Abstract
A new protein expression vector design utilizing an N-terminal six-histidine tag and tobacco etch virus protease cleavage site upstream of the hepatitis C virus NS5A sequence has resulted in a more straightforward purification method and improved yields of purified NS5A domain I protein. High-resolution diffracting crystals of NS5A domain I (amino acids 33 to 202) [NS5A(33-202)] were obtained by using detergent additive crystallization screens, leading to the structure of a homodimer which is organized differently from that published previously (T. L. Tellinghuisen, J. Marcotrigiano, and C. M. Rice, Nature 435:374-379, 2005) yet is consistent with a membrane association model for NS5A. The monomer-monomer interface of NS5A(33-202) features an extensive buried surface area involving the most-highly conserved face of each monomer. The two alternate structural forms of domain I now available may be indicative of the multiple roles emerging for NS5A in viral RNA replication and viral particle assembly.
Hepatitis C virus (HCV) is a member of the Flaviviridae family of enveloped, positive-strand RNA viruses (23). It is responsible for persistent infections in humans, with associated risk of chronic liver diseases, including cirrhosis and hepatocellular carcinoma. Nearly 3% of the global population is chronically infected with HCV, and there are no clinically proven vaccines. Antiviral therapeutic agents are at an early stage of clinical evaluation, and standard treatments (interferon and ribavirin combinations) are associated with suboptimal response rates and/or high incidence of side effects. Complicating the discovery of new therapies is the highly complex and incompletely understood nature of the viral life cycle. The HCV genome consists of a single strand of RNA of about 9,600 nucleotides encoding a polypeptide precursor of about 3,000 amino acids (26). Co- and posttranslational proteolytic cleavage of this precursor by cellular and viral enzymes yields structural proteins involved in viral assembly, along with nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B, which are required for membrane-associated RNA replication (14).
Nonstructural protein NS5A is a critical component of HCV replication and is involved in several cellular processes, such as interferon resistance (3, 13) and apoptotic regulation (9). It is a phosphoprotein of 447 residues with three domains (35), and while no clear enzymatic functions have been assigned, it appears to function through interactions with other HCV proteins and host cell factors (17). Domain I (residues 1 to 213) contains a zinc-binding motif (35) and an amphipathic N-terminal helix which promotes membrane association (4, 12, 30), possibly through specific interaction of the helix with target membrane proteins (8). Domain II (residues 250 to 342) has regulatory functions, such as interactions with protein kinase PKR and PI3K (13), as well as NS5B (32); contains the interferon sensitivity-determining region (13); and appears to lack major elements of secondary structure (22). Recent studies have demonstrated that domain III (residues 356 to 447) plays a critical role in infectious virion assembly but not in RNA replication (1, 34) and that the former role is modulated by phosphorylation within the domain (33). High-throughput screening of small-molecule inhibitors using HCV replicon cell systems has identified NS5A as a promising therapeutic target (31).
A crystal structure of domain I lacking the amphipathic helix and spanning residues 25 to 215 showed two subdomains and a homodimeric association and was interpreted as having a potential role in RNA binding (36). Although specific binding to domain I was not described, RNA binding to full-length NS5A has been reported, using, for example, the 3′ nontranslated region of HCV (15). Efforts in our laboratory to study the structure of NS5A have yielded an alternative arrangement of the domain I homodimer (residues 33 to 202) that differs substantially from that previously described. The observation that the NS5A domain I homodimer can exist in alternate configurations may have ramifications for the role of NS5A in the viral life cycle.
MATERIALS AND METHODS
Materials.
Ultra Yield protein expression flasks (5) were purchased from Thomson Instrument Company (Carlsbad, CA). Benzonase nuclease was obtained from Novagen, and Terrific Broth (TB) was purchased from Teknova (12.00 g/liter casein peptone, 2.31 g/liter KH2PO4, 24.00 g/liter yestolate, 16 ml/liter glycerol; catalog number T7100). Tobacco etch virus (TEV) protease containing the S219V mutation was produced in-house by using the TEV expression vector pRK793 (16) in Escherichia coli strain BL21(DE3)-pRIL (catalog number 230245; Stratagene).
Construction of an NS5A(33-202) protein expression vector.
The production of the recombinant NS5A domain I protein spanning residues 33 to 202 [NS5A(33-202)] was accomplished without the requirement to express NS5A as a yeast ubiquitin proprotein chimera (16, 35, 36). Our strategy utilized a custom T7-inducible TOPO vector, pLJEC1 (modified pET24a vector from EMD), which features an N-terminal TEV protease-cleavable polyhistidine purification tag (MKHHHHHHDYGIPTTENLYFQAL) that, upon cleavage of the recombinant protein, releases NS5A(33-202) with an additional Ala-Leu sequence at the N terminus (the Ala-Leu codons encode the TOPO cloning sequence). A blunt-ended DNA fragment encompassing amino acids 33 to 202 of NS5A was amplified by PCR by using the His-Δ-NS5A construct (16) (HCV genotype 1b, subtype Con1) as template and by using primers NS5A-P5 (5′-GGA GTC CCC TTC TTC TCA TGT CAA CG-3′) and NS5A-P3 (5′-CTA CTT GTC GTC GTC GTC CAT GGA AGT GAG CAC-3′) as the 5′ and 3′ amplification primers, respectively. The 3′ primer includes nucleotides that append the NS5A sequence with a sequence encoding (Asp)4-Lys, which is the enterokinase recognition sequence believed to improve the solubility of the NS5A domain I construct reported previously (36), followed by a translation stop codon. The blunt-ended PCR product was TOPO ligated into pLJEC1 and transformed into E. coli strain BL21(DE3) for the production of recombinant NS5A(33-202).
Production of recombinant NS5A(33-202) protein.
The production of NS5A(33-202) protein was carried out by using 2.5-liter Ultra-Yield flasks (5) containing 1 liter of TB (see above) supplemented with 100 μg/ml kanamycin and 100 μl/liter of antifoam that had been inoculated with 10 ml of an overnight culture grown in selective LB medium. Cultures were grown at 37°C and 250 rpm for 6 h prior to lowering the temperature to 16°C. After equilibration for approximately 1 h, the cultures were induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and growth continued overnight at 16°C (approximately 16 h). Typically, cultures reached an optical density (A600) of between 12 and 14. The cells were harvested by centrifugation at 7,000 × g for 10 min, and the pellets stored at −80°C for up to 1 month prior to processing. Two to 4 liters of culture were sufficient to produce multimilligram quantities of purified protein.
Purification of recombinant NS5A(33-202) protein.
All steps were performed on ice or at 4°C. The paste from 2 to 4 liters of cell culture producing recombinant NS5A(33-202) was resuspended in lysis buffer (100 mM Tris-HCl, pH 8.0, containing 50 mM NaCl, 5 mM imidazole-HCl, pH 8.0, and 100 μl/liter benzonase, pH 8.0) by using approximately 10 ml buffer per gram of cell paste. The resuspended material was twice passed through a microfluidizer (110Y; Microfluidics) at 90 lb/in2, and the lysed extract centrifuged at 5,000 × g for 1 h. The supernatant fraction was incubated with 5 ml of ProBond resin (Invitrogen) for approximately 2 h with mixing. The resin containing the bound NS5A protein was captured by passage through a gravity flow column, and the resin washed twice with 4 column volumes of wash buffer (100 mM Tris-HCl, pH 8.0, containing 200 mM NaCl and 40 mM imidazole-HCl, pH 8.0). The bound protein was step eluted from the column by using 4 column volumes of elution buffer (100 mM Tris-HCl, pH 8.0, containing 400 mM NaCl, 500 mM imidazole-HCl, pH 8.0, and 10% glycerol). The eluted protein was passed over 5 ml of Q-Sepharose resin (preequilibrated with elution buffer) by using a gravity-flow column, and the flowthrough fraction collected. The protein solution was subsequently dialyzed overnight against 4 liters of dialysis buffer A (100 mM Tris-HCl, pH 8.0, containing 400 mM NaCl, 40 mM imidazole-HCl, pH 8.0, and 10% glycerol) in the presence of TEV protease (18) at 1 mg TEV per 20 mg NS5A(33-202). The dialyzed protein was passed over 5 ml of fresh Probond resin (preequilibrated with the post-dialysis buffer A), and the flowthrough collected. The protein in the flowthrough was concentrated to approximately 5 mg/ml and further purified by high-performance liquid chromatography size exclusion chromatography (SEC; 300- by 21.2-mm Phenomenex BioSep S3000 column) in SEC buffer (25 mM Tris-HCl, pH 8.0, containing 250 mM NaCl and 10% glycerol). The peak monodisperse fractions were pooled and concentrated to approximately 26 mg/ml by using an Amicon Ultracel (Millipore) centrifugal concentrator. Protein measurements were determined by using Coomassie plus protein reagent (Pierce) with bovine serum albumin (2 mg/ml ampoule; Pierce) as the standard. The purified NS5A(33-202) protein was used soon after preparation or following flash freezing in liquid nitrogen.
Crystallization of NS5A(33-202).
Protein was diluted to 13 mg/ml in SEC buffer (see above) prior to crystallization trials. Initial crystals (small needles) were obtained with hanging-drop vapor diffusion at 4°C by mixing 2 μl each of protein and well solution on siliconized glass coverslips and suspending them over 1-ml reservoirs in 24-well Linbro plates. The reservoir buffer was 22% polyethylene glycol 3350, 0.1 M HEPES, pH 7.5, and 10% (vol/vol) isopropanol. The subsequent application of Hampton detergent additive screens nos. 1 to 3, also in a hanging-drop vapor diffusion format, yielded crystals with improved and similar morphology (hexagonal plates), although the majority remained too small for X-ray analysis. Hampton protocols were followed for volume ratios of protein solution, reservoir buffer, and stock detergent, such that the final detergent concentration was slightly below the critical micelle concentration. In this procedure, 2 μl of protein (13 mg/ml), 0.5 μl of supplied detergent solution, and 2.5 μl of reservoir buffer (same as above) were mixed on a coverslip and suspended over a reservoir of 0.5 ml. After approximately 2 weeks at 4°C, the best crystals reached a final size of about 0.1 mm and grew in the presence of the detergents N-nonylphosphocholine (FOS-choline-9; Hampton screen 3, no. 18) and 2,6-dimethyl-4-heptyl-β-d-maltopyranoside (Hampton screen 2, no. 3). These detergents are hereinafter abbreviated as “PC-9” and “maltose.” The crystals belong to space group P3121, with unit cell a = b ∼57 Å, c = 195 to 197 Å.
X-ray data collection and structure determination.
Crystals were mounted in Hampton loops and flash-frozen in liquid nitrogen following rapid immersion in a cryoprotectant composed of reservoir buffer with 20% glycerol (vol/vol). Diffraction data were collected at the Lawrence Berkeley Laboratory Advanced Light Source (ALS) for maltose detergent crystals and Argonne National Laboratory Advanced Photon Source (APS) for PC-9 detergent crystals, in both cases using ADSC Quantum-210 detectors. Data were integrated/scaled using HKL2000 (29). Molecular replacement was performed with CNX (6), using one monomer from the previously published NS5A domain I structure (36). The molecular replacement solution yielded a single dimer featuring a novel intermolecular configuration, which was confirmed by difference electron density maps displaying strong positive density at the position of the expected zinc atoms (not included in the starting model). The two dimer configurations observed thus far are unique, i.e., the monomer-monomer interface found in one dimer does not appear as a crystal lattice contact in the other case.
Refitting proceeded using MIFit (Rigaku Americas Corp.), and atomic refinement was performed with REFMAC (27). Data and refinement statistics are shown in Table 1. The final model consists of residues 32 to 191 in monomer A and 33 to 191 in monomer B. Residue 32 of monomer A (leucine) was visible, but this originates from the TEV cleavage site and is not part of the endogenous HCV sequence (which would be proline for this genotype). PROCHECK indicated only one disallowed residue, Phe 36, which lies in a highly constrained region next to the zinc site. However, this residue's dihedral angles fall within the allowed gamma-turn region of updated Ramachandran plots (24). Electron density was present for a portion of the maltose detergent (bound to monomer A only, remote from the dimer interface), and therefore, a partial model was built which lacks the second sugar ring. Density was not observed for the PC-9 detergent. One glycerol molecule was fit into extended density found at the dimer interface in both the PC-9 and maltose detergent cases. The two models of the domain I dimer obtained from these two detergents are essentially equivalent, with a root mean square deviation (RMSD) on all Cα atoms of only 0.25 Å; the PC-9 case is used henceforth to represent NS5A(33-202).
TABLE 1.
Crystallographic data and refinement statistics
| Parameter | Detergent additive
|
|
|---|---|---|
| PC-9 | Maltose | |
| Crystal data | ||
| Space group | P3121 | P3121 |
| Unit cell a & b, c (Å) | 57.2, 195.4 | 57.1, 197.4 |
| Diffraction dataa | ||
| X-ray source | APS 17ID | ALS 5.0.1 |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution range (Å) | 50-1.9 | 50-2.2 |
| Redundancy of data | 7.1 (6.7) | 6.2 (4.0) |
| Completeness (%) | 99.3 (98.6) | 99.0 (92.0) |
| <I/σI> | 44 (3.6) | 17.7 (2.0) |
| Rsym (%)b | 5.4 (39.8) | 10.8 (53.5) |
| Structure refinement | ||
| No. of reflections used | 29,345 | 19,213 |
| R factor, Rfree (%)c | 24.7, 28.8 | 22.2, 25.5 |
| No. of protein atoms (including Zn) | 2,464 | 2,470 |
| No. of solvent atoms | 254 | 203 |
| No. of other atoms (glycerol, detergent) | 6 | 28 |
| Average B factor (Å2) | 33.9 | 29.8 |
| RMSD bond lengths (Å) | 0.009 | 0.009 |
| RMSD bond angles (°) | 1.22 | 1.24 |
Values in parentheses refer to the highest-resolution shell.
Rsym = Σ|<I> − I|/ΣI, where I is measured intensity for reflections with indices hkl.
R factor = 100 × Σ|Fo − Fc|/Σ|Fo|; Rfree = free R factor based on random 2% of all data.
Solvent-accessible-surface calculations were performed using CNX with a default probe radius of 1.4 Å. Surface complementarity was calculated using CCP4 (7) with a default probe radius of 1.7 Å. Surface complementarity measures correlation of directions (unit vectors normal to the surface) and is relatively insensitive to the precise atomic radii used in defining the molecular surface (21). Figures were prepared using PyMol (10) and GRASP (for Fig. 3) (28).
FIG. 3.
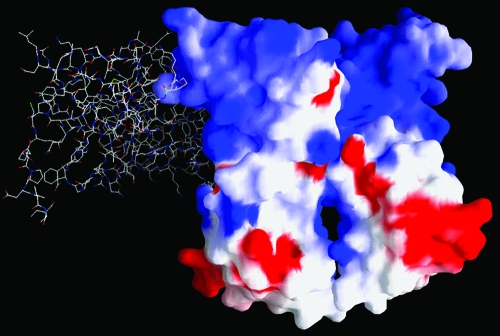
Electrostatic surface potential for the NS5A(33-202) dimer, with blue denoting positive and red negative values. The N termini of both monomers are located at the top, and the C termini are located at the bottom. For reference, the location of the second monomer in the NS5A(25-215) dimer is included as a stick representation, with overall orientation similar to that depicted in Fig. 2A.
Protein structure accession numbers.
Coordinates for the NS5A(33-202) dimer have been deposited in the Protein Data Bank with codes 3FQM (PC-9 case) and 3FQQ (maltose case).
RESULTS
Production of recombinant NS5A(33-202).
To date, recombinant forms of NS5A consisting of domain I alone or the near-full-length protein containing domains I, II, and III have been produced in E. coli as an initial chimeric species with yeast ubiquitin that, following translation, undergoes processing in vivo by yeast ubiquitin protease (Ubp1) generated from the coexpression plasmid pCG1 (16, 35, 36). In order to move away from this dependency on ubiquitin and ubiquitinase as coexpression partners, the production of NS5A in the current study utilized an N-terminal TEV-cleavable polyhistidine purification tag that allowed for efficient overexpression in E. coli.
Our initial work with NS5A domain I utilized the same HCV boundaries (residues 25 to 215) as described previously (35, 36). However, endogenous proteolysis in E. coli led to a mixture of protein species in the final preparation as a result of incomplete cleavage of NS5A at the Met 202-Leu 203 junction (determined by mass spectrogram analysis) (data not shown). Although mutagenesis of these two residues to Ala eliminated this proteolytic event, the alternate construct terminating at Met 202 was adopted for further studies. Truncation of the N terminus at Gly 33 improved the recombinant protein expression levels. The recombinant protein was appended at its C terminus with the cleavage recognition sequence for enterokinase (Asp)4-Lys since it was postulated that this charged motif probably increases the solubility of the recombinant NS5A products (35, 36). In summary, the final construct, following tag removal, produced for the crystallization studies reported herein was Ala-Leu-(HCV NS5A:33-202)-(Asp)4-Lys. This may be compared with the earlier crystallization construct of (HCV NS5A:25-215)-(X)-(Asp)4-Lys, where the reported cloning procedures did not allow X to be specifically defined (35, 36).
The purification of NS5A(33-202) takes advantage of serial immobilized metal affinity chromatography columns with tag removal by TEV protease between chromatographies. The second immobilized metal affinity chromatography column binds the same impurities as the first; however, the recombinant NS5A protein passes through as a result of tag removal. Additional steps utilizing ion exchange and SECs serve to remove nucleic acids and to provide a final purification step, respectively.
Overall structure of NS5A(33-202).
The crystal structure of NS5A(33-202) is shown in Fig. 1. The polypeptide fold within each monomer of the dimer of NS5A(33-202) is very similar to that reported previously for NS5A(25-215) (36), with only 1.0-Å RMSD on Cα atoms (Fig. 2). In addition, the zinc-binding site displays the same coordination geometry as observed earlier. These similarities likely reflect the absence of any amino acid differences between the two constructs within the overlapping residue range of 33 to 202. One notable difference is that the disulfide bond between Cys 142 and Cys 190 reported previously is not present in the current structure. While these side chains remain adjacent, the distance between thiols is too great (3.61 Å and 3.43 Å in monomers A and B, respectively) to allow covalent bond formation, and only very weak electron density exists between the side chains. No reducing agents were present during the purification or crystallization of this construct. Site-directed mutagenesis has shown that this disulfide is not essential for HCV replication (35).
FIG. 1.
Overall structure of the NS5A(33-202) dimer. (A and B) Ribbon (A) and molecular surface (B) representations, with monomer long axes vertical, N termini at the top, C termini at the bottom, and Zn atoms. IA and IB show subdomains of each monomer. (C and D) Views similar to those in panels A and B but looking down the long axes of the monomers.
FIG. 2.
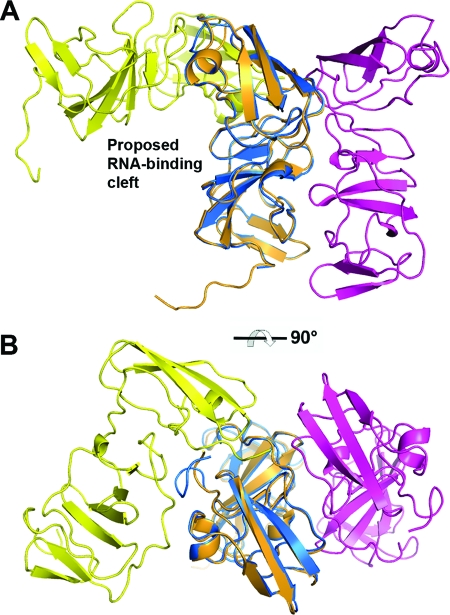
Comparison of the two known structures of NS5A domain I. (A) Side view. (B) Top view. The dimer of NS5A(33-202) (blue/magenta) is aligned with that from the previously reported NS5A(25-215) dimer (yellow/orange) following superposition of one monomer from each structure. Transformation between these two configurations requires complete separation of the monomers along with twisting of the long axes.
The ordered residues of NS5A(33-202) consist of residues 33 to 191, whereas residues 36 to 198 were observed in the previous structure of NS5A(25-215). The stabilization of a few extra residues at the N terminus in NS5A(33-202) appears to arise from crystal packing, since the relatively hydrophobic residues Phe 36/Phe 37 associate with the same residues of symmetry-related molecules (this applies to either monomer of the dimer). At the C terminus of NS5A(33-202), amino acids beyond residue 191 are disordered; their presence in NS5A(25-215) could be attributed to more-extensive crystal contacts but may also reflect added stability provided by a disulfide bond at Cys 190.
The association of the monomers in the dimer of NS5A(33-202) is significantly different from that in the previous structure of NS5A(25-215) (Fig. 1 and 2). In NS5A(33-202), the long axes of the monomers are nearly parallel, with numerous interactions along the entire side of each monomer, giving rise to the appearance of side-by-side cylinders. The monomers are related by a twofold-symmetry axis that runs parallel to their length. The two N termini are found on the same end of the dimer, implying a colocalization of the two amphipathic N-terminal helices, and this is a feature shared with the previous structure. However, there is no overlap between the two monomer-monomer interface surfaces, and in fact the two regions are on opposite sides of the monomer, as seen in Fig. 2B. Thus, the two dimer configurations of domain I are not related by a simple rearrangement, such as a slight twisting of one monomer. The transformation requires a complete separation of monomers and the translocation of one monomer to the other side of its initial partner, followed by a tilting of each monomer's long axis relative to the other.
The electrostatic surface potential for the NS5A(33-202) dimer is shown in Fig. 3. There is a predominance of basic charge at the N-terminal end of the dimer and more-acidic charge at the C-terminal end. In both of the domain I dimers, there is a presentation of hydrophobic and basic residues at the end presumably closest to the membrane surface, and in NS5A(33-202) this involves Val 34, Pro 35, Phe 36, Phe 37, and Trp 84 (these positions being highly conserved as hydrophobic amino acids), along with Arg 73 and Arg 78 (positions highly conserved as basic amino acids). For comparison, the location of a second monomer from the NS5A(25-215) dimer is shown by the stick representation in Fig. 3. Basic residues comprising the proposed RNA-binding cleft of NS5A(25-215) (36) are fully exposed in NS5A(33-202) and distant from the latter dimer's monomer-monomer interface.
NS5A(33-202) dimer interface.
At the NS5A(33-202) monomer-monomer interface, there is an extensive buried surface area, although a number of water molecules and one glycerol are trapped and make hydrogen bonds with surface residues (Fig. 4). A total of 1,820 Å2 of solvent-accessible surface area becomes buried during dimerization, suggesting a contact area of 910 Å2 per monomer; however, this is not contiguous van der Waals contact but includes several pockets or “gaps” with buried water molecules. The same calculation using the NS5A(25-215) dimer gives 1,738 Å2 of buried surface, with a contact area of 829 Å2. Thus, the two dimer modes provide a similarly large buried surface area, even though this is mapped across different faces of each monomer.
FIG. 4.
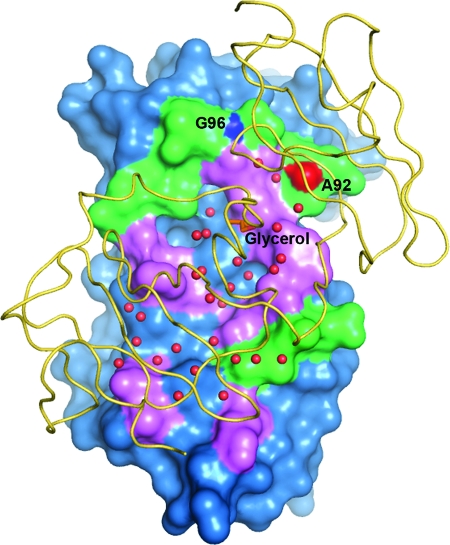
Dimer interface of NS5A(33-202) as seen looking through monomer B (yellow ribbon) onto monomer A (blue surface). Residues of monomer A in contact with monomer B are colored either magenta or green. Magenta signifies amino acid conservation above 95% based on 35 sequences spanning genotypes 1 to 6 from the Los Alamos database (20). Water molecules at the interface are shown as red spheres, and the trapped glycerol is an orange stick model. Intermolecular main-chain-main-chain H bonds formed between Gly 96 N and Ala 92 O are colored and labeled separately.
A shape correlation (SC) statistic was used to measure the geometric surface complementarity of the protein-protein interaction within each dimer. SC depends both on the relative shapes of the surfaces with respect to each other and on the extent to which the interaction brings individual elements of the opposing surfaces into proximity (21). SC values of 0.68 and 0.69 were obtained for the NS5A(33-202) and the NS5A(25-215) dimers, respectively. These two numbers are essentially equivalent but slightly below the typical range of 0.70 to 0.76 found for oligomeric proteins and for protein-protein inhibitor interfaces (21).
The dimer interface of NS5A(33-202) is composed of about 20 (mostly equivalent) residues from each monomer, due to the twofold character of the dimer. To a large extent the contact surfaces are formed by residues 92 to 99, 112 to 116, 139 to 143, 146 to 149, and 160 to 161 from each monomer. Approximately half of this surface is composed of highly conserved residues (Fig. 4). Interestingly this face of the monomer is fully exposed in the NS5A(25-215) structure and was noted for its large patch of conserved residues which might represent an important molecular interaction surface (36).
Intermolecular hydrogen bonds in the NS5A(33-202) dimer that are formed by conserved residues or main-chain atoms are shown in Fig. 5A. Of note are the salt bridge formed by the highly conserved Glu 148-Arg 112 pair and the main-chain-main-chain hydrogen bond between Ala 92 O and Gly 96 NH. Arg 48 (not shown) appears to stabilize the position of Glu 148 (through an intramolecular salt bridge); however, the former is not highly conserved. Finally, there is an electrostatic interaction between Arg 48 and Glu 116 across the dimer interface, although these two positions are not conserved as charged amino acids.
FIG. 5.
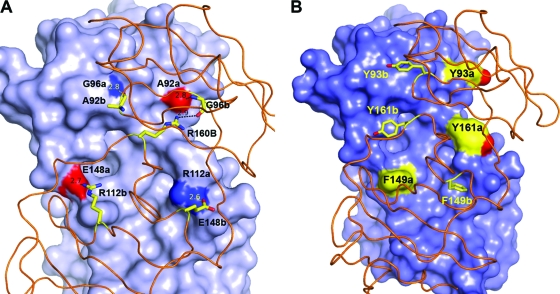
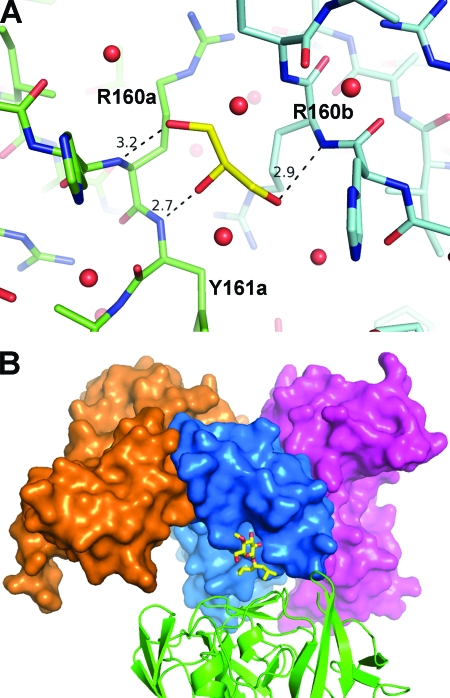
Key intermolecular interactions at the dimer interface in the NS5A(33-202) structure. (A) Intermolecular hydrogen bonds include highly conserved Arg 112 (occupied rarely by Lys) paired with invariant Glu 148 and main-chain atom Ala 92 O interacting with Gly 96 N. Invariant Arg 160 forms intermolecular contact with Ala 92 O, along with an intramolecular hydrogen bond to Gly 96. (B) Aromatic side chains at the interface. At position 93 (least buried), Tyr is predominant, although His and Thr sometimes arise. At position 161, Phe and Tyr are found about equally but His occurs on occasion. At position 149 (most buried), Phe is predominant, with the rare exception of Leu. Monomer A has a surface with highlighted colors for residues denoted “a.” Monomer B is displayed as a ribbon with stick sidechains for residues denoted “b.” Distances shown are in angstroms.
Aromatic amino acids found at the interface include Tyr 93, Tyr 161, and Phe 149 (Fig. 5B). The most buried side chains are Tyr 161 and Phe 149, positions at which an aromatic residue is highly conserved (>99% Phe or Tyr at position 161 and >99% Phe at position 149). These two side chains are solvent exposed on the surface of the NS5A(25-215) dimer. Least buried in NS5A(33-202) is Tyr 93, although the side chain rests against the neighboring monomer; at this position tyrosine is generally conserved, although His and Thr may occur. In the NS5A(25-215) structure, Tyr 93 is also partially exposed but interacts with the neighboring monomer's Phe 37 side chain.
Finally, a glycerol molecule lies on approximately the twofold axis between monomers, trapped within a cavity that also contains several ordered water molecules (Fig. 4 and 6A). The glycerol's hydroxymethyl groups form hydrogen bonds with Arg 160 N from each monomer, while the central hydroxyl interacts with Tyr 161 N of monomer A.
FIG. 6.
Small molecules observed in crystals of NS5A(33-202). (A) Glycerol molecule (yellow) found at the dimer interface, with hydrogen bond distances in Å. Glycerol was present in the protein and crystallization buffer, and this binding is probably a fortuitous event rather than a determinant of dimer formation. Red spheres show ordered water molecules at the interface. (B) Maltose detergent molecule (yellow) bound to surface of monomer A (blue surface), with its second sugar ring disordered. Also shown is monomer B of the dimer (magenta surface), along with the expected location for a second monomer in the NS5A(25-215) configuration (orange surface). The detergent is distant from either dimer interface and appears to facilitate contacts between an NS5A(33-202) dimer and a symmetry-related dimer (green ribbon).
DISCUSSION
The dimer observed in the crystal structure of either NS5A(33-202) or NS5A(25-215) suggests a mechanism of membrane anchoring for NS5A via two N-terminal, amphipathic alphahelical “arms” which extend away from the core of each monomer. The dimer interface of NS5A(33-202) incorporates the most highly conserved surface region of the domain, with several interlocking aromatic and hydrogen bonding elements. These features would tend to promote consistent dimer formation across HCV genotypes. However, the NS5A(25-215) dimer interface also contains a number of conserved residues and a similarly large buried surface area. Neither dimer incorporates shape complementarity to the same degree as most proteins known to function as homodimers (21), yet this characteristic may allow the monomers to dissociate and reassemble in alternate dimer configurations or to interact readily with other proteins. The propensity for domain I of NS5A to dissociate and reform while tethered to a membrane is difficult to assess, given the imprecise definition and unknown flexibility of “linker” residues (residues 26 to 35) which reside between the N-terminal amphipathic helix and the core of domain I. While it is of course possible that either or both of the current dimer structures represent an artifact arising from the preparation and crystallization of isolated domains, we propose that both may reflect a physiological state of NS5A. There is limited evidence for NS5A homodimerization from in vitro cell systems (11).
As noted earlier, the NS5A(25-215) dimer features a net-positively charged cleft that has been proposed to serve an RNA-binding function (36), whereas in the NS5A(33-202) structure, this cleft is absent and the proposed RNA-binding residues are exposed and distributed between two physically remote, flat surfaces. To date, RNA binding has been reported only for three-domain NS5A(25-447) (15). This observation was confirmed in the present study, but only weak affinity for RNA by either construct of domain I was found (results to be published elsewhere). Since the dimeric state of NS5A during our binding experiments was unclear (see below), it is difficult to interpret the results of RNA binding by reference to either crystal structure of domain I.
What factors might influence the formation of either domain I dimer, at least with the isolated protein in solution? There are no amino acid differences within the interface regions of these two dimers to suggest contributions from dissimilar surface features. Other possibilities include differences between the two constructs at their amino and carboxy termini and alternative protein preparation and crystallization procedures.
The N terminus of NS5A(25-215) contains eight additional authentic NS5A residues (residues 25 to 32) in comparison to the N terminus of NS5A(33-202), and the latter features non-HCV residues Ala-Leu at 31 and 32 (instead of HCV Leu-Pro), which are derived from the cloning strategy. While this difference may influence dimer configuration in solution, the crystal structures do not reveal a potential mechanism because residues 25 to 35 of NS5A(25-215) are disordered. There is significant interaction between ordered and adjacent N-terminal residues 36 to 38 at the NS5A(25-215) dimer interface, but the equivalent residues in the NS5A(33-202) dimer are far apart and do not interact. Any potential for non-HCV amino acids 31 and 32 in NS5A(33-202) to preclude NS5A(25-215) dimer formation is unpredictable, again due to the uncertainty of residues 25 to 35 in the latter case. At the C termini, ordered polypeptides in either dimer are well separated from one another and do not appear likely to impact dimer formation, at least for these isolated domain I proteins. There are additional disordered C-terminal residues, 192 to 202 in NS5A(33-202) and 199 to 215 in NS5A(25-215), which may interact in a manner not discernible from the crystal structures. In summary, differences of terminal residues between the two domain I constructs, as observed in the crystallized proteins, do not offer an obvious explanation for the alternate dimer configurations.
Another consideration is the additive used during crystallization of each domain I, i.e., nondetergent sulfobetaine 201 (NDSB) for NS5A(25-215) and the phosphocholine- or sugar-based detergents for NS5A(33-202), since each might give rise to a homogeneous population of dimers prior to crystal nucleation. The two detergents used in the present study belong to very different chemical classes (phosphocholine versus sugar core), and thus, the dimer configuration reported here is not likely a direct consequence of the particular detergent employed. While a binding site for the maltose detergent molecule on one monomer of NS5A(33-202) was observed (Fig. 6B), it does not sit near the interface defined by either dimer, and thus, this particular additive probably does not promote formation of the NS5A(33-202) dimer nor sterically preclude formation of the NS5A(25-215) dimer. Instead, the maltose detergent appears to stabilize an interaction between preformed NS5A(33-202) dimers. While NDSB might favor formation of the NS5A(25-215) dimer, our NS5A(33-202) protein would not crystallize under conditions involving NDSB (36). Finally, the presence of a bound glycerol molecule at the dimer interface of NS5A(33-202) (Fig. 6A) is an unlikely explanation for that particular configuration. Glycerol was present in the protein buffer during the preparation and crystallization of both constructs, and its presence here may simply reflect a fortuitous proximity of backbone amides. Ordered glycerol is commonly found in crystal structures.
A complication of this survey is that the oligomeric state of our purified NS5A in solution (both versions of domain I and the three-domain construct), alone or with additives, could not be determined unambiguously despite a number of attempts using a variety of biophysical measurements. Sedimentation velocity and equilibrium analysis, dynamic light scattering, and small-angle X-ray scattering (SAXS) all indicated that, over a variety of concentrations, the oligomeric state is heterogeneous, with a wide range of molecular-weight species (data not shown). Interestingly, the SAXS analysis suggested an oligomerization mode more reminiscent of an ordered assembly rather than random aggregation, since the latter has its own unique and distinctive scattering profile.
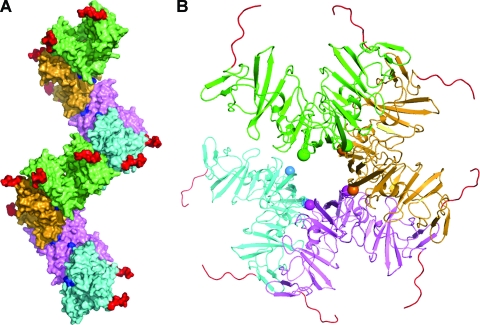
One implication of the two large and nonoverlapping dimerization interfaces on opposite faces of each monomer of domain I is the potential for polymerization. Such a phenomenon would help to explain the propensity for higher oligomerization observed in preparations of either domain I or three-domain constructs (lacking the N-terminal helix). The solution state of purified NS5A may thus represent an equilibrium between the two known dimer states and various linear assemblies of dimers. It is theoretically possible to build an assembly of domain I proteins through the use of alternating interfaces, as shown in Fig. 7. This can be described as a polymerization of preformed dimers in either the NS5A(33-202) or NS5A(25-215) configuration, since in either case a second surface patch is always accessible and remote from the buried interface. This oligomerization model of domain 1 has a superhelical appearance, with the C termini directed outward, which would sterically accommodate domains II and III. The N-terminal amphipathic helices (not included in this model) would be directed inward toward the central axis of the superhelix and could theoretically interact with one another. However, the resultant nonplanar spatial arrangement of these N termini would preclude association of this assembly with a flat membrane surface. Other associations with cellular or viral components might be possible, but there is currently no evidence for such an assembly during the HCV life cycle.
FIG. 7.
Theoretical assembly of NS5A domain I proteins based on simultaneous recruitment of the two monomer-monomer interfaces, resulting in a superhelical array. (A) Side view. In this depiction, NS5A(25-215) dimers are distinguished by separate colors and are joined by means of the interface identified in the NS5A(33-202) dimer. One full “turn” of the superhelix consists, for example, of dimers colored green, orange, magenta, and cyan, and then green again. The two C termini of each dimer are colored red and project outward, implying that domains II and III of NS5A could be accommodated sterically. The location of each emerging N-terminal residue 36 is colored blue. (B) View down long axis of superhelix, with only four dimers displayed for clarity, colored green (nearest), orange, magenta, and cyan (farthest). The two N termini of each dimer (residue 36) are indicated by a sphere with color matching the dimer. The C termini are in red. This arrangement suggests that the N-terminal amphipathic helices (not modeled) would be directed inward toward the assembly's central axis, where they might interact with one another. Any membrane associated with these amphipathic helices could not adopt a planar geometry but instead would be highly twisted. Currently there is no evidence for such an assembly in the cellular environment, but this polymerization may explain the properties of isolated NS5A in solution, which include a highly heterogeneous oligomeric state.
In recent years, permissive cell cultures that support the production of infectious HCV particles have provided further insight into the multiple roles of NS5A, which now includes participation in virion particle assembly by means of association of NS5A with intracellular lipid storage compartments termed lipid droplets (1, 19, 25). Phosphorylation of NS5A appears to modulate its various functions, possibly serving as a switch between viral RNA replication and viral assembly (17). It is conceivable that the two dimer conformations observed thus far in crystallized domain I reflect two different functional states of membrane-tethered NS5A and that other configurations (even monomeric) could exist. One might speculate that the NS5A(25-215) dimeric state, with its suggestive binding cleft, promotes RNA binding during NS5A's participation in the replication phase, while the NS5A(33-202) dimeric state is a prerequisite for NS5A roles in lipid droplet association and/or particle assembly. The differential exposure of surface features of the NS5A(33-202) and NS5A(25-215) dimers would allow an alternative set of interactions between domain I and other HCV or host proteins. For example, NS5A residues 163 to 167, implicated in binding to NS4A, with consequences for enhanced NS5A phosphorylation (2), are buried in the dimer of NS5A(25-215) but exposed in the NS5A(33-202) dimer. In any case, the large difference in relative monomer orientation between these two domain I dimers would likely result in dissimilar positioning of downstream domains II and III, with potentially major functional consequences.
In summary, to date two possible modes of NS5A dimerization have been identified through determination of the structure of the isolated domain I protein. Both dimer structures support the notion of membrane-bound NS5A proteins associating through contact of domain I surfaces. Further studies of full-length NS5A and of the protein in a membrane environment will be needed to elucidate the potential contribution of such dimers to the HCV life cycle.
Acknowledgments
We are grateful to Craig E. Cameron for providing a sample of the His-Δ-NS5A construct. We thank Dan Knighton for collection of diffraction data for the NS5A(33-202) crystals at the ALS and APS beamlines. We also thank Michal Hammel, Greg Hura, and John Tainer, for assistance with SAXS analysis at the SIBYLS beamline at ALS.
All funding for these studies was provided by Pfizer Global Research and Development.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asabe, S. I., Y. Tanji, S. Satoh, T. Kaneko, K. Kimura, and K. Shimotohno. 1997. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J. Virol. 71790-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2778130-8139. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky, O., and C. N. Cronin. 2006. Economical parallel protein expression screening and scale-up in Escherichia coli. J. Struct. Funct. Genomics 7101-108. [DOI] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54(Pt. 5)905-921. [DOI] [PubMed] [Google Scholar]
- 7.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 8.Cho, N. J., K. H. Cheong, C. Lee, C. W. Frank, and J. S. Glenn. 2007. Binding dynamics of hepatitis C virus' NS5A amphipathic peptide to cell and model membranes. J. Virol. 816682-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, Y. L., M. L. Sheu, and S. H. Yen. 2003. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int. J. Cancer 10765-73. [DOI] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA.
- 11.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 775401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 776055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230217-227. [DOI] [PubMed] [Google Scholar]
- 14.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 671385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 28036417-36428. [DOI] [PubMed] [Google Scholar]
- 16.Huang, L., E. V. Sineva, M. R. Hargittai, S. D. Sharma, M. Suthar, K. D. Raney, and C. E. Cameron. 2004. Purification and characterization of hepatitis C virus non-structural protein 5A expressed in Escherichia coli. Protein Expr. Purif. 37144-153. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Y., K. Staschke, R. De Francesco, and S. L. Tan. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 3641-9. [DOI] [PubMed] [Google Scholar]
- 18.Kapust, R. B., J. Tozser, J. D. Fox, D. E. Anderson, S. Cherry, T. D. Copeland, and D. S. Waugh. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14993-1000. [DOI] [PubMed] [Google Scholar]
- 19.Kaul, A., I. Woerz, P. Meuleman, G. Leroux-Roels, and R. Bartenschlager. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 8113168-13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21379-384. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, M. C., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234946-950. [DOI] [PubMed] [Google Scholar]
- 22.Liang, Y., H. Ye, C. B. Kang, and H. S. Yoon. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 4611550-11558. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 24.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50437-450. [DOI] [PubMed] [Google Scholar]
- 25.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 26.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5453-463. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., A. A. Bagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likehood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11281-296. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski, Z., and W. Minor. 1997. Processing X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. Carter, Jr., and R. M. Sweet (ed.), Methods in enzymology, vol. 276. Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 30.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 27940835-40843. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz, U., and S. L. Tan. 2008. NS5A: from obscurity to new target for HCV therapy. Recent Patents Anti-Infect. Drug Disc. 377-92. [DOI] [PubMed] [Google Scholar]
- 32.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRp) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 27711149-11155. [DOI] [PubMed] [Google Scholar]
- 33.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 821073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 27948576-48587. [DOI] [PubMed] [Google Scholar]
- 36.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]