Abstract
Successful viral replication is dependent on a conducive cellular environment; thus, viruses must be sensitive to the state of their host cells. We examined the idea that an interplay between viral and cellular regulatory factors determines the switch from Kaposi's sarcoma-associated herpesvirus (KSHV) latency to lytic replication. The immediate-early gene product K-Rta is the first viral protein expressed and an essential factor in reactivation; accordingly, this viral protein is in a key position to serve as a viral sensor of cellular physiology. Our approach aimed to define a host transcription factor, i.e., host sensor, which modulates K-Rta activity on viral promoters. To this end, we developed a panel of reporter plasmids containing all 83 putative viral promoters for a comprehensive survey of the response to both K-Rta and cellular transcription factors. Interestingly, members of the NF-κB family were shown to be strong negative regulators of K-Rta transactivation for all but two viral promoters (Ori-RNA and K12). Recruitment of K-Rta to the ORF57 and K-bZIP promoters, but not the K12 promoter, was significantly impaired when NF-κB expression was induced. Many K-Rta-responsive promoters modulated by NF-κB contain the sequence of the RBP-Jκ binding site, a major coactivator which anchors K-Rta to target promoters via consensus motifs which overlap with that of NF-κB. Gel shift assays demonstrated that NF-κB inhibits the binding of RBP-Jκ and forms a complex with RBP-Jκ. Our results support a model in which a balance between K-Rta/RBP-Jκ and NF-κB activities determines KSHV reactivation. An important feature of this model is that the interplay between RBP-Jκ and NF-κB on viral promoters controls viral gene expression mediated by K-Rta.
Kaposi's sarcoma-associated herpesvirus (KSHV), also designated human herpesvirus 8, has been linked to Kaposi's sarcoma (KS) (8) as well as primary effusion lymphoma (or body cavity B-cell lymphoma [BCBL]), and a subset of multicentric Castleman's disease (54). Although infrequent in the United States, Europe, and Asia, this virus shows a significant prevalence in the Mediterranean area and is common in several countries in sub-Saharan Africa (9). KS has emerged as the major malignancy in human immunodeficiency virus-infected AIDS patients coinfected with KSHV (48). Similar to other oncogenic herpesviruses, malignant transformation requires that KSHV enter into a latent state, in which all but a few viral genes involved in latency and/or transformation are silenced. Viral replication, which recruits target cells and creates a unique paracrine environment, is a critical prelude to disease development (11). Importantly, KSHV also encodes genes that govern cellular transformation, evasion of apoptosis, aberrant angiogenesis, and an inflammatory tumor microenvironment (3, 16). These genes are important for enabling the virus to establish chronic infection by various mechanisms, such as evasion of host immune responses.
Epidemiological and clinical data support the concept that chronic inflammation potentiates or promotes tumor development, growth, and progression. Proinflammatory gene products involved in these oncogenic processes include tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), IL-8, and vascular endothelial growth factor. Expression of these immunomodulatory proteins is mainly regulated by members of the NF-κB family of transcription factors that function as either dimers or heterodimers (21). The classical transactivating form of NF-κB is composed of RelA (p65)/p50 heterodimers. NF-κB dimers and heterodimers are kept sequestered in the cytoplasm by interaction with one of several inhibitors, which are designated IκB proteins (21). Upon cell stimulation, IκB is phosphorylated and degraded in a proteosome-dependent manner, and the active dimer or heterodimer NF-κB is released and translocates to the nucleus (21). Because IκB expression is regulated by NF-κB, new IκB protein is synthesized and accumulates to serve as negative feedback for the NF-κB signal. Several KSHV gene products activate the NF-κB pathway; these include the viral G protein-coupled receptor, K15 protein, viral FLICE/caspase 8 inhibitory protein (v-FLIP), and K1 protein (16, 52, 55, 59). Importantly, activation of NF-κB is required for direct and paracrine mechanisms of viral neoplasia as well as cell transformation by this virus (16, 43). Ironically, recent studies showed that NF-κB repressed lytic viral replication (1, 15). These findings, however, implicated a model that inflammation caused by KSHV infection may be a key to maintaining the latent viral state by inhibiting viral reactivation. This model was further supported by the recent observation that the latent protein v-FLIP suppressed viral reactivation through activation of NF-κB (63).
Accumulating evidence suggests that K-Rta (open reading frame 50 [ORF50]) is a key KSHV transcription factor that plays an essential role in the initiation of viral replication (42, 46, 56, 62). K-Rta is a strong transactivator and thereby influences expression of many viral genes as well as several cellular genes (5, 11). Overexpression of K-Rta activates a transcription cascade of KSHV genes that leads to lytic replication of virus (14, 41, 46, 56). In addition, this cascade is also induced by various chemicals or physiological stimuli, all of which are believed to function through the main trigger of viral lytic replication, K-Rta (6, 10, 40). The broad promoter specificity of this viral regulatory protein comes from its ability both to bind DNA directly (7) and to associate with cellular transcription factors that bind DNA such as C/EBPα (22) and RBP-Jκ (36).
RBP-Jκ is a DNA binding protein that has the capacity to regulate NF-κB and participate in the control of cytokines and NF-κB proteins (23, 28, 47). Structural analysis shows that RBP-Jκ contains a Rel homology domain which is shared by members of the NF-κB family (30). In the presence of Notch, RBP-Jκ behaves as a bifunctional transcription factor by switching from repressor to activator (24, 61). Repression by RBP-Jκ has been described for several cellular genes, including Hes1, IL-6, and NF-κB2 (29, 47, 51), and this repression is mediated, at least in part, by interaction with nuclear corepressors SMRT and N-CoR (29). Mechanistic similarities are observed between Notch and K-Rta, both of which activate the Notch pathway by converting RBP-Jκ from the repressive form to the active form. Accordingly, the Notch intracellular domain has the potential to activate K-Rta target genes (4). In some cases, the Notch intracellular domain triggers the KSHV lytic replication cycle (34).
Our previous work identified K-Rta binding sites on the KSHV genome during reactivation and revealed the transactivation potency of K-Rta for many viral promoters in transient-transfection assays (11a). Based on a KSHV genome-wide analysis, we showed that K-Rta was preferentially recruited to two promoters, K12 and Ori-RNA, during reactivation. Interestingly, viral promoters, which are highly responsive to K-Rta in the transient-transfection reporter assay (e.g., the ORF57 and K-bZIP promoters), were not the major binding targets of K-Rta. Accordingly, the aim of the current study was to search for cellular cofactors that regulate K-Rta transactivation by studying transcription factor binding sites on KSHV promoters regulated by K-Rta. The focus was on the key cellular transcription factor NF-κB. Results of these studies support a model in which a balance between K-Rta and NF-κB activities regulates viral reactivation through the interplay between NF-κB and RBP-Jκ on viral promoters.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney epithelial 293 cells were grown in monolayer culture in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) in the presence of 5% CO2. The TREx-RelA-IRES-K-Rta and TREx-IκBα-IRES-K-Rta BCBL-1 cell lines were cultured in RPMI 1640 supplemented with 15% FBS, 100 μg/ml of blasticidin (Invitrogen), and 100 μg/ml of hygromycin (Invitrogen).
Plasmids.
The plasmid encoding the full-length K-Rta gene was described previously (26). This cloning introduced a CpoI site and a Flag tag or hemagglutinin (HA) tag at the N terminus of the K-Rta gene as described previously (39). Similarly, a recombinant baculovirus transfer vector, pFAST-BAC (Invitrogen), was modified by inserting a Flag tag and CpoI site. Full-length forms of RBP-Jκ, RelA, RelB, c-Rel, IκBα, active NF-κB1 (p50), and active NF-κB2 (p52) were amplified by PCR using Pfu Turbo (Stratagene). Amplified DNA fragments were cloned into pFAST-BAC-Flag, pcDNA-Flag vector, or pcDNA-HA vector (25, 26). Primers for cloning are listed in Table 1. Plasmid clones were confirmed by sequence analyses. The NF-κB reporter was constructed by inserting three repeats of NF-κB binding sites (5′-aggGGACTTTCCcaggc-3′) in a position 5′ to the E1B minimal TATA box sequence. The pGL3 E1B TATA plasmid, which contains the E1B TATA box upstream of the luciferase gene, was a generous gift from Dan Robinson (UC Davis Cancer Center).
TABLE 1.
Primers used for cloning
| Primer | Sequence (5′→3′)a |
|---|---|
| RBP-Jκ CpoI, F | aaaCGGTCCGATGGGGGGCTGCAGGAAATTTGGTGA |
| RBP-Jκ CpoI, R | aaaCGGACCGTTAGGATACCACTGTGGCTGTAG |
| NF-κB1 CpoI, F | aaaCGGTCCGATGGCAGAAGATGATCCATATTTG |
| NF-κB1 433 CpoI, R | aaaCGGACCGTTACATGGTTCCATGCTTCATCCCAG |
| NF-κB2 CpoI, F | aaaCGGTCCGATGGAGAGTTGCTACAACCCAGGT |
| NF-κB2 473 CpoI, R | aaaCGGACCGTTACAGCGCGCGCGCGTCCGCGGTGA |
| RelA CpoI, F | aaaCGGTCCGATGGACGATCTGTTTCCCCTCAT |
| RelA CpoI, R | aaaCGGACCGTTAGGAGCTGATCTGACTCAAA |
| RelB CpoI, F | aaaCGGTCCGATGCCGAGTCGCCGCGCTGCCAG |
| RelB CpoI, R | aaaCGGACCGCTACGTGGCTTCAGGCCCTGGA |
| c-Rel CpoI, F | aaaCGGTCCGATGGCCTCCGGTGCGTATAACCCGTA |
| c-Rel CpoI, R | aaaCGGACCGTTATACTTGAAAAAATTCATATGGAA |
| IκBα CpoI, F | aaaCGGTCCGATGTTCCAGGCGGCCGAGCG |
| IκBα CpoI, R | aaaCGGACCGTCATAACGTCAGACGCTGGCCTC |
Underlined nucleotides represent restriction enzyme (CpoI) sites used for cloning the PCR products; lowercase letters indicate nucleotides added to enhance recognition by the restriction enzyme. F, forward; R, reverse.
Generation of dual inducible BCBL-1.
DNA fragments of the internal ribosome entry site (IRES) sequence were amplified by PCR and cloned in front of the K-Rta coding sequence in a pcDNA5 FRT/TO vector (Invitrogen). The coding sequence of RelA or IκBα was then cloned upstream of the IRES sequence. The resulting transfer plasmid was denoted pcDNA/FRT/TO-RelA-K-Rta or pcDNA5-IκBα-K-Rta. To establish the cell lines, which inducibly expressed RelA/IκBα and Rta (TREx-RelA-IRES-K-Rta or TREx-IκBα-IRES-K-Rta BCBL-1 cell line), the transfer plasmid was cotransfected with pOG44 (Flp recombinase expression plasmid [Invitrogen]) into 3 × 107 total TREx-BCBL-1 cells (46) by electroporation. At 48 hours after electroporation, the cell cultures were selected with 200 μg of hygromycin B (Invitrogen)/ml for 3 to 4 weeks as described elsewhere (46). After generating stable clones, protein expression was confirmed by immunoblotting. To avoid variation associated with cell clones, 10 inducible clones were combined and used for this study.
Immunoprecipitation and immunoblot analyses.
Cells were rinsed in ice-cold phosphate-buffered saline (PBS), and 1 × 107 cells were lysed in EBC lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Roche). After centrifugation (15,000 × g for 10 min at 4°C), 20-μl volumes of protein A- and protein G-Sepharose beads (Upstate) were added to the supernatants and preincubated overnight at 4°C. A 500-μg aliquot of each of the cleared supernatants was reacted with respective antibodies for 3 h at 4°C with gentle rotation. The immune complex was captured by the addition of 20 μl of a protein A- and protein G-Sepharose bead mixture and rocked for an additional 2 h at 4°C. Beads were washed four times with EBC buffer and boiled for 5 min in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromphenol blue).
293T cells were cotransfected with 2 μg of p3HA-RBP-Jk and 2 μg of pFlag-RelA or pFlag-empty expression plasmids, using FuGENE 6 (Roche) according to the supplier's recommendations. A reciprocal experiment with 2 μg of pHA-RelA and pFlag-RBP-Jκ or pFlag-empty was similarly performed. The cells were harvested 48 h after transfection and lysed in EBC buffer. Aliquots of 500 μg of cell lysates were immunoprecipitated with the addition of 25 μl of anti-Flag antibody-conjugated agarose (Sigma). Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer. Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred as described below.
For immunoblotting, cells were rinsed in ice-cold PBS, and 1 × 107 cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.5], 1.0% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented with a protease inhibitor cocktail (Roche). After centrifugation (15,000 × g for 10 min at 4°C), protein concentration was measured by using a bicinchoninic acid protein assay kit (Pierce), and equal amounts of total cell lysates were subjected to a SDS-PAGE and subsequently transferred to a polyvinylidene fluoride membrane (Millipore) using a semidry transfer apparatus (Bio-Rad). Final dilution of primary antibody was 1:4,000 for anti-K-Rta immunoglobulin G (IgG), 1 μg/ml of anti-K-bZIP IgG′ at 1:1,000 for anti-Flag antibody (Sigma), 1:500 for anti-K8.1 antibody (Advanced Biotechnology), 1 μg/ml of anti-RelA (Cell Signaling), and 1:1,000 for anti-RBP-Jκ (Abcam).
In vitro interaction assay.
Purified proteins (100 ng each) were mixed in the binding buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 0.02% NP-40, 10% glycerol supplemented with 1 mg/ml bovine serum albumin [BSA], 0.5 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktails) and then incubated for 30 min at 4°C. After 10-fold dilution with the binding buffer, 3 μg of anti-RelA antibody was added to the mixture and incubated for another 1 h at 4°C to form immunocomplex. The immunocomplex was captured with 20 μl of a protein A and protein G beads mixture. Beads were washed four times with binding buffer and boiled for 5 min in 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromphenol blue) and subjected to SDS-PAGE.
ChIP assay.
TREx-RelA-IRES-K-Rta BCBL-1 cells or TREx-IκBα-IRES-K-Rta cells (3 × 10 7/time point) were seeded in RPMI plus 15% serum, at a density of 5 × 10 5/ml, allowed to recover for 15 h, and pulsed with 100 ng/ml doxycycline for 4 h, after which time the medium was changed to prevent overexpression of K-Rta. At various time points following the treatment, the cultures were cross-linked and fixed with formaldehyde solution (0.1 M NaCl, 1 mM EDTA, 50 mM HEPES, pH 7.5, and 11% HCHO with CH3OH) for 10 min at room temperature. A chromatin immunoprecipitation (ChIP) assay was performed as described previously (25). Chromatin fragments were immunoprecipitated with 5 μg of anti-K-Rta IgG or preinoculated rabbit IgG overnight at 4°C with gentle rotation. After reverse cross-linking at 65°C overnight, the DNA fragments were purified with a QIAquick PCR purification kit (Qiagen).
Real-time qPCR for ChIP analysis.
Eluted DNA from each ChIP assay was diluted 1:50 in double-distilled H2O and used as a template for quantitative PCR (qPCR) with the primer sets listed in Table 2. Five microliters of template was used per 25-μl reaction mixture, and all samples were analyzed in triplicate using SYBR green 2x master mix (Bio-Rad) on an iCycler iQ thermal cycler (Bio-Rad). After an initial denaturation and activation incubation of 3 min at 95°C, 45 cycles of three-step cycling were performed with an annealing temperature of 60°C. Melt curve analysis was performed to verify product specificity. Relative fold induction over IgG for each immunoprecipitate and time point was determined by applying the ΔCt as a power of 2, with standard deviations calibrated to account for variances in IgG and IP-DNA signal, and then propagated logarithmically. All primer sets were assayed for all time points from at least two independent experimental replicates. Primers used for qPCR are listed in Table 2.
TABLE 2.
Primers used for real-time PCR
| Primer name and orientation | Sequence (5′→3′) |
|---|---|
| ORF6, F | GCTTCGACAAAGGAGCAATC |
| ORF6, R | GCTCTGGCTATCCTGACCTG |
| ORF57, F | TCTGCAATGCGTTTGTTACC |
| ORF57, R | CGCTACCAAATATGCCACCT |
| ORF59, F | CTATGCCAGCGTCGAGTACA |
| ORF59, R | GGAAGGCAGTGGAGACGTTA |
| K12, F | GTTGCAACTCGTGTCCTGAA |
| K12, R | AGTTCATGTCCCGGATGTGT |
| LANA, F | TGGAGATGGGAGATGTAGGC |
| LANA, R | TCCCCCTAGATGTGACTTCG |
| ORF57p, F | TTCCATTAGGGTGAGCGAAG |
| ORF57p, R | CCACTGGTACCACAAACGAA |
| K-bZIPp, F | CAGTTTGGTGCAAAGTGGAG |
| K-bZIPp, R | TCGACAACGGAGGAAATACC |
| K12p, F | GTCGGTCTCCCCTCTTCTTT |
| K12p, R | CTAGGTCCACGCTCACCTAT |
Quantification of encapsidated KSHV genome.
Two hundred microliters of cell culture supernatant was treated with 12 μg of DNase I for 15 min at room temperature to degrade unencapsidated DNA. This reaction was stopped by addition of EDTA to 2 mM and heat inactivation at 70°C for 15 min. Viral genomic DNA was purified from the digested supernatant using a QIAmp MinElute virus spin kit (Qiagen) and eluted in 100 μl of AVE buffer supplied by the manufacturer. In order to quantify viral copy number, 2 μl of the eluate was amplified in triplicate by qPCR using an iCycler iQ thermal cycler (Bio-Rad). The PCR mix contained primers targeting the coding region of ORF73 (K-ORF73 Fw, 5′-CGAATACCGCTATGTACTCAGA-3′; K-ORF73 Rev, 5′-CGCCTCATACGAACTCCAG-3′), flanking the hybridization site of a 5′-6-carboxyfluorescein-labeled probe with a 3′ Black Hold quencher (K-ORF73 probe, 5′-6-carboxyfluorescein-TCAGAACATCACCACCCCACAGAC-Black Hole quencher-3′ [Integrated DNA Technologies]) as well as 2× qPCR master mix (Eurogentec). Threshold cycle number for each sample was used in comparison to a standard curve generated using an ORF73 expression plasmid to determine absolute copy number in the reaction mixture, from which the number of copies/ml of the initial supernatant was inferred.
Generation of recombinant baculoviruses and protein purification.
Spodoptera fugiperda Sf9 cells were maintained EX-CELL 420 medium (JRH Biosciences), and recombinant baculoviruses were generated according to the manufacturer's protocol (Invitrogen). Recombinant baculovirus bacmid DNA was transfected into Sf9 cells by using FuGENE6 (Roche), and recombinant viruses were subsequently amplified twice. Expression of recombinant proteins was confirmed by immunoblotting with anti-Flag monoclonal antibody (Sigma). A large-scale culture of Sf9 cells (100 ml) was infected with recombinant baculovirus at a multiplicity of infection of 0.1 to 1.0, and cells were harvested 48 h after infection. Recombinant proteins were purified as described previously (25). Purity and amount of protein was measured by SDS-PAGE and Coomassie blue staining with BSA as a standard.
EMSA.
A 50 nM concentration of 32P-labeled oligonucleotides was used in 20-μl reaction mixtures. Probe sequences for the electrophoretic mobility shift assay (EMSA) were as follows: ORF57 promoter probe, 5′-AATAATGTTCCCACGGCCCATTTTT-3′; K12 promoter probe, 5′-CCCGGGAAATGGGTGGCTAACCCCTACATAAGCAGTTTG-3′. Labeled probes were incubated with purified proteins in binding buffer (20 mM HEPES [pH 7.9], 60 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 0.2 mg/ml BSA, 100 ng dI-dC, 2% glycerol). The final protein concentration (total volume, 20 μl) was 100 nM for RBP-Jκ, 400 nM for K-Rta, and 100 nM or 400 nM for NF-κB (RelA/p50). Samples were electrophoresed at 140 V in 4% TG0E (25 mM Tris [pH 8.3], 190 mM glycine) gels. Gels were dried and visualized by autoradiography with a PhosphorImager (Bio-Rad).
Reporter assays in transient-transfection experiments.
Reporter plasmid was constructed by inserting a promoter region upstream of the firefly luciferase coding region (Luc) in the pGL3-Basic vector (Promega). The KSHV promoter library was created based on published work where available; otherwise putative KSHV promoter regions of approximately 500-bp DNA fragments upstream of the AUG of the respective open reading frame were amplified by PCR using Pfu Turbo (Stratagene) with appropriate primers. Sequences and locations for these primers are available upon request (11a) (Table 1). 293 cells were seeded in 12-well plates at 1 × 10 5/well in 1.0 ml of Dulbecco's modified Eagle's medium supplemented with 10% FBS and incubated at 37°C with 5% CO2. For each well, an equal amount of plasmid DNA, including the reporter and the control or expression plasmid, was transfected using the Fugene6 reagent following the manufacturer's protocol (Roche). Transfected cells were treated with TNF-α (Chemicon) at final concentration of 20 ng/ml where indicated. Cell lysates were prepared 48 h after transfection with 1× passive lysis buffer (Promega). A luciferase assay was performed according to the manufacturer's protocol using a Lumat LB 9501 luminometer (Wallac Inc., CA). At least three independent determinations were performed at each setting.
RESULTS
Identification of a cellular cofactor of K-Rta.
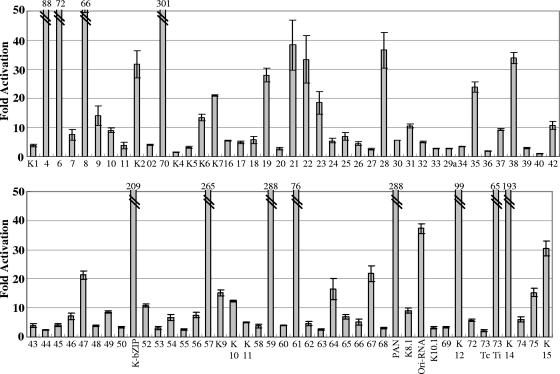
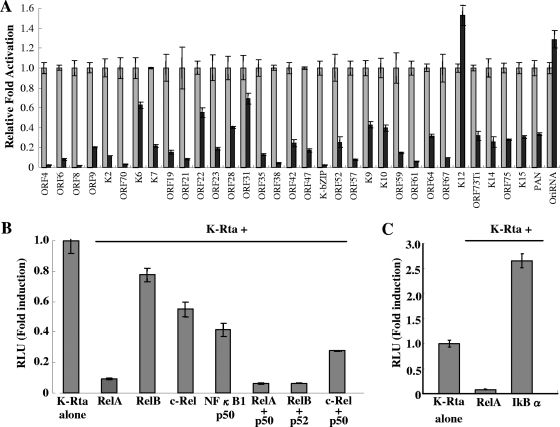
A functional approach, based on transient-expression assays of test promoters in reporter plasmids, was used to identify potential K-Rta target promoters. Reporter plasmids, each containing one of the 83 KSHV promoters, were cotransfected with the K-Rta expression plasmid in 293 cells (Fig. 1A). Thirty-four promoters were activated at least 10-fold. To implicate potential cofactors for K-Rta, these promoter sequences were analyzed with a weight matrix-based computer tool, Match, to search for putative transcriptional factor binding sites. This analysis revealed sites for several cellular transcription factors, including C/EBPα, AP-1, Oct-1, HNF4, and NF-κB/Rel. DNA binding motifs for these cellular proteins were frequently conserved in K-Rta-responsive viral promoters compared to nonresponsive promoters. Plasmid vectors designed to express these cellular transcription factors were prepared to investigate a potential coregulatory role in K-Rta-responsive promoters. In reporter assays, NF-κB was identified as a strong negative regulator of K-Rta (Fig. 2A). Importantly, NF-κB repressed K-Rta activity in over 90% of K-Rta-responsive promoters (Table 3). In particular, strong repression was observed for the early lytic gene promoters, such as K-bZIP, ORF70/K3, ORF59, and ORF57. Interestingly, the K12 and the Ori-RNA promoters were immune from this repression (Fig. 2A). Although many of the K-Rta-responsive promoters modulated by NF-κB contain NF-κB consensus motifs, others do not, suggesting multiple mechanisms involved in regulation. Alternatively, NF-κB may have a broader binding specificity than previously recognized (see below).
FIG. 1.
Identification of K-Rta-responsive KSHV promoters. Individual cultures of 293 cells were cotransfected with the K-Rta expression plasmid vector and a luciferase reporter plasmid containing 1 of each of 83 putative KSHV gene promoters. Fold activation over plasmid vector control is shown. Thirty-four KSHV promoters were activated over 10-fold.
FIG. 2.
NF-κB is a repressor of K-Rta. (A) 293 cells were cotransfected with K-Rta, K-Rta-responsive promoter, and NF-kB (RelA). Luciferase activity of K-Rta alone was normalized to a value of 1. (B) Effects of canonical and noncanonical pathways of NF-κB on K-Rta activity. 293 cells were cotransfected with different combinations of NF-κB proteins, K-Rta, and ORF57 promoter. (C) Enhancement of K-Rta activity by NF-κB inhibitor. 293 cells were cotransfected with K-Rta, ORF57 promoter, and the indicated plasmid. RelA significantly inhibits K-Rta transactivation, and IκBα enhances K-Rta activity. RLU, relative light units.
TABLE 3.
Inhibition of K-Rta-mediated gene expression by RelAa
| Promoter | Fold activation
|
|
|---|---|---|
| K-Rta + vector (% CV) | K-Rta + RelA (% CV) | |
| ORF4 | 88.3 (9.3) | 2.3 (21.7) |
| ORF6 | 72.3 (17.8) | 6.0 (13.2) |
| ORF8 | 66.4 (15.3) | 1.4 (3.5) |
| ORF9 | 14.0 (12.6) | 2.8 (3.3) |
| K2 | 31.7 (14.6) | 3.8 (1.8) |
| ORF70 | 301.8 (11.5) | 9.0 (17.8) |
| K6 | 13.4 (8.0) | 8.4 (5.4) |
| K7 | 21.0 (0.9) | 4.6 (6.0) |
| ORF19 | 28.0 (8.2) | 4.3 (13.9) |
| ORF21 | 38.3 (22.6) | 3.2 (8.9) |
| ORF22 | 33.4 (24.6) | 18.3 (8.9) |
| ORF23 | 18.5 (21.2) | 3.5 (6.3) |
| ORF28 | 36.6 (16.8) | 14.7 (2.5) |
| ORF31 | 10.5 (6.3) | 7.2 (8.2) |
| ORF35 | 23.9 (7.0) | 3.1 (6.9) |
| ORF38 | 33.9 (5.6) | 1.4 (21.1) |
| ORF42 | 10.7 (12.5) | 2.6 (14.0) |
| ORF47 | 21.3 (6.3) | 3.7 (9.1) |
| K8 (K-bzip) | 209.5 (8.1) | 5.3 (6.8) |
| ORF52 | 10.8 (4.1) | 2.7 (19.4) |
| ORF57 | 265.1 (8.4) | 20.4 (8.9) |
| K9 | 15.2 (6.9) | 6.5 (7.4) |
| K10 | 12.5 (2.0) | 5.0 (7.5) |
| ORF59 | 288.3 (9.3) | 43.0 (5.2) |
| ORF61 | 76.00 (18.5) | 4.6 (7.1) |
| ORF64 | 16.4 (22.1) | 5.2 (5.0) |
| ORF67 | 21.8 (12.1) | 2.1 (0.5) |
| K12 | 99.2 (3.4) | 151.4 (6.6 |
| ORF73Ti | 65.6 (10.5) | 20.7 (13.9) |
| K14 | 193.7 (16.2) | 49.5 (19.4) |
| ORF75 | 15.3 (9.6) | 4.3 (1.8) |
| K15 | 30.4 (8.4) | 9.3 (4.8) |
| PAN | 288.9 (18.4) | 96.9 (4.7) |
| OriRNA | 37.3 (4.5) | 48.1 (6.7) |
The fold activation over the vector-transfected luciferase value is shown. The covariances (as percentages) of triplicate wells are also shown.
Canonical and noncanonical forms of NF-κB inhibit K-Rta-mediated gene expression.
NF-κB functions as a hetero- or homodimer which can be produced from the five NF-κB subunits: NF-κB1 (p50 and its precursor p105), NF-κB2 (p52 and its precursor p100), RelA (p65), RelB, and c-Rel. In response to diverse cell stimuli, NF-κB is rapidly activated through either the canonical or noncanonical pathway (50). For this study, the form(s) of NF-κB that represses K-Rta was determined. The well-studied ORF57 promoter was used as a model for this assay. Cultures of 293 cells were cotransfected with the K-Rta expression plasmid and different combinations of plasmids expressing the five NF-κB subunits. This experiment showed that both the canonical (RelA/p50) and noncanonical (RelB/p52) forms repressed K-Rta efficiently (Fig. 2B). Transfection of RelA alone also resulted in the repression of K-Rta, which is likely due to the high level of endogenous p50 in 293 cells as well as physical association with RBP-Jκ (see below). Importantly, similar results were also observed for K-bZIP and ORF8 promoters (data not shown). If the NF-κB pathway specifically represses K-Rta-mediated gene expression, a prediction is that inhibition of this pathway might enhance K-Rta activation. Accordingly, a plasmid expressing the NF-κB inhibitor, IκBα, was cotransfected with both the K-Rta expression plasmid and the ORF57 promoter reporter plasmid. Indeed, IκBα enhanced K-Rta-mediated gene expression over twofold. In contrast to IκBα, RelA strongly inhibited K-Rta-mediated gene expression (Fig. 2C). These results demonstrate that NF-κB is a strong repressor of K-Rta, and importantly, most of the K-Rta-responsive promoters are repressed by NF-κB.
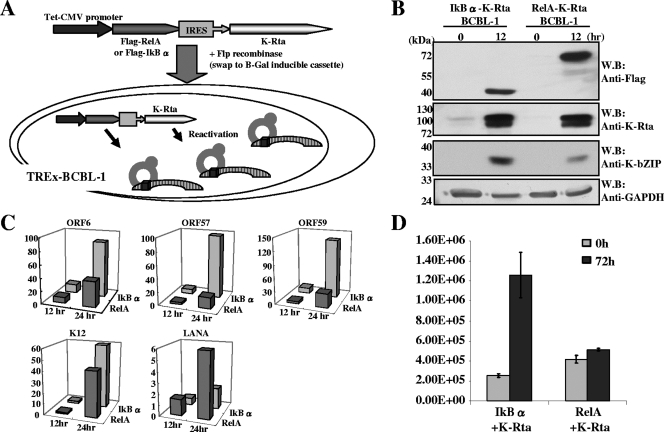
NF-κB inhibits KSHV reactivation.
NF-κB inhibits K-Rta-mediated gene expression in over 90% of K-Rta-responsive promoters in 293 cells. This finding prompted an analysis of whether NF-κB is a negative regulator of KSHV reactivation, because K-Rta is the trigger of lytic replication. Previous work by Brown et al. and Grossmann et al. showed that NF-κB inhibitors trigger the reactivation of several herpesvirus latent genomes, in a cell context-dependent manner (1, 15). In this study, the BCBL-1 cell line, harboring latent KSHV, was utilized to prepare derivative cell lines for dual induction of K-Rta and either NF-κB or IκBα (Fig. 3A). This approach obviated the complications of using chemicals (i.e., phorbol ester or sodium butylate) to induce reactivation, because such treatments also influence many cell signaling cascades, including the NF-κB pathway (50, 57). For induction, cells were treated with doxycycline for 4 hours to trigger lytic viral replication. Four hours of induction was confirmed to be sufficient for producing lytic reactivation, as verified by measuring levels of expression of the late viral K8.1 protein at 72 h after induction (Fig. 4A). Importantly, this approach, based on a short induction period, also avoids accumulation of K-Rta protein, which may produce nonspecific binding. Twelve hours after reactivation, one of the K-Rta targets, K-bZIP, was probed with specific antibody. Consistent with the reporter assay, the level of K-bZIP expression was significantly impaired in RelA-K-Rta-inducible cells compared to IκBα-K-Rta-inducible cells (Fig. 3B). Furthermore, transcript levels of five other K-Rta target genes were measured by qPCR (Fig. 3C). Early lytic genes were chosen to avoid the potential complications associated with viral DNA replication. As before, RelA significantly inhibited K-Rta-mediated gene expression. Consistent with the reporter assay data, K12 gene expression was relatively unaffected compared to other viral genes. Importantly, the expression of LANA, a latent gene whose expression is suppressed by K-Rta, is affected by RelA and IκBα in an opposite way. Finally, KSHV replication was analyzed by measuring viral DNA copy numbers in culture supernatant at 96 h after reactivation. Supernatant was collected, pretreated with DNase, and measured for encapsidated KSHV DNA molecules. Overexpression of NF-κB significantly reduced levels of virions accumulated in the culture supernatant. These results indicate that NF-κB is a strong, general repressor of lytic viral replication.
FIG. 3.
NF-κB inhibits KSHV reactivation. (A) Schematic diagram of the dual inducible cell lines. The cassette (for dual induction of IκBα and K-Rta or RelA and K-Rta) was transduced into TREx-BCBL-1 cells by cotransfecting with Flp recombinase expression vector. This cassette integrates at the identical site because of the design of the recombination system. Therefore, the copy number and the transcription levels remain the same. By adding doxycycline, K-Rta triggers reactivation, and the effects of NF-κB on KSHV reactivation can be examined. (B) RelA inhibits K-bZIP expression. After 12 h of induction, total cell protein was prepared and the indicated proteins were probed with specific antibodies. RelA reduced K-bZIP expression. W.B., Western blot. (C) RelA inhibits KSHV gene expression. Total RNA was prepared at 0, 12, and 24 h after induction. Viral gene expression was analyzed by measuring levels of transcripts by real-time qPCR. Cellular actin gene expression was used as an internal control. All values are expressed as the fold increase over the 0-h (uninduced) time point. Triplicate wells were used for each time point, and at least two independent experiments were conducted. (D) RelA inhibits KSHV replication. Viral DNA copy number in culture supernatant, as a measure of virion levels, was analyzed by real-time qPCR.
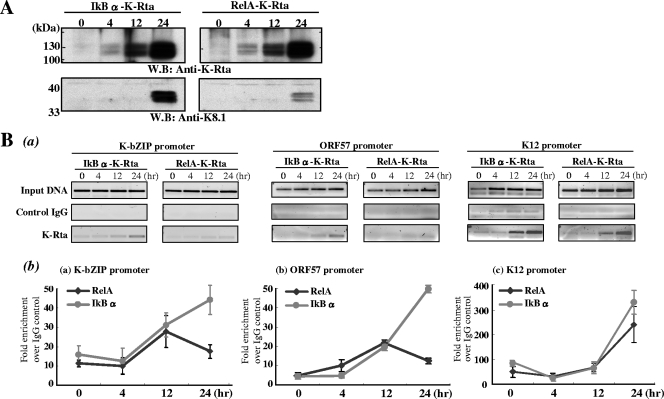
FIG. 4.
NF-κB inhibits K-Rta recruitment to the promoter. (A) Confirmation of KSHV reactivation. Total cell lysates were prepared at the indicated time points. For SDS-PAGE and immunoprobing, 20 μm of lysate per lane was used to test for the anti-K-Rta antibody, and 100 μg per lane was used for the K8.1 antibody. Comparable amounts of K-Rta expression were confirmed, and induction of KSHV lytic replication was assessed by measuring K8.1 expression. W.B., Western blot. (B) Time course analysis of K-Rta recruitment to selected KSHV promoters during reactivation. (a) Low-cycle PCR analysis was performed with ChIP DNA. (b) All values are expressed as fold increase over IgG control pull-down results, assayed by real-time PCR at 4, 12, and 24 h after doxycycline treatment. Each sample was tested in triplicate, and average values are shown in the figure. The ChIP elutions were assayed directly by real-time PCR after a 1:50 dilution. Two independent experiments were performed with similar results.
NF-κB inhibits K-Rta recruitment to viral promoters.
K-Rta activates viral promoters through at least two different mechanisms: (i) direct binding to the promoters and (ii) indirect recruitment by other cellular factors such as RBP-Jκ (36, 37). Previous studies showed that both the Ori-RNA and K12 promoters, which are not repressed by NF-κB, are the major binding targets of K-Rta, presumably through direct binding of K-Rta to sequence targets in these promoters (Ellison and Izumiya, unpublished). Close inspection of viral promoters regulated by K-Rta revealed that a core binding sequence of RBP-Jκ (5′-TGGGAA-3′) also partially overlapped the target site for NF-κB (5′-GGRNNTYCC-3′). In addition, the KSHV genome contains a large number of potential RBP-Jκ binding sites, which are not detected in the Match program. We therefore propose that NF-κB mediates K-Rta repression by antagonizing RBP-Jκ binding to target promoter. This can occur via competitive binding of the DNA target and/or formation of an inactive complex between NF-κB and RBP-Jκ. Thus, an experiment was designed to determine whether K-Rta recruitment to RBP-Jκ target promoters, such as ORF57 and K-bZIP, is inhibited by NF-κB expression. Cultures of dual inducible cell lines were treated with doxycycline for 4 hours and harvested at different time points. First, comparable amounts of K-Rta and induction of reactivation were confirmed by immunoblotting (Fig. 4A). Subsequently, ChIP was performed with highly purified anti-K-Rta antibody. The background (negative control) antibody was preimmune rabbit IgG from the same rabbit. ChIP DNAs from independent experiments were quantified by qPCR and low-cycle PCR (Fig. 4B). The results showed that recruitment of K-Rta to both the ORF57 and K-bZIP promoters was significantly inhibited in RelA-K-Rta-inducible cells, particularly at the 24-h time point. In contrast, K-Rta recruitment to the K12 promoter was relatively unaffected (Fig. 4B). These results demonstrate that NF-κB represses K-Rta through inhibition of K-Rta binding to target promoters. In addition, ChIP analysis with RelA antibody was performed. RelA was recruited to the ORF57 promoter in RelA-K-Rta-inducible cells but not IκB-K-Rta-inducible cells (data not shown).
NF-κB inhibits binding of RBP-Jκ to target DNA.
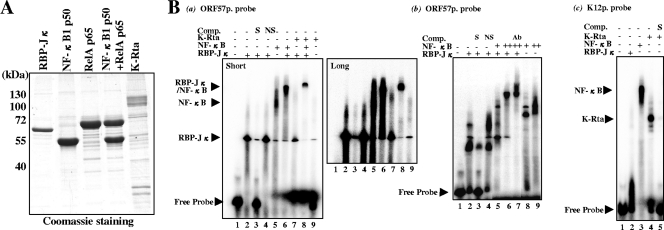
The previous ChIP analysis suggested that the mechanism of K-Rta repression may involve association between RBP-Jκ and NF-κB on the target viral promoter. To further examine this possibility, a gel mobility shift assay (i.e., EMSA) was performed with purified proteins. The RBP-Jκ, RelA, NF-κB1 p50, and K-Rta were expressed with recombinant baculoviruses and purified from infected insect cells by affinity to the Flag tag. Bound proteins were gently eluted by competition with 3× Flag peptide (Fig. 5A). Double-stranded oligonucleotides representing K-Rta-responsive elements from the ORF57 or K12 promoters were used as probes. As expected, RBP-Jκ bound the K-Rta-responsive elements of the ORF57 promoter (Fig. 5B, gel a, lane 2); the interaction was competed with the specific sequence competitor but not with the AP-1 binding sequence (i.e., a nonspecific competitor control) (Fig. 5B, gel a, lanes 3 and 4). Importantly, the RBP-Jκ band shifted in the presence of NF-κB; this shifted band with both proteins was different from the one with NF-κB alone, indicating a possible ternary complex (Fig. 5B, gel a, lanes 5 and 6). Interestingly, at a lower concentration of NF-κB (equimolar with RBP-Jκ), NF-κB directly forms a complex with the target sequence replacing RBP-Jκ (Fig. 5B, gel b, lane 5). At a higher concentration, NF-κB and RBP-Jκ form a ternary complex with the DNA probe (Fig. 5B, gel b, lane 6). Interaction between NF-κB and RBP-Jκ was confirmed by supershift with anti-RBP-Jκ antibody (Fig. 5B, gel b, lane 7). These results indicate that NF-κB is not only able to compete with RBP-Jκ for DNA binding but also to form a complex with RBP-Jκ on target DNA. Although the band intensity was very low, the slower-migrating band was observed when K-Rta was incubated with RBP-Jκ (Fig. 5B, gel a, lane 7). Importantly, incubation with NF-κB completely eliminated the band corresponding to the RBP-Jκ/K-Rta complex, indicating that NF-κB is able to perturb the formation of the K-Rta/RBP-Jκ-DNA complex. Binding of K-Rta alone to the ORF57 promoter was not observed even with an eightfold excess of K-Rta to the probe in this setting (Fig. 5B, gel a, lane 9). However, when the amount of K-Rta was further increased to a 20-fold excess of the probe, or when dI-dC was eliminated from the binding reaction mixture, a band shift was detected (data not shown); this result is likely due to nonspecific binding. In the same setting, K-Rta alone but not RBP-Jκ efficiently bound to the K12 probe (Fig. 5G, gel b, lanes 2 and 4), and this binding was effectively outcompeted by specific competitor bearing the target sequence (Fig. 5B, gel b, lane 5). These results provide evidence for two distinguishable mechanisms of K-Rta binding to the promoters. Taken together, the data support a model in which repression of K-Rta by NF-κB is caused by its ability to dislodge the active K-Rta/RBP-Jκ complex from the target promoter by either directly competing for DNA binding or by the formation of an inactive NF-κB/RBP-Jκ complex.
FIG. 5.
Competition of promoter binding between NF-κB and RBP-Jκ. (A) Purified proteins used in the gel shift assay. The RBP-Jκ, RelA, NF-κB1 (p50), and K-Rta proteins were produced in baculovirus expression systems. (B) NF-κB inhibits RBP-Jκ binding to the promoter. A gel shift assay was performed with the ORF57 promoter probe (gels a and b) or K12 promoter probe (gel c). The final concentration of RBP-Jκ was kept 100 nM (+). K-Rta was used at 400 nM, and NF-κB (RelA/p50 complex) was used at either a 100 nM (+) or 400 nM (++) final concentration. A supershift experiment was conducted with anti-RBP-Jκ andtibody (Ab). Specific (S) or nonspecific (NS; AP-1) competitor was used at a 100-fold excess of the labeled probe. Short, short exposure; long, long exposure.
NF-κB physically associates with RBP-Jκ.
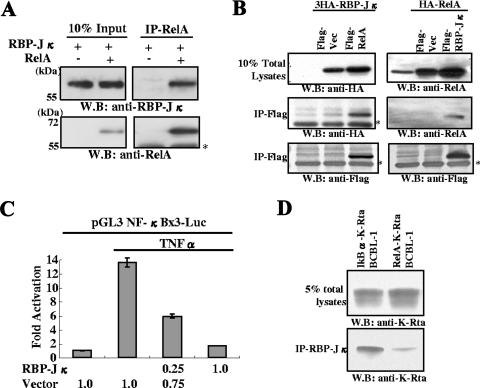
Analysis by EMSA suggested a physical interaction between NF-κB and RBP-Jκ. Accordingly, the interaction between NF-κB and RBP-Jκ was further examined. Purified forms of these two proteins were incubated in binding buffer, and the interaction was analyzed by immunoprecipitation with specific antibody. The results demonstrated direct association between RelA and RBP-Jκ (Fig. 6A). The interaction was further confirmed in vivo by the detection of NF-κB and RBP-Jκ complex in 293T cells. The Flag-tagged RelA was cotransfected with HA-tagged RBP-Jκ into 293T cells. Coprecipitation of RelA and RBP-Jκ was detected by immunoprecipitation with Flag antibody followed by Western blotting with HA antibody and vice versa (Fig. 6B). To determine the physiological relevance of this interaction, an NF-κB reporter plasmid was constructed, and the effect of RBP-Jκ on NF-κB-mediated gene expression was examined. The NF-κB reporter plasmid was cotransfected with RBP-Jκ expression plasmid, and NF-κB was activated by adding TNF-α to the culture medium. RBP-Jκ strongly inhibited NF-κB-mediated gene expression in a dose-dependent manner. This result shows that RBP-Jκ and NF-κB antagonize each other, and RBP-Jκ may have significant effects on NF-κB-mediated cellular gene expression. Finally, the effect of NF-κB on the RBP-Jκ/K-Rta complex was examined by coimmunoprecipitation. After induction of RelA-K-Rta or IκBα-K-Rta expression in BCBL-1, endogenous RBP-Jκ protein was immunoprecipitated with specific antibody and the amount of coprecipitated K-Rta was measured by immunoblotting. The result demonstrated that expression of RelA reduced the amount of K-Rta associated with RBP-Jκ, suggesting NF-κB may compete against K-Rta for binding to RBP-Jκ.
FIG. 6.
RBP-Jκ interactions with Rel-A and functional consequences. (A) Association between RelA and RBP-Jκ. Purified RelA (0.1 μg) and RBP-Jκ (0.1 μg) were mixed in binding buffer, incubated at 4°C, and immunoprecipitated with anti-RelA antibody. The immunoprecipitate was probed with the indicated antibodies. The asterisk shows the position of the IgG heavy chain. W.B., Western blot. (B) Association between RelA and RBP-Jκ in transfected 293T cells. The indicated plasmids were cotransfected into 293T cells. Total cell lysates were immunoprecipitated with agarose beads conjugated with anti-Flag antibody. Immunoprecipitates were probed with the indicated antibodies. Endogenous RelA was also detected with anti-RelA rabbit IgG (right upper panel). (C) Suppression of NF-κB activation by RBP-Jκ. The NF-κB reporter was cotransfected with the RBP-Jκ expression plasmid. NF-κB was activated by treating the cultures with TNF-α at 24 h posttransfection. Forty-eight hours after transfection, cells were harvested, and levels of luciferase were determined. Fold activation over the control transfected value is shown. The amount of plasmid used is indicated at the bottom of the panel. (D) RelA expression reduced K-Rta binding to RBP-Jκ. Dual expression of IκBα and K-Rta or RelA and K-Rta was induced in BCBL-1 by adding doxycycline. After 12 h of induction, cells were harvested and subjected to immunoprecipitation with anti-RBP-Jκ antibody. Coimmunoprecipitated K-Rta was measured by immunoblotting with anti-K-Rta antibody. Due to similar molecular masses, the presence of rabbit IgG heavy chain obscured the detection of the immunoprecipitated RBP-Jκ.
DISCUSSION
KS is a multifocal angioproliferative neoplasm characterized by inflammation, edema, neoangiogenesis, and spindle cell proliferation. These pathological effects are associated with NF-κB activation. Inflammation associated with KSHV infection could contribute to maintenance of the latent state by inhibiting the activity of the viral “switch protein,” K-Rta, which plays the central role in viral reactivation and lytic replication. It has previously been reported that NF-κB negatively regulates K-Rta transactivation potential (1), but the detailed mechanism was not pursued in that study. A KSHV genome-wide search revealed that transcription of 32 K-Rta-responsive viral genes was repressed by NF-κB. This finding set the stage for analysis of the mechanism by which NF-κB inhibits K-Rta-mediated expression of viral genes. Because of this broad repression involving promoters of many viral genes, we first considered a sequestration model, in which overexpression of NF-κB removes K-Rta from its active pool or a cellular coactivator(s) from K-Rta. Both K-Rta and NF-κB have been shown to interact with coactivators, including CBP and p300 (13, 19). This model was tested by overexpression of CBP in a transient-expression reporter assay; however, CBP did not rescue K-Rta transactivation (data not shown). Second, we examined the physical interaction between K-Rta and RelA by cotransfection in 293T cells; this experiment also did not provide sufficient evidence for this interaction. Based on these negative results, we proposed that K-Rta inhibition by NF-κB is due to competition for DNA binding between NF-κB and a cellular factor(s) which K-Rta utilizes to bind to viral promoters. This idea was supported by our earlier findings, in which two promoters immune from NF-κB-mediated repression were major targets for direct binding by K-Rta during reactivation (11a). Interestingly, repression was also observed in viral promoters that lack the typical NF-κB consensus motif, 5′-GGRNNTYCC-3′. However, this was partly explained by a recent observation that the majority of RelA-bound loci in the human genome do not contain the RelA binding motif (38, 44). In fact, neither the ORF57 promoter nor the K-bZIP promoter contains a consensus target sequence for NF-κB. Nonetheless, we demonstrated NF-κB binding to the promoter sequence by both gel shift and ChIP assay, suggesting that NF-κB may have a previously unrecognized, broader specificity of DNA binding.
If NF-κB competes DNA binding with other cellular factors, what is the target cellular transcriptional factor? K-Rta associates with a large number of transcription regulators, including RBP-Jκ, C/EBPα, Oct-1, STAT3, SWI/SNF, CBP, and TRAP/Mediator (2, 18-20, 22, 36). Close analysis revealed that the RBP-Jκ binding core sequence (5′-TGGGAA-3′) was conserved at 113 sites in the KSHV genome. This sequence is contained in 37 of 83 KSHV promoters and in 22 of 34 K-Rta-responsive promoters. The genome of this virus contains 20 sites that exactly match the RBP-Jκ consensus site (5′-CGTGGGAA-3′), and 11 of these sites are located in K-Rta-responsive promoter regions. Furthermore, this analysis even excluded the K-bZIP promoter, which contains a similar sequence (5′-GTGAGAA-3′); this sequence also binds RBP-Jκ (60). Considering the large number of possible RBP-Jκ binding sites on the viral genome, this cellular protein is likely a key regulatory factor for most of the K-Rta-mediated transactivation events. Therefore, we examined interactions of RBP-Jκ and NF-κB. Importantly, a coregulatory role between RBP-Jκ and NF-κB has been established for several cellular promoters that contain overlapping DNA recognition motifs for these two proteins (28, 35, 47). Furthermore, a functional interplay between Notch and NF-κB signaling pathways has been established. For example, Notch can interact with NF-κB1 p50, thus modulating NF-κB-dependent gene expression (17, 58). Conversely, NF-κB synergizes with Notch to activate Notch target gene expression by cytoplasmic translocation of the transcriptional corepressor, N-CoR (12). These findings indicate that NF-κB influences Notch signaling (RBP-Jκ protein complex) both directly and indirectly. Our results demonstrated that NF-κB affects K-Rta recruitment through inhibition of RBP-Jκ binding to the viral promoters. Interestingly, we also observed physical interaction between RelA and RBP-Jκ protein in vitro and in vivo. We further demonstrated that the interaction sequesters K-Rta from the RBP-Jκ protein. Thus, there are at least two mechanisms for inhibition of K-Rta by NF-κB: (i) competition between RBP-Jκ and NF-κB for binding the DNA target and (ii) sequestration of K-Rta from RBP-Jκ. These mechanisms are not mutually exclusive and may depend on the relative amount of NF-κB expressed (Fig. 5). Our results are also consistent with a previous study, which demonstrated that both the Rel homology domain and DNA binding domain of RelA are important for K-Rta inhibition (1). Although ChIP analyses were performed with RBP-Jκ antibodies to demonstrate competition of DNA binding to KSHV promoters in vivo, we could not detect RBP-Jκ binding on KSHV promoters, even at the 0-h time point in the induction period. This is likely due to the low affinity of the antibodies to RBP-Jκ, which are unable to precipitate chromatin-associated RBP-Jκ.
The viral K12 promoter is not subject to NF-κB negative regulation, and its transcript is the most abundant transcript expressed in latent KSHV infection; this transcript is also induced during lytic replication. The K12 transcript produces as many as three variants of the viral kaposin protein (53), as well as a microRNA (49), which has been linked to the tumorigenic potential of KSHV. Interestingly, one of the K12 products, kaposin B, was shown to increase the expression of cytokines (45); thus, it is possible that K12 may activate NF-κB by an indirect mechanism. This suggests that the expression of K12 may initiate viral latency by providing a steady supply of NF-κB without feedback regulation. Our study also showed that NF-κB activates expression of latent viral genes (i.e., LANA, v-FLIP, and v-Cyclin) (Fig. 3C). Such activation of LANA could reinforce the negative feedback loop which downmodulates K-Rta expression; in addition, activation of NF-κB through v-FLIP expression may further contribute to inhibition of K-Rta (16, 32, 33, 55). Interestingly, expression of the Epstein-Barr virus latent protein, LMP1, is also upregulated by NF-κB activation (27), indicating a conserved mechanism of latent gene activation for gammaherpesviruses. The most important question is why KSHV suppresses its own lytic replication. We suggest that maintaining latency might be the strategy for this virus to survive in the infected host. A similar conclusion was reached by other investigators who showed that KSHV immediately establishes latency after initial infection without expressing the majority of lytic genes, except a few genes that are associated with immunomodulatory functions (31).
In summary, we propose that the molecular switch of KSHV, between latency and lytic replication, is rooted in the balance between the host cell factor, NF-κB, and the viral factor, K-Rta. In addition, we have defined the mechanism of NF-κB-mediated repression of K-Rta and shown that this repression is mediated through an interaction between NF-κB and a coactivator RBP-Jκ. In our model, the ratio of these two transcription regulatory proteins will determine the fate of virus in the infected individual. Because maintaining latency is one of the most effective strategies for herpesviruses to hide from the host immune system, disrupting the balance of NF-κB and K-Rta activity could be the basis for an effective antiviral therapeutic strategy. Thus, NF-κB has emerged as a key sensor and regulator of both KSHV replication and host immune responses to this virus.
Acknowledgments
This work was supported by grants from the California HIV/AIDS Research Program (ID07-D-165 to Y.I.) and the National Institutes of Health (CA111185 to H.-J.K.). Additional funding was provided by the Cancer Center Support Grant (P30 CA93373-04 to Y.I.). This research was also supported by NCRR grant UL-RR024146, which funds programs of the NIH Roadmap for Medical Research.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Brown, H. J., M. J. Song, H. Deng, T. T. Wu, G. Cheng, and R. Sun. 2003. NF-κB inhibits gammaherpesvirus lytic replication. J. Virol. 778532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll, K. D., F. Khadim, S. Spadavecchia, D. Palmeri, and D. M. Lukac. 2007. Direct interactions of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation. J. Virol. 818451-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caselli, E., S. Fiorentini, C. Amici, D. Di Luca, A. Caruso, and M. G. Santoro. 2007. Human herpesvirus 8 acute infection of endothelial cells induces monocyte chemoattractant protein 1-dependent capillary-like structure formation: role of the IKK/NF-κB pathway. Blood 1092718-2726. [DOI] [PubMed] [Google Scholar]
- 4.Chang, H., D. P. Dittmer, Y. C. Shin, Y. Hong, and J. U. Jung. 2005. Role of Notch signal transduction in Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 7914371-14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, H., Y. Gwack, D. Kingston, J. Souvlis, X. Liang, R. E. Means, E. Cesarman, L. Hutt-Fletcher, and J. U. Jung. 2005. Activation of CD21 and CD23 gene expression by Kaposi's sarcoma-associated herpesvirus RTA. J. Virol. 794651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, M., H. J. Brown, A. Collado-Hidalgo, J. M. Arevalo, Z. Galic, T. L. Symensma, L. Tanaka, H. Deng, J. A. Zack, R. Sun, and S. W. Cole. 2005. β-Adrenoreceptors reactivate Kaposi's sarcoma-associated herpesvirus lytic replication via PKA-dependent control of viral RTA. J. Virol. 7913538-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 763168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chatlynne, L. G., and D. V. Ablashi. 1999. Seroepidemiology of Kaposi's sarcoma-associated herpesvirus (KSHV). Semin. Cancer Biol. 9175-185. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, A., C. Brodie, and R. Sarid. 2006. An essential role of ERK signalling in TPA-induced reactivation of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 87795-802. [DOI] [PubMed] [Google Scholar]
- 11.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 768252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Ellison, T. J., Y. Izumiya, C. Izumiya, P. A. Luciw, and H.-J. Kung. 6 March 2009, posting date. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi's sarcoma-associated herpesvirus. Virology. doi: 10.1016/j.virol.2009.02.016. [DOI] [PMC free article] [PubMed]
- 12.Espinosa, L., S. Santos, J. Ingles-Esteve, P. Munoz-Canoves, and A. Bigas. 2002. p65-NFκB synergizes with Notch to activate transcription by triggering cytoplasmic translocation of the nuclear receptor corepressor N-CoR. J. Cell Sci. 1151295-1303. [DOI] [PubMed] [Google Scholar]
- 13.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 942927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 746207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossmann, C., and D. Ganem. 2008. Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology 37594-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossmann, C., S. Podgrabinska, M. Skobe, and D. Ganem. 2006. Activation of NF-κB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J. Virol. 807179-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, E., J. Wang, J. Laborda, M. Norcross, P. A. Baeuerle, and T. Hoffman. 1996. T cell leukemia-associated human Notch/translocation-associated Notch homologue has I kappa B-like activity and physically interacts with nuclear factor-kappa B proteins in T cells. J. Exp. Med. 1832025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 232055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 751909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 2776438-6442. [DOI] [PubMed] [Google Scholar]
- 21.Hayden, M. S., and S. Ghosh. 2008. Shared principles in NF-κB signaling. Cell 132344-362. [DOI] [PubMed] [Google Scholar]
- 22.Hayward, G. S. 2003. Initiation of angiogenic Kaposi's sarcoma lesions. Cancer Cell 31-3. [DOI] [PubMed] [Google Scholar]
- 23.Honjo, T. 1996. The shortest path from the surface to the nucleus: RBP-Jκ/Su(H) transcription factor. Genes Cells 11-9. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumiya, Y., C. Izumiya, A. Van Geelen, D. H. Wang, K. S. Lam, P. A. Luciw, and H. J. Kung. 2007. Kaposi's sarcoma-associated herpesvirus-encoded protein kinase and its interaction with K-bZIP. J. Virol. 811072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 771441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson, P., A. Jansson, U. Ruetschi, and L. Rymo. 2009. Nuclear factor-κB binds to the Epstein-Barr Virus LMP1 promoter and upregulates its expression. J. Virol. 831393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kannabiran, C., X. Zeng, and L. D. Vales. 1997. The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol. Cell. Biol. 171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 122269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovall, R. A., and W. A. Hendrickson. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 233441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 783601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 793468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 786585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan, K., M. Murakami, T. Choudhuri, D. A. Kuppers, and E. S. Robertson. 2006. Intracellular-activated Notch1 can reactivate Kaposi's sarcoma-associated herpesvirus from latency. Virology 351393-403. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. H., X. Wang, and J. DeJong. 2000. Functional interactions between an atypical NF-κB site from the rat CYP2B1 promoter and the transcriptional repressor RBP-Jκ/CBF1. Nucleic Acids Res. 282091-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 161977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. USA 1008490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, C. A., F. Yao, J. J. Wong, J. George, H. Xu, K. P. Chiu, W. K. Sung, L. Lipovich, V. B. Vega, J. Chen, A. Shahab, X. D. Zhao, M. Hibberd, C. L. Wei, B. Lim, H. H. Ng, Y. Ruan, and K. C. Chin. 2007. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol. Cell 27622-635. [DOI] [PubMed] [Google Scholar]
- 39.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 731909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 7711425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 739348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252304-312. [DOI] [PubMed] [Google Scholar]
- 43.Martin, D., R. Galisteo, Y. Ji, S. Montaner, and J. S. Gutkind. 2008. An NF-κB gene expression signature contributes to Kaposi's sarcoma virus vGPCR-induced direct and paracrine neoplasia. Oncogene 271844-1852. [DOI] [PubMed] [Google Scholar]
- 44.Martone, R., G. Euskirchen, P. Bertone, S. Hartman, T. E. Royce, N. M. Luscombe, J. L. Rinn, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA 10012247-12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCormick, C., and D. Ganem. 2005. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 307739-741. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 774205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oswald, F., S. Liptay, G. Adler, and R. M. Schmid. 1998. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol. Cell. Biol. 182077-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagano, J. S., M. Blaser, M. A. Buendia, B. Damania, K. Khalili, N. Raab-Traub, and B. Roizman. 2004. Infectious agents and cancer: criteria for a causal relation. Semin. Cancer Biol. 14453-471. [DOI] [PubMed] [Google Scholar]
- 49.Pearce, M., S. Matsumura, and A. C. Wilson. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 7914457-14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins, N. D. 2007. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 849-62. [DOI] [PubMed] [Google Scholar]
- 51.Plaisance, S., W. Vanden Berghe, E. Boone, W. Fiers, and G. Haegeman. 1997. Recombination signal sequence binding protein Jκ is constitutively bound to the NF-κB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol. Cell. Biol. 173733-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prakash, O., O. R. Swamy, X. Peng, Z. Y. Tang, L. Li, J. E. Larson, J. C. Cohen, J. Gill, G. Farr, S. Wang, and F. Samaniego. 2005. Activation of Src kinase Lyn by the Kaposi sarcoma-associated herpesvirus K1 protein: implications for lymphomagenesis. Blood 1053987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 735722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 861276-1280. [PubMed] [Google Scholar]
- 55.Sun, Q., H. Matta, G. Lu, and P. M. Chaudhary. 2006. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene 252717-2726. [DOI] [PubMed] [Google Scholar]
- 56.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9510866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vertegaal, A. C., H. B. Kuiperij, S. Yamaoka, G. Courtois, A. J. van der Eb, and A. Zantema. 2000. Protein kinase C-alpha is an upstream activator of the IκB kinase complex in the TPA signal transduction pathway to NF-κB in U2OS cells. Cell Signal. 12759-768. [DOI] [PubMed] [Google Scholar]
- 58.Wang, J., L. Shelly, L. Miele, R. Boykins, M. A. Norcross, and E. Guan. 2001. Human Notch-1 inhibits NF-kappa B activity in the nucleus through a direct interaction involving a novel domain. J. Immunol. 167289-295. [DOI] [PubMed] [Google Scholar]
- 59.Wang, L., M. M. Brinkmann, M. Pietrek, M. Ottinger, O. Dittrich-Breiholz, M. Kracht, and T. F. Schulz. 2007. Functional characterization of the M-type K15-encoded membrane protein of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 881698-1707. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jκ in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 35919-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson, J. J., and R. A. Kovall. 2006. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124985-996. [DOI] [PubMed] [Google Scholar]
- 62.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 793479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye, F. C., F. C. Zhou, J. P. Xie, T. Kang, W. Greene, K. Kuhne, X. F. Lei, Q. H. Li, and S. J. Gao. 2008. Kaposi's sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-κB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 824235-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]