cells/ml) | 7.4 ± 0.91 | 0 | 2.9 ± 1.2 |
| 293 (104 GFP+ cells/ml) | 1.9 ± 1.3 | 0 | 1.4 ± 0.45 |
| Primary B (% EBNA2+) | 18 ± 12 | 0 | 11 ± 2.5 |
| SVK-CR2 (% GFP+ cells) | 55 ± 18 | 0 | 68 ± 20 |
| Primary sEC (% GFP+ cells) | 7.8 ± 1.2 | 0 | 13 ± 2.8 |
Supernatants from induced 293/ΔBALF4, 293/ΔBALF4-C, or 293/EBV-wt cells were used to infect various cell lines or primary cells. Raji and 293 cells were infected using a serial-limiting dilution of the various supernatants to determine the concentration of infectious EBV, whereas the remaining samples were infected at a fixed MOI of 10 (primary B cells) or at an MOI of 100 (SVK-CR2 and primary sphenoidal sinus epithelial cells [sEC]). The efficacy of infection was monitored by assessing the percentage of EBNA2-positive (EBNA2+) primary B cells (2 days after infection) or of GFP-positive (GFP+) cells (3 days after infection; all other cell types).
gp110 is required for postbinding events.
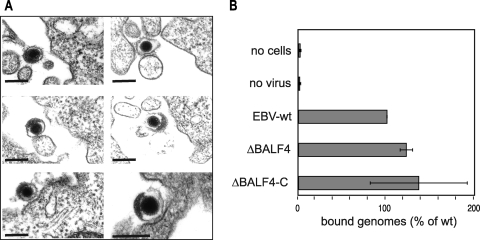
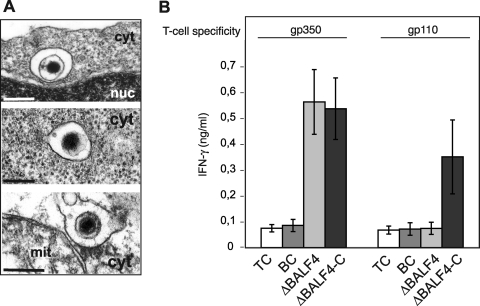
The finding that ΔBALF4 viruses are noninfectious prompted us to determine at which step(s) EBV entry was blocked in the absence of gp110. Filtered supernatants from lytically induced 293/ΔBALF4 cells were therefore incubated with primary B cells on ice to allow binding of putative cell-free viral particles. We found no obvious difference in the numbers of ΔBALF4 mutant and wild-type particles bound to B cells as assessed by electron microscopy (Fig. 3A). In an attempt to quantify binding more accurately, we incubated supernatant containing ΔBALF4 viruses with primary B cells and measured the number of EBV particles bound to the cells by qPCR. This assay demonstrated that binding was not impaired and even increased slightly compared to that of the wild-type virus (Fig. 3B). This showed that binding to primary B cells is not dependent upon gp110 expression. We then monitored postbinding events by examining primary B cells exposed to ΔBALF4 virus supernatants on ice and switched to room temperature for 20 min to allow infection. Electron microscopy revealed the presence of rare virions internalized into vesicles, suggesting that transport to the endosomal compartment was not impaired in the absence of gp110 (Fig. 4A). We did not find evidence for a reduced frequency of internalization in the ΔBALF4 mutant virus compared to that in wild-type viruses, but the paucity of these events precluded any meaningful statistical analysis. We sought additional support for this claim by monitoring the abilities of ΔBALF4 virus particles to elicit activation of EBV antigen-specific T cells. B cells have previously been found to transport viruses bound to their surfaces to the endosome-lysosome compartment, where viral structural proteins are subjected to proteolysis followed by presentation on major histocompatibility complex class II molecules to cytotoxic CD4+ T cells. Activated antigen-specific T cells release soluble gamma interferon (IFN-γ), which can be quantified with an enzyme-linked immunosorbent assay (1). Thus, the ability of a virus to elicit activation of T cells and to cause release of IFN-γ depends on its ability to enter the endosome. We found that the ΔBALF4 virus activated gp350-specific T cells to the same extent as wild-type viruses did (Fig. 4B). As expected, ΔBALF4 viruses failed to elicit activation of gp110-specific T cells, an observation that further confirmed the absence of gp110 from these virions. We therefore conclude that internalization of EBV particles into the endosome was not affected in the absence of gp110.
FIG. 3.
ΔBALF4 viruses efficiently bind to primary B cells. Primary human B cells were incubated with supernatants from induced 293/ΔBALF4 cells. (A) Six representative examples of virions bound to the cell membrane are shown. Bars, 0.15 μm. (B) The numbers of EBV-wt, ΔBALF4, and ΔBALF4-C virions bound to primary B cells were determined by qPCR.
FIG. 4.
ΔBALF4 viruses reach the endosomal and endosomal-lysosomal cellular compartments. (A) ΔBALF4 viruses are efficiently internalized into primary B cells. Electron micrographs show three virions surrounded by intracytoplasmic vesicles. cyt, cytoplasm; nuc, nucleus; mit, mitochondrium. Bars, 0.15 μm. (B) Primary B cells exposed to ΔBALF4 virions present viral antigens to CD4+ cytotoxic T cells. ΔBALF4 mutant-infected B cells were used as targets for gp350- and gp110-specific T cells. IFN-γ concentration was measured as a read-out for antigen-specific recognition. Supernatants from B cells (BC) or T cells (TC) were only used as negative controls.
EBV-wt- and ΔBALF4 mutant-immortalized B cells produce low numbers of virions upon induction.
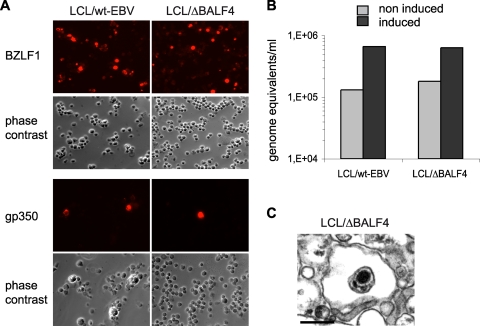
The data gathered so far suggested that BALF4 is not required for virus maturation and egress in 293 cells. We wished to determine whether these observations extend to B cells. We therefore incubated primary B cells from the same donor either with supernatants from induced and transiently complemented 293/ΔBALF4 cells or with those from its wild-type counterparts, and the resulting cell lines were named LCL/ΔBALF4 or LCL/EBV-wt. ΔBALF4 viruses do not carry the BALF4 gene; LCL/ΔBALF4 cells therefore represent a suitable tool to evaluate the BALF4 gene's functions. Induction of LCLs required expression of BZLF1 from an expression plasmid. However, direct transfection in B cells, or in LCLs, is very inefficient. We therefore cotransfected LCL/ΔBALF4 or LCL/EBV-wt cells with a plasmid that encodes BZLF1 from a doxycyclin-inducible bidirectional promoter and with a plasmid that encodes a truncated version of the NGFR under the control of the CMV promoter (2). Cells that contained these plasmids could then be purified with an NGFR-specific antibody. We therefore added doxycyclin to LCL/ΔBALF4 or LCL/EBV-wt cells and evaluated lytic replication by immunostaining cells for BZLF1 and gp350. Ten percent of LCL/ΔBALF4 cells and 22% of LCL/EBV-wt cells were BZLF1 positive, and 0.2% of LCL/ΔBALF4 cells and 0.9% of LCL/EBV-wt cells stained positively for gp350, showing that these two types of LCLs support lytic replication with weak but comparable efficiency (Fig. 5A). We then gauged viral DNA replication in the supernatants by qPCR. This assay evidenced a modest but significant fivefold increase in the concentration of DNase I-resistant EBV DNA upon induction of the lytic cycle (Fig. 5B). Finally, 30 ml of supernatants from induced LCL/ΔBALF4 or LCL/EBV-wt cells was ultracentrifuged and pellets were examined by electron microscopy. Exceedingly rare mature virions could be identified in both types of supernatants (Fig. 5C).
FIG. 5.
LCL/ΔBALF4 cells or LCL/EBV-wt cells produce virions with low efficiency. (A) Immunostaining with antibodies against BZLF1 or gp350 was carried out on induced LCL/ΔBALF4 or LCL/EBV-wt cells. (B) Supernatants from induced LCL/ΔBALF4 or LCL/EBV-wt cells were assessed for DNase I-resistant EBV DNA. Noninduced LCLs provided a negative control. (C) Electron micrograph of pelleted LCL/ΔBALF4 virions.
DISCUSSION
EBV gp110, an envelope glycoprotein encoded by the BALF4 gene, shows marked structural homology to the gB proteins from other herpesviruses (8, 13, 32). Both gp110 of EBV and gB of HSV-1 comprise a large extracellular domain, a hydrophobic domain of approximately 50 amino acids, and a short cytoplasmic tail (104 and 109 amino acids for gp110 and gB, respectively). Despite these genomic and structural similarities, gBs from different herpesviruses fail to complement the phenotype of EBV and HSV-1 gB deletion mutants (25). This justifies independent studies of both gp110 and the other gB and their cognate cellular receptors.
Herein we report construction of a ΔBALF4 EBV mutant and the description of its phenotype. Virus lytic DNA replication and lytic protein synthesis were unaffected by the BALF4 deletion. Virus maturation and egress, monitored by electron microscopy, were indistinguishable for mutant and wild-type viruses; viral titers in supernatants from 293/ΔBALF4 cells, 293/ΔBALF4-C cells, and 293/EBV-wt cells, monitored by qPCR, were very similar and perfectly compatible with efficient virion production. ΔBALF4 virus infection was, however, strictly abortive in all tested target cell types. These included primary epithelial cells with squamous differentiation, primary B lymphocytes, and various cell lines of lymphoid and epithelial origin (Table 1). Therefore, gp110 appears to be strictly required for target cell infection but not for maturation in 293 cells. These findings fit with our previous observation that overexpression of gp110 in 293/EBV-wt cells increases the efficiency of infection without altering viral titers (31). We further attempted to identify the bottleneck in the ΔBALF4 virus infection pathway. We found strong evidence for normal binding, virus uptake, and internalization into cytoplasmic vesicles, demonstrating that the infection block was located further downstream, presumably during transport from the endosomal compartment to the nucleus (Fig. 3 and 4). Such an assumption would fit with reported evidence that gp110 is essential for virus-cell fusion. In such a context, our data would indicate that fusion with both the plasma membrane and the endosomal membrane absolutely requires gp110.
Our data are congruent with the phenotypic traits previously ascribed to another BALF4 null mutant constructed in an LCL and from which no infectious virus could be recovered upon induction of the lytic cycle (19). There is, however, a discordance regarding the mechanisms that led to a loss of the infectious potential; the present work reports efficient production of ΔBALF4 viruses that are unable to complete a successful round of infection, whereas the authors of work describing the other BALF4 knockout suggested that gp110 plays an essential role during viral maturation (19, 26). In the absence of gp110, these authors found that the viral replicative cycle was abortive at the stage of capsid formation and assembly (26). This discrepancy can reflect the different cellular backgrounds in which these experiments were conducted (293 cells versus B cells). Clearly, the very high level of virus production in 293 cells relative to that in LCL cells very easily allows identification and characterization of virion progeny. We further readdressed this issue by infecting primary B cells with the ΔBALF4 mutant or EBV-wt. The resulting LCL/ΔBALF4 cells or LCL/EBV-wt cells were induced to assess virus replication. Both LCLs were found to produce gp350 upon induction; their supernatants contained DNase I-resistant linear viral DNA in equal amounts as well as rare virions (Fig. 5). These results concur to indicate that the BALF4 gene is required neither for virus maturation nor for viral egress. It should, however, be stated that replication in LCLs is usually very weak and that we had to enrich LCL/ΔBALF4 or LCL/EBV-wt cells for plasmids encoding BZLF1 in order to obtain a visible level of lytic replication; attempts to induce LCL/ΔBALF4 or LCL/EBV-wt cells with tetradecanoyl phorbol acetate and sodium butyrate proved unsuccessful (data not shown). Altogether, we could find only one or two viruses per section from pellets formed after centrifugation of 30-ml supernatants from LCL/EBV-wt and LCL/ΔBALF4 cells. Clearly, such a low level of replication prevents any meaningful comparison in the efficiency of virus replication between wild-type and ΔBALF4 viruses in B cells, and it is perfectly possible that, even if not absolutely required for virus maturation, gp110 plays an ancillary role during this process that could not be detected in our assays.
Further support for our results is provided by previous observations made with HSV-1 or pseudorabies viruses that showed that gB is not required for virus maturation (3, 4, 23). However, it should be noted that HSV-1 and pseudorabies viruses appear to differ in that a double HSV-1 mutant lacking both gB or gH proteins shows impaired primary nuclear egress, whereas a pseudorabies virus carrying the same deletions displays perfectly normal virus maturation and production (9, 23). Furthermore, human herpesvirus 8 (HHV-8) gB has been found to be essential for virus egress at 293T cell plasma membranes (24). HHV-8 gB, however, was not required for HHV-8 capsid assembly or nuclear egress. A similar role has been ascribed to the C-terminal sequences of varicella-zoster virus; mutant viruses lacking the C-terminal 36 amino acids show impaired virus egress into the extracellular milieu (18). Despite a high level of genomic homology, gB from different herpesviruses seems to serve highly variable functions during virus maturation (including none), irrespective of its membership in the different HSV subfamilies.
We further found that the BALF4 gene is not required for effective virion antigen presentation by infected B cells. We therefore conclude that sequestration of the virion within the endosome does not preclude mounting of an efficient immune response against its components. These data fit with a model that posits that some virion antigens, such as gp350, gp110, or the tegument protein BNRF1, are processed within the endosome-lysosome before being associated with HLA class II molecules and presented to CD4+ T cells at the cell surface (1). It will be interesting to determine whether the trapping of ΔBALF4 virions within the endosome as a result of a failed fusion might even enhance the amplitude of the T-cell response against capsid or core proteins that normally escape from the endosome to allow capsid migration toward the nucleus.
A potentially interesting consequence of the present work is that it is now in principle possible to induce a CD4+ EBV-specific cytotoxic T-cell response with a noninfectious viral mutant. Such an attenuated strain would fulfill the safety criteria expected for a live vaccine. Whether the elicited T-cell response would be sufficient to induce protective immunity, however, remains to be seen.
Acknowledgments
We thank B. Sugden for the p91.D2 plasmid.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Adhikary, D., U. Behrends, A. Moosmann, K. Witter, G. W. Bornkamm, and J. Mautner. 2006. Control of Epstein-Barr virus infection in vitro by T helper cells specific for virion glycoproteins. J. Exp. Med. 203995-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bornkamm, G. W., C. Berens, C. Kuklik-Roos, J. M. Bechet, G. Laux, J. Bachl, M. Korndoerfer, M. Schlee, M. Holzel, A. Malamoussi, R. D. Chapman, F. Nimmerjahn, J. Mautner, W. Hillen, H. Bujard, and J. Feuillard. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201575-588. [DOI] [PubMed] [Google Scholar]
- 4.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume, G., M. Amasio, E. Avitabile, A. Cerretani, C. Forghieri, T. Gianni, and L. Menotti. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 17313-326. [DOI] [PubMed] [Google Scholar]
- 6.Clement, C., V. Tiwari, P. M. Scanlan, T. Valyi-Nagy, B. Y. Yue, and D. Shukla. 2006. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J. Cell Biol. 1741009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 958245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emini, E. A., J. Luka, M. E. Armstrong, P. M. Keller, R. W. Ellis, and G. R. Pearson. 1987. Identification of an Epstein-Barr virus glycoprotein which is antigenically homologous to the varicella-zoster virus glycoprotein II and the herpes simplex virus glycoprotein B. Virology 157552-555. [DOI] [PubMed] [Google Scholar]
- 9.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 10410187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feederle, R., B. Neuhierl, G. Baldwin, H. Bannert, B. Hub, J. Mautner, U. Behrends, and H. J. Delecluse. 2006. Epstein-Barr virus BNRF1 protein allows efficient transfer from the endosomal compartment to the nucleus of primary B lymphocytes. J. Virol. 809435-9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feederle, R., B. Neuhierl, H. Bannert, K. Geletneky, C. Shannon-Lowe, and H. J. Delecluse. 2007. Epstein-Barr virus B95.8 produced in 293 cells shows marked tropism for differentiated primary epithelial cells and reveals interindividual variation in susceptibility to viral infection. Int. J. Cancer 121588-594. [DOI] [PubMed] [Google Scholar]
- 12.Gardella, T., P. Medveczky, T. Sairenji, and C. Mulder. 1984. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, M., T. Ooka, T. Matsuo, and E. Kieff. 1987. Epstein-Barr virus glycoprotein homologous to herpes simplex virus gB. J. Virol. 61499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 3659-74. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, B. E., E. Bjorck, G. Bjursell, and T. Lindahl. 1981. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J. Virol. 4011-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290106-114. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55427-433. [DOI] [PubMed] [Google Scholar]
- 18.Heineman, T. C., and S. L. Hall. 2002. Role of the varicella-zoster virus gB cytoplasmic domain in gB transport and viral egress. J. Virol. 76591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 702049-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 817825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 7410142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempkes, B., D. Pich, R. Zeidler, B. Sugden, and W. Hammerschmidt. 1995. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J. Virol. 69231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 826299-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan, H. H., N. Sharma-Walia, L. Zeng, S. J. Gao, and B. Chandran. 2005. Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 7910952-10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. K., T. Compton, and R. Longnecker. 1997. Failure to complement infectivity of EBV and HSV-1 glycoprotein B (gB) deletion mutants with gBs from different human herpesvirus subfamilies. Virology 237170-181. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. K., and R. Longnecker. 1997. The Epstein-Barr virus glycoprotein 110 carboxy-terminal tail domain is essential for lytic virus replication. J. Virol. 714092-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q. X., L. S. Young, G. Niedobitek, C. W. Dawson, M. Birkenbach, F. Wang, and A. B. Rickinson. 1992. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 356347-350. [DOI] [PubMed] [Google Scholar]
- 28.Miller, N., and L. M. Hutt-Fletcher. 1992. Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 663409-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moosmann, A., N. Khan, M. Cobbold, C. Zentz, H. J. Delecluse, G. Hollweck, A. D. Hislop, N. W. Blake, D. Croom-Carter, B. Wollenberg, P. A. Moss, R. Zeidler, A. B. Rickinson, and W. Hammerschmidt. 2002. B cells immortalized by a mini-Epstein-Barr virus encoding a foreign antigen efficiently reactivate specific cytotoxic T cells. Blood 1001755-1764. [PubMed] [Google Scholar]
- 30.Nemerow, G. R., and N. R. Cooper. 1984. Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology 132186-198. [DOI] [PubMed] [Google Scholar]
- 31.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. USA 9915036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellett, P. E., M. D. Biggin, B. Barrell, and B. Roizman. 1985. Epstein-Barr virus genome may encode a protein showing significant amino acid and predicted secondary structure homology with glycoprotein B of herpes simplex virus 1. J. Virol. 56807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulvertaft, R. J. V. 1964. Cytology of Burkitt's lymphoma (African lymphoma). Lancet i238-240. [DOI] [PubMed] [Google Scholar]
- 34.Reske, A., G. Pollara, C. Krummenacher, B. M. Chain, and D. R. Katz. 2007. Understanding HSV-1 entry glycoproteins. Rev. Med. Virol. 17205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rey, F. A. 2006. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 71000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seigneurin, J. M., M. Vuillaume, G. Lenoir, and G. De-The. 1977. Replication of Epstein-Barr virus: ultrastructural and immunofluorescent studies of P3HR1-superinfected Raji cells. J. Virol. 24836-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16869-871. [DOI] [PubMed] [Google Scholar]
- 38.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 6401-410. [DOI] [PubMed] [Google Scholar]