Abstract
Swine influenza was first recognized as a disease entity during the 1918 “Spanish flu” pandemic. The aim of this work was to determine the virulence of a plasmid-derived human 1918 pandemic H1N1 influenza virus (reconstructed 1918, or 1918/rec, virus) in swine using a plasmid-derived A/swine/Iowa/15/1930 H1N1 virus (1930/rec virus), representing the first isolated influenza virus, as a reference. Four-week-old piglets were inoculated intratracheally with either the 1930/rec or the 1918/rec virus or intranasally with the 1918/rec virus. A transient increase in temperature and mild respiratory signs developed postinoculation in all virus-inoculated groups. In contrast to other mammalian hosts (mice, ferrets, and macaques) where infection with the 1918/rec virus was lethal, the pigs did not develop severe respiratory distress or become moribund. Virus titers in the lower respiratory tract as well as macro- and microscopic lesions at 3 and 5 days postinfection (dpi) were comparable between the 1930/rec and 1918/rec virus-inoculated animals. In contrast to the 1930/rec virus-infected animals, at 7 dpi prominent lung lesions were present in only the 1918/rec virus-infected animals, and all the piglets developed antibodies at 7 dpi. Presented data support the hypothesis that the 1918 pandemic influenza virus was able to infect and replicate in swine, causing a respiratory disease, and that the virus was likely introduced into the pig population during the 1918 pandemic, resulting in the current lineage of the classical H1N1 swine influenza viruses.
As the 1918 “Spanish flu” pandemic spread through the central United States, a swine respiratory disease was concurrently observed in this region. The swine disease was highly contagious; it had high morbidity with fever, anorexia, dyspnea, cough, and prostration, with sudden onset and fast recovery within 2 to 6 days after first clinical signs, and low mortality (between 1 and 4%). Due to a strong resemblance of the clinical signs to the human influenza disease, a clinical name of “hog flu” was given by J. S. Koen to this new disease of pigs (15, 27). Similar swine respiratory diseases suspected to be influenza were reported at about the same time in Europe and China (2).
Following the human pandemic, hog flu or, in today's terminology, swine influenza was reported intermittently in the Midwest of the United States. In 1930 swine H1N1 influenza virus (A/swine/Iowa/14 and A/swine/Iowa/15/1930) was isolated from diseased pigs and was demonstrated to play a critical role in the disease although severity often depended on secondary bacterial infections (27, 28).
Early serological studies linked the first human H1N1 influenza virus isolates (e.g., PR8/1934) and, even more so, the swine H1N1 1930 virus isolate to the 1918 pandemic virus (5, 30). Laidlaw (15) suggested that the swine influenza virus could be the 1918 pandemic influenza virus which became established in pigs. Recent phylogenetic analyses of the 1930 swine flu virus, the first human H1N1 influenza virus isolates, the classical H1N1 swine influenza viruses, and the reconstructed 1918 human influenza virus (1918/rec virus) (37) strongly support the originally proposed hypothesis as all these viruses appear to be derived from a common source, the 1918 pandemic virus (7, 34, 41). Interestingly, the 1930 swine influenza virus may still be circulating in swine (1). Although the origin of the 1918 virus is not known, it has been suggested that the virus came from an avian reservoir and either entered the human population directly or indirectly through an intermediate host (34).
Swine have been proposed as an intermediate host in the indirect transmission of influenza A viruses from an avian reservoir to humans, based on the unique distribution in pigs of 2,3- and 2,6-linked sialic acid moieties that are considered to be avian- and human-specific receptors for influenza A viruses, respectively. The presence of the avian and human receptors in the swine respiratory tract can enable the pigs to become infected with either avian or human influenza A viruses, setting the stage for reassortant events between swine, avian, and human viruses or for adaptation of an avian virus to a mammalian receptor (20). Support for this hypothesis can be found in the isolation of entirely avian or human viruses from swine, as well as reassortant viruses that contain swine, human, and avian genes (2, 13, 22, 42, 43). Reports also document interspecies transmission from pigs to people (21, 42). However, even though the 1957 and 1968 human pandemic viruses were human-avian reassortants, there is no evidence that the reassortment occurred in pigs. This classical theory was based on contemporary knowledge of receptor distribution in different host species, and it has been recently questioned, particularly in light of the proven direct transmission of avian influenza virus to humans (40).
Recently, the 1918/rec virus (14, 37) was demonstrated to be highly pathogenic in mice (37), ferrets (38), and nonhuman primates (14). This report describes experimental infection of swine with the 1918 influenza virus with the hemagglutinin of the 1918 South Carolina influenza virus isolate in comparison to the H1N1 swine influenza virus isolate from the year 1930. The A/swine/Iowa/15/1930 was chosen as a reference virus because it is thought to be a descendant of the 1918 pandemic influenza virus. The aim of this work was to gain some insight into influenza infections of swine during the 1918 influenza pandemic by determining whether the 1918 human influenza virus can infect and replicate in swine and cause clinical disease and lesions in the infected animals.
MATERIALS AND METHODS
All live virus work with the 1918/rec pandemic influenza virus was performed in the biosafety level 4 laboratory and animal cubicle at the National Centre for Foreign Animal Disease (NCFAD).
Viruses.
Viruses used in this study were rescued using reverse genetics. Plasmids for the influenza viruses were transfected into cocultures of MDCK and 293T cells using Lipofectamine 2000 (Invitrogen) as described by Schickli et al. (25). The generation of infectious influenza virus from the transfections was assessed by plaque assay of the culture supernatants on MDCK cells.
(i) H1N1 1918/rec virus.
Although the H1N1 1918/rec virus was previously rescued at the CDC (37), in order to perform the studies at NCFAD, the virus was rerescued using the same pPol-I rescue plasmids directing the synthesis of negative-sense (virion RNA) genomic-length transcripts of the eight genomic segments of the 1918 influenza virus and supporting pCAGGS expression plasmids encoding the A/WSN/33 polymerase subunits (PB1, PB2, and PA) and nucleocapsid protein NP. The rescue plasmid used for hemagglutinin (HA) encoded the HA gene of the 1918 South Carolina influenza virus isolate with a receptor binding specificity for α2,6-linked sialic acids (6, 37). Reverse transcription-PCR (RT-PCR) and sequencing confirmed that the genome of the rescued virus was identical in sequence to the cDNA in the plasmids used for its rescue.
(ii) H1N1 1930/rec virus (A/swine/Iowa/15/1930).
The plasmid pHW2000/pDZ (9) that directs the synthesis of negative-sense (virion RNA) and positive-sense (cRNA) genomic-length influenza virus transcripts was utilized to create a set of eight rescue plasmids for the A/swine/Iowa/15/1930 (H1N1) influenza virus (17). Novel restriction enzyme sites were introduced into each gene segment in order to differentiate the rescued 1930/rec virus from the wild-type virus.
Virus titers were determined by endpoint titration on monolayers of MDCK cells in 96-well microtiter plates (Costar; Corning). Briefly, 50 μl of 10-fold serial dilutions of virus samples in alpha-minimal essential medium-0.3% bovine serum albumin supplemented with 10 U/ml of TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (Sigma) was incubated on the cells for 1 h at 30°C in 5% CO2. Following the virus adsorption, an additional 50 μl of alpha-minimal essential medium-0.3% bovine serum albumin supplemented with 10 U/ml of TPCK-trypsin was added to each well, and the plates were incubated as described above for 5 days. The 50% tissue culture infective dose (TCID50) was determined using the method of Reed and Muench.
Animal inoculations. (i) Mice.
Female BALB/c mice, 6 to 7 weeks old (Charles River Laboratories, Wilmington, MA), were anesthetized with an intraperitoneal injection of 0.2 ml of 2,2,2-tribromoethanal in tert-amyl alcohol (Avertin; Aldrich Chemical Co., Milwaukee, WI.). Ten mice per each treatment group were inoculated intranasally with 50 μl/104 TCID50/animal of either 1918/rec or 1930/rec virus, or mock inoculated with phosphate-buffered saline (PBS). Individual body weights and clinical signs were recorded each day postinfection (dpi) up to 8 dpi. At the end of the experiment, surviving mice were euthanized by intraperitoneal injection of an overdose of ketamine hydrochloride (200 mg/kg). Whole lungs were collected for virus RNA detection.
(ii) Swine.
The pregnant sows and then the piglets were tested on the farm for antibodies against H3N2 and H1N1 by Idexx enzyme-linked immunosorbent assay to confirm the swine influenza virus-free status of the high-health status herd at Designed Genetics, Lockport, Manitoba (Canada). The 3-week-old piglets were retested upon arrival at the NCFAD for influenza virus-specific antibodies using the HerdCheck Swine Influenza Virus (H1N1) Antibody Test Kit (Idexx Laboratories) according to the manufacturer's instructions. The animals were acclimatized for 7 days and inoculated either intratracheally (1 ml) or intranasally (2 ml) with 105.4 TCID50 of 1918/rec or 1930/rec influenza virus/animal or mock inoculated with PBS. Six piglets were inoculated intratracheally, and 12 were inoculated intranasally with the 1918/rec virus (a total of 18 piglets). Twelve piglets were inoculated intratracheally with the same dose of the 1930/rec virus, and 12 control piglets were mock inoculated with PBS. Body temperatures and clinical signs were recorded daily. Piglets inoculated with the 1918/rec virus were sampled and euthanized at 3, 5, and 7 dpi or at 3, 5, 7, 12, and 17 dpi; 1930/rec virus- and PBS-inoculated pigs were euthanized and sampled at 3, 5, and 7 dpi. Blood/serum, oropharyngeal swabs, nasal swabs or washes, bronchoalveolar lavage fluid (BALF), and the following tissues were collected: heart, brain, digestive tract samples, kidney, urine, liver, muscles, tonsil, thymus, spleen, cerebrospinal fluid, feces, submandibular and bronchial lymph nodes, trachea, and lung. The experimental design is summarized in Table 1.
TABLE 1.
Experimental design: route of inoculation and animals euthanized
| Animal group and treatmenta | Route of inoculationb | Animal(s) (no.) euthanized at:
|
||||
|---|---|---|---|---|---|---|
| 3 dpi | 5 dpi | 7 dpi | 12 dpi | 17 dpi | ||
| A | ||||||
| Control | T | 73, 74 | 75, 76 | 77, 78 | ||
| 1930/rec | T | 66, 67 | 69, 71 | 70, 72 | ||
| 1918/rec | T | 88, 89 | 90, 91 | 92, 93 | ||
| B | ||||||
| Control | N | 79, 94 | 80, 95 | 81, 96 | ||
| 1930/rec | T | 82, 83 | 84, 85 | 86, 87 | ||
| 1918/rec | N | 97, 98 | 99, 100 | 101, 102 | ||
| 1918/rec | N | 103 | 104 | 105 | 106 | 107, 108 |
Due to space constraints in the biosafety level 4 facility, only 6 piglets could be housed in the animal cubicle at the same time, and the experiments had to be performed in series.
T, intratracheal; N, intranasal.
All animal manipulations were approved by the Animal Care Committee of the Canadian Science Centre for Human and Animal Health, according to the guidelines of the Canadian Council on Animal Care.
Gross pathology.
Pathological changes were evaluated in all major organs of all piglets at the time of necropsy. Photographic images of lungs were taken from most of the piglets. The extent of the macroscopic lesions on the lung surfaces was visually evaluated based on the photographic images or directly during the necropsy and is expressed as a percentage of the entire lung using the Ambico, Inc. Lung Score Sheet.
Histopathology and immunohistochemistry (IHC).
Tissue samples were fixed in 10% neutral phosphate-buffered formalin, routinely processed, and stained with hematoxylin and eosin (H&E) following standard protocols.
Formalin-fixed paraffin-embedded tissue sections were pretreated with proteolytic enzyme (DakoCytomation) for 15 min prior to immunostaining. Monoclonal antibody specific for the nucleoprotein of influenza A virus (ME Clone 1331; Biodesign Sasco) was used at a dilution of 1:5,000 to detect the viral antigen and visualized using the Envision anti-mouse (horseradish peroxidase labeled) polymer kit (DakoCytomation) reacted with the chromogen diaminobenzidine. Tissues were counterstained with Gill's hematoxylin. IHC slides were scored from 0 to 4+ as follows: 1+, occasional positive bronchiolar epithelial cells; 2+, <25% of bronchioles having positive cells; 3+ 25 to 75% of bronchioles having positive cells; 4+, >75% of bronchioles having positive cells.
Virus isolation.
Virus isolation on MDCK cells was attempted from BALF and from clarified 10% (wt/vol) lung homogenates in PBS as described above in the paragraph on virus titer determinations.
Nine-day-old embryonated eggs were inoculated via the allantoic cavity with 0.2 ml of undiluted clarified BALF and incubated until embryo death or up to 5 dpi.
Real-time RT-PCR.
Probe and primers targeting the matrix protein were used in the real-time RT-PCR detection of viral RNA (23, 31) on all samples listed in the paragraph on swine inoculations above. Viral RNA was extracted from samples according to the manufacturer's instructions using TriPure Isolation Reagent (Roche Applied Science, Indianapolis, IN).
Hemagglutination inhibition was used to characterize the rescued viruses and to determine serum antibodies in the infected piglets. The microtiter HA and hemagglutination inhibition (HI) assays were performed by standard protocols using a 0.5% suspension of chicken red blood cells in PBS. Sera serially diluted twofold were tested against 4 HA units of the respective H1N1 virus. Serum against influenza A/mallard/ON/629/2005 (H1N1) virus that cross-reacted with the influenza 1930/rec virus to a titer of 80 to 160 was used as a positive control.
The HA identities of the rescued 1918/rec and the 1930/rec viruses were verified with mouse monoclonal antibodies using the same assay format as above. The monoclonal antibody 5D3 recognized both 1918 and 1930 HA, and 58F4E10 recognized 1918 HA (unpublished data).
RESULTS
Infection of mice.
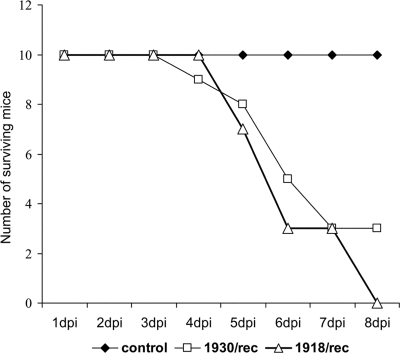
The virulence of the rescued viruses was initially confirmed in mice before inoculation of swine. Mice were inoculated intranasally with 104 TCID50/animal with either the 1918/rec or 1930/rec virus or mock inoculated with PBS. All mice inoculated with the 1918/rec virus died by 8 dpi, as expected based on the previous characterization of the 1918/rec virus (37). The 1930/rec virus was also highly virulent for mice. Seven out of 10 inoculated mice died by 6 dpi; the remaining animals survived until the end of the experiment (8 dpi) (Fig. 1). The three surviving mice continued losing weight, their respiratory rate remained elevated, and their fur was ruffled. Activity and water and food intake were decreased. The control PBS-inoculated mice did not exhibit clinical signs.
FIG. 1.
Lethality of the rescued viruses in mice, indicated by the number of surviving animals. Mice were inoculated intranasally with 104 TCID50 per animal of the 1930/rec or the 1918/rec virus. Controls were inoculated intranasally with PBS.
The lungs from the 1918/rec and the 1930/rec virus-infected mice collected at necropsy were severely hemorrhagic in appearance and friable while lungs from the control mice were normal. Virus replication in lungs of the infected mice was confirmed by real-time RT-PCR targeting the matrix gene. There was no significant difference in copy numbers between the 1918/rec and the 1930/rec virus-infected groups. Comparable threshold cycle numbers (approximately 23 to 24) indicated the presence of approximately 200,000 to 250,000 copies/g of the lung tissue (data not shown).
Infection of swine.
There were no significant differences found between the animals inoculated intratracheally or intranasally with the 1918/rec virus, and for the purposes of this report, the two groups are discussed as one.
(i) Clinical signs.
Clinical signs and rectal temperatures were recorded daily for all animals. Transient increase in body temperature and mild respiratory signs developed starting at 1 dpi in both influenza virus-inoculated groups but not in the control animals. Body temperatures above 39.9°C were considered above normal.
In both groups (1918/rec and 1930/rec virus infections), body temperatures above normal were observed postinoculation at 1 dpi and in some animals also between 4 to 6 dpi. Respiratory signs were observed in only the first group of piglets inoculated with the 1930/rec virus (animals no. 66 to 72) at 2 and at 5 dpi. Two pigs displayed abdominal breathing, and one of them was also sneezing at 2 dpi. None of the 1918/rec virus-inoculated animals, control animals, or the second group of the 1930/rec virus-inoculated animals displayed apparent clinical signs.
(ii) Gross pathology.
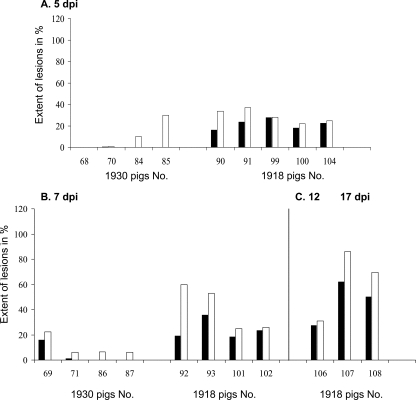
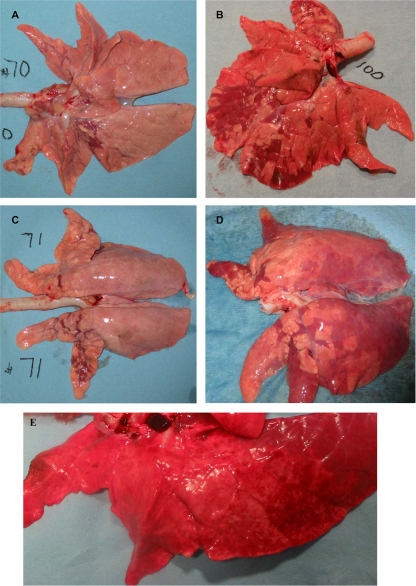
Gross lung lesions, accompanied by enlarged tracheobronchial lymph nodes, were observed in both influenza virus-infected groups. The extent of the macroscopic lesions was comparable between the 1930/rec and 1918/rec virus-inoculated animals at 3 dpi. Starting at 5 dpi, the lung lesions were more severe in the 1918/rec virus-inoculated piglets than in the 1930/rec virus-inoculated animals, and appeared to be progressively more widespread at later time points (Fig. 2). Initially, the lesions were mostly located in the apical lobes, with some involvement of the cardiac lobes. In general, the lesions can be described as irregularly distributed, plum-colored consolidated depressions, corresponding to individual lobules. At 7 dpi lobular septae of the 1918/rec virus-inoculated piglets were prominent, suggesting mild edema. Some piglets had reticulated, gray-white bulging foci in several lobules, suggesting areas of inflammation centered on bronchioles. (Fig. 3) At later time points, the lung lesions in piglets inoculated with the 1918/rec virus spread also to the diaphragmatic lobes; at 12 and 17 dpi petechia and extensive multiple small spider-like hemorrhages were noted on the surfaces of the lungs, and blood was observed in the larger lung airways (trachea and bronchi). At 17 dpi the lungs had uniform purple discoloration of the diaphragmatic lobes, suggesting hypostatic congestion.
FIG. 2.
Extent of gross lung lesions in the infected piglets. Lung lesions at 5 and 7 dpi in the 1930/rec and 1918/rec virus-infected animals and in the 1918/rec virus-inoculated pigs also at 12 (pig 106) and 17 (pigs 107 and 108) dpi. Total extent of the lung lesions (white columns) and the extent of the lesions in the diaphragmatic lobes (black columns) are given as the total lung surface percentage for individual animals. While the macroscopic lesions in the lungs of the 1930/rec virus-infected animals did not appear to be spreading, the lung lesions in the 1918/rec virus-infected animals were more extensive later times postinfection (after 7 dpi).
FIG. 3.
Gross lung lesions in the infected piglets. A few plum-colored, depressed lobules can be observed in the apical, cardiac, and anterior diaphragmatic lobes of piglets inoculated with the 1930/rec influenza virus. The extent of the lesions appeared to be the same at 3 and 5 dpi (Fig. A and B, respectively). The 1918/rec virus-inoculated piglets had more extensive lesions, and the plum-colored depressed lobules could be observed in the diaphragmatic and cardiac lobes at 5 dpi (C) and 7 dpi (D), while the lesions in 1930/rec virus-inoculated piglets remained localized in the apical lobes. At 17 dpi, plum-colored, depressed lobules were observed throughout the entire lungs of both 1918/rec virus-infected piglets although both lungs also had uniform purple discoloration of the dorsal portion of the diaphragmatic lobe, suggesting hypostatic congestion, likely developed during euthanasia. Panel E illustrates the spider-like/petechial hemorrhages present both on the ventral and dorsal side of lungs at 17 dpi.
(iii) Histopathology.
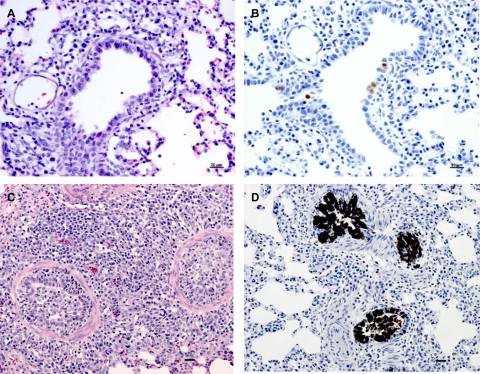
Microscopic lesions and presence of viral antigen in lungs of either the 1918/rec or 1930/rec virus-inoculated piglets corresponded with previous reports on swine influenza (10, 12, 22, 23, 24, 32). Lesions were centered on bronchioles and followed a lobular pattern. Mononuclear cell infiltration surrounding small bronchioles and extending into septal areas was observed at 3 and 5 dpi in all infected piglets. In addition, there was moderate to severe inflammation of bronchioles characterized by intraepithelial edema and neutrophil infiltration, epithelial necrosis and attenuation, and peribronchiolar cuffing of mononuclear cells. Accumulation of large macrophages and proteinaceous exudate was observed within alveolar spaces. In the severely affected lungs of the 1918/rec virus-infected pigs, hyperplasia of bronchiolar epithelium with pronounced mixed inflammatory cell reaction, often occluding the lumen of small bronchioles and alveoli, was noted at 5 dpi (Fig. 4A). In 1918/rec virus-infected pigs at 7 dpi, severe necrotizing inflammatory lesions involving bronchioles and alveoli as well as bronchiolitis obliterans and hyperplasia of terminal bronchioles were noted. In contrast, microscopic lesions in the 1930/rec virus-infected animals were resolved at 7 dpi. In the 1918/rec virus-infected pigs, mild perivascular and parabronchiolar mononuclear cell cuffing was scattered throughout the lungs at 12 and 17 dpi with occasional perivascular hemorrhage.
FIG. 4.
Microscopic lesions and antigen presence in lungs of infected animals. Microscopic lesions in lungs of pigs inoculated with the 1918/rec or the 1930/rec influenza virus at 5 dpi, using serial lung sections with H&E and IHC staining. H&E staining reveals mononuclear cells infiltrating the small bronchioles in the 1930/rec virus-infected pig (A) compared to the severely affected lungs of the 1918/rec virus-infected piglet with bronchiolitis and pneumonitis and hyperplasia of the bronchiolar epithelium (C). Panel D shows positive IHC staining for influenza virus antigen in bronchiolar epithelial cells in the 1918/rec virus-infected piglet; lower-intensity staining in only few cells of the same type in the 1930/rec virus-infected animal is shown in panel B. Note intense staining both in nuclei and cytoplasm in the 1918/rec virus-infected animal. Bar, 20 μm.
In addition to the lungs, lesions were also observed in trachea and nasal turbinates. At 3 dpi there were mild mononuclear cell infiltrates within the submucosa and neutrophils within the epithelial layer of the trachea in both 1918/rec and 1930/rec virus-infected pigs, whereas at 5 dpi epithelial attenuation and loss of cilia were noticed in both groups. Mild inflammatory reaction in the submucosa of the turbinates was noted only in the piglets inoculated intranasally with the 1918/rec virus at 3 dpi and 5 dpi.
(iv) IHC.
Viral antigen (nucleoprotein) was detected in lungs, trachea, and nasal turbinates of infected animals. At 3 dpi, viral nucleoprotein was detected in moderate amounts (score of 1+ to 2+) in epithelial cells of medium to small bronchioles and occasionally in mucous gland epithelial cells and alveolar macrophages in both groups of influenza virus-infected piglets. High numbers of positively staining cells (score of 3+ to 4+) were observed at 5 dpi in the bronchiolar epithelium as well as alveolar macrophages in the 1918/rec virus-infected pigs (Fig. 4B). As BALF was harvested during the necropsies, only few luminal cells remained in the lungs. Positive staining was detected in macrophages transiting the epithelial layer as well as in macrophages in bronchiolar lumens and in septal areas. Low numbers (1+ to 2+) of these same types of cells were immunopositive in the 1930/rec virus-infected pigs at this dpi. At 7 dpi, only a few, scattered positive epithelial cells were detected in the lung in both groups of infected animals. No positive signal was detected at 12 and 17 dpi in the 1918/rec virus-inoculated pigs.
At all dpi, the staining of the bronchiolar epithelial cells in piglets inoculated with the 1930/rec virus was less intense and localized in the nuclei, whereas in the 1918/rec virus-inoculated pigs, the entire bronchiolar epithelial cells stained very intensely (Fig. 4B).
In the 1918/rec and 1930/rec virus-infected animals, positive immunolabeling was found in tracheal epithelial cells as well as in the tracheal subepithelial mononuclear cells/macrophages at 3 and 5 dpi. At 7 dpi, only few scattered positively stained epithelial cells were found in the trachea. Occasional turbinate respiratory and serous gland epithelial cells were positive at 3, 5, and 7 dpi in the 1918/rec virus-inoculated pigs, both intratracheally or intranasally, but not the 1930/rec virus-infected piglets. No positive signal was detected past 7 dpi. None of the PBS-inoculated control animals displayed microscopic lesions or the presence of influenza virus antigen in their respiratory tracts.
(v) Virus RNA detection.
Full genomic sequence was obtained for virus reisolated from BALF of six pigs infected with the rescued 1918/rec virus, confirming that the virus remained stable in the infected animals for the course of the experimental work.
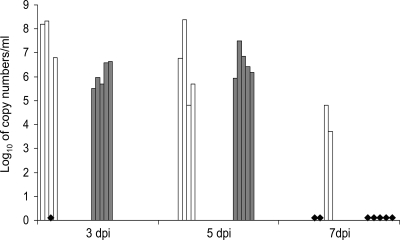
Real-time RT-PCR targeting the matrix protein was used to detect viral RNA in a wide range of tissues. The heart, brain, digestive tract samples, kidney, urine, liver, muscles, tonsil, thymus, spleen, cerebrospinal fluid, and feces were all negative. Influenza virus RNA was detected in samples from the respiratory tract of both groups of experimentally infected animals. The results are summarized in Fig. 5 and Tables 2, 3, and 4. The RNA copy numbers obtained for both groups did not significantly differ and ranged from 105.3 to 108.3/ml of BALF at 3 and 5 dpi. At 7 dpi, only BALFs from the 1930/rec virus-inoculated pigs were positive for virus RNA (Fig. 5). Major differences in RNA copy numbers were noted for lung samples collected from normal-looking and diseased areas within one individual lung. For example, pig 104 at 5 dpi infected with the 1918/rec virus had 107.1 copies in samples collected from macroscopic lesions in lung, compared to 106.3 copies per g of “healthy” lung tissue. Similarly, the lung of pig 69 infected with the 1930/rec virus, and euthanized at 5 dpi, had 106.5/g copies in one sample, while samples collected from different sections of the lungs were negative. These results are consistent with the notion that influenza virus replication in swine lungs is compartmentalized, as also suggested by the macroscopic lung lesions located in individual, distinct lobules (Fig. 3).
FIG. 5.
Detection of viral RNA in BALFs of infected pigs using real-time RT-PCR. Copy numbers in log10 per 1 ml of BALFs obtained by real-time RT-PCR targeting the matrix protein gene are given for individual animals. The white columns represent piglets inoculated with the 1930/rec influenza virus at 3 dpi (pigs 66, 67, and 83) 5 dpi (pigs 69, 71, 84, and 85), and at 7 dpi (pigs 86 and 87). The gray columns represent piglets inoculated with the 1918/rec virus at 3 dpi (pigs 88, 89, 97, 98, and 103) and at 5 dpi (pigs 90, 91, 99, 100, and 104). Black diamonds indicate samples where no viral RNA was detected: at 7 dpi, pigs 92, 93, 101, 102, and 105 inoculated with the 1918/rec virus and piglets 70 and 72 inoculated with the 1930/rec virus; at 3 dpi, pig 82 inoculated with the 1930/rec virus. Viral RNA was not detected in the 1918/rec virus-inoculated piglets after 7 dpi.
TABLE 2.
Results of the real-time RT-PCR targeting the matrix protein in animals infected with 1930/rec
| Sample source | Avg RNA copy number after intratracheal inoculation of the indicated animal at:a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 3 dpi
|
5 dpi
|
|||||||
| No. 66 | No. 67 | No. 82 | No. 83 | No. 69 | No. 71 | No. 84 | No. 85 | |
| Trachea | 4.33 | 5.53 | 4.52 | 5.53 | 5.7 | |||
| Bronchial lymph node | 5.54 | 5.53 | 5.5 | 5.5 | 5.36 | |||
Average copy numbers from three samples are given in log10 per ml of sample.
TABLE 3.
Results of the real-time RT-PCR targeting the matrix protein in animals infected with 1918/rec
| Sample source | Avg RNA copy number after inoculation of the indicated animal (route) at:a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 dpi
|
5 dpi
|
|||||||||
| No. 88 (T) | No. 89 (T) | No. 97 (N) | No. 98 (N) | No. 103 (N) | No. 90 (T) | No. 91 (T) | No. 99 (N) | No. 100 (N) | No. 104 (N) | |
| Trachea | 5.72 | 4.95 | 6.9 | 6.3 | ||||||
| Nasal turbinate | 5.9 | 4.48 | 5.68 | 6.6 | ||||||
Average copy numbers from three samples are given in log10 per ml of sample. T, intratracheal inoculation; N, intranasal inoculation.
TABLE 4.
Results of the real-time RT-PCR targeting the matrix protein in animals infected with 1918/rec by intranasal inoculation
| Pig no. | Avg RNA copy number at the indicated time postinoculationa
|
||
|---|---|---|---|
| 3 dpi | 5 dpi | 7 dpi | |
| 103 | NA | NA | |
| 104 | 6.8 | 6.4 | NA |
| 105 | 5.4 | 5.04 | |
| 106 | 4.55 | ||
| 107 | 5.47 | 5.09 | 5.5 |
| 108 | 5.33 | ||
Average copy numbers from three nasal wash samples are given in log10 per ml of sample. NA, not available.
Viral RNA was detected in tracheal samples from both inoculated groups. Interestingly viral RNA was detected in the tracheobronchial lymph nodes of the 1930/rec virus-inoculated pigs only. Viral RNA was detected at 3 dpi and 5 dpi in nasal turbinates of pigs inoculated intranasally with the 1918/rec virus. Oropharyngeal and nasal swabs, however, were negative by real-time RT-PCR, even from pigs inoculated intranasally. Therefore, nasal washes were collected instead from the last group of pigs inoculated intranasally with the 1918/rec virus, and the viral RNA was detected in some animals up to 7 dpi (Table 4). Viral RNA was not detected in any of the samples collected at 12 and 17 dpi.
(vi) Virus isolation.
At 3 and 5 dpi, virus was isolated from BALFs of all 1918/rec virus-infected pigs. In contrast, the virus isolation attempts were successful only in 5 out of 8 animals infected by the 1930/rec virus. The lung tissue samples were inconsistently virus positive, which is similar to the results of the real-time RT-PCR. Influenza virus was isolated from trachea, turbinates (4 out of 6 animals), and nasal washes (6 out of 11 samples) of pigs intranasally infected with the 1918/rec virus at 3 and 5 dpi (Tables 5, 6, and 7). Starting with samples collected at 7 dpi, all the virus isolation attempts were negative.
TABLE 5.
Virus isolation in of piglets inoculated intranasally with 1918/rec
| Pig no. | Virus titer (log10 PFU/ml) from the indicated sample type after intranasal inoculation with 1918/rec
|
||||
|---|---|---|---|---|---|
| Nasal wash at:a
|
Trachea | Nasal turbinate | |||
| 3 dpi | 5 dpi | 7 dpi | |||
| 103 | NA | NA | 5.6 | 4.6 | |
| 104 | 5.4 | 4.1 | NA | 3.75 | 3.6 |
| 105 | 4.36 | ||||
| 106 | |||||
| 107 | 4 | 2.75 | |||
| 108 | 4.6 | ||||
NA, not available.
TABLE 6.
Virus isolation in lung and BALF of pigs inoculated with 1918/rec
| Sample source | Virus titer (log10 PFU/ml) after inoculation of the indicated animal (route) at:a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 dpi
|
5 dpi
|
|||||||||
| No. 88 (T) | No. 89 (T) | No. 97 (N) | No. 98 (N) | No. 103 (N) | No. 90 (T) | No. 91 (T) | No. 99 (N) | No. 100 (N) | No. 104 (N) | |
| BALF | 2.8 | 2.8 | 2.8 | 3.8 | 3.25 | 3.8 | 4.8 | 4.3 | 3.9 | 5.5 |
| Lung | 4.3 | 5.3 | ||||||||
T, intratracheal inoculation; N, intranasal inoculation.
TABLE 7.
Virus isolation in of piglets inoculated intratracheally with 1930/rec
| Sample source | Virus titer (log10 PFU/ml) after intratracheal inoculation of the indicated animal at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 3 dpi
|
5 dpi
|
|||||||
| No. 66 | No. 67 | No. 82 | No. 83 | No. 69 | No. 71 | No. 84 | No. 85 | |
| BALF | 4.3 | 4.7 | 3 | 1.6 | 4.6 | |||
| Lung | 1 | 1.5 | ||||||
(vii) Antibody detection.
Antibody titers in final bleed sera were determined by an HI test using heterologous and homologous virus. Sera from only 3 out of 8 animals infected with the 1930/rec virus had detectable HI antibody titers ranging from 8 to 64 against the homologous virus at 7 dpi. All three sera with antibodies against 1930/rec virus cross-reacted with the 1918/rec virus with titers between 4 and 8. Interestingly, all the animals infected with the 1918/rec virus developed influenza virus-specific antibodies by 7 dpi, with titers reaching 128, and cross-reacted with the 1930/rec virus (Table 8). We were able to detect HI in sera from some of the 1918/rec virus-inoculated pigs already at 5 dpi. Development of antibodies against the 1918/rec virus appears to be somewhat faster than with other swine influenza viruses (8, 16). The control A/mallard/ON/629/2005 (H1N1) antiserum cross-reacted with the 1918/rec virus at an HI titer of 20 and with the 1930/rec virus at a titer of 80.
TABLE 8.
Determination of HI antibodies
| Virus | HI titer in serum from the indicated animal at:a
|
HI titer in control serumb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 7 dpi
|
12 dpi
|
17 dpi
|
|||||||
| No. 92 | No. 93 | No. 101 | No. 102 | No. 105 | No. 106 | No. 107 | No. 108 | ||
| 1918/rec | 32 | 128 | 128 | 64 | 32 | 128 | 128 | 64 | 20 |
| 1930/rec | 32 | 64 | 32 | 16 | 8 | 32 | 32 | 32 | 80 |
HI assay with sera obtained from the 1918/rec-infected animals with homologous 1918/rec and heterologous 1930/rec virus.
Control serum was raised in ducks against influenza A/mallard/ON/629/2005 (H1N1) virus. Current avian (mallard) serum also cross-reacted with 1918/rec and 1930/rec.
DISCUSSION
The experimental work presented in this report demonstrated that the human 1918 influenza virus can infect and replicate in pigs and cause clinical disease and lesions in the infected animals. Combined with the original swine influenza outbreak descriptions, the early serological work, and the recent phylogenetic analyses, these results support the hypothesis that the 1918 human influenza virus and the virus causing the hog flu during the 1918 pandemic were the same.
The virulence of both the 1918 and the 1930 rescued recombinant viruses was first tested in mice to ensure that these viruses were phenotypically identical with the virus previously reconstructed at CDC, in the case of the 1918/rec virus (37), or with the original isolate as described by Shope, in the case of the 1930/rec virus (29). The high lethality observed in the inoculated mice confirmed the anticipated phenotypes of the viruses.
Interestingly, the 1918/rec virus, which is known to induce lethal infection upon experimental inoculation in ferrets and macaques (14, 38), was not highly virulent in pigs, indicating a potential resistance of swine to highly virulent influenza viruses. Experimental infections of pigs with highly pathogenic H5N1 avian viruses are also consistent with this notion (3, 19). Host factors appear to be critical in the resistance to virulent influenza viruses: BALB/c mice that carry a deletion in the Mx gene (Mx protein is a mediator of cellular resistance to influenza virus) (33) were highly susceptible to the infection with the reconstructed 1918 influenza virus (37), while mice carrying a wild-type Mx1 gene were able to restrict the virus replication (39).The functional Mx protein is expressed in the lungs of pigs experimentally infected with swine influenza virus (11), but our work does not exclude the possibility that additional host factors may have contributed to the high resistance against the 1918 pandemic influenza in pigs.
The early pathological changes and virus titers in both the 1918/rec and 1930/rec virus-infected pigs were considered consistent with typical swine influenza virus infections. No differences were observed between animals inoculated intratracheally (somewhat simulating aerosol inoculation by delivery of the virus into the lower respiratory tract) or intranasally with the 1918/rec virus; for example, the virus titers recovered from BALFs were within the same range for both inoculation routes. Interestingly, human volunteers experimentally inoculated by aerosol developed disease resembling natural influenza virus infections while the ones inoculated via nasal drops did not (36).
The relatively rapid antibody response (HI titers of 128 at 7 dpi) in the 1918/rec virus-infected animals, however, was a somewhat surprising finding as this is not usually reported for swine influenza virus. The more widespread and atypical lesions observed in lung of the 1918/rec virus-infected animals at later times postinfection, such as perivascular hemorrhages along with presence of blood in the airways and the gross lesions in the diaphragmatic lobes, were not expected and may be related to differences in immune response to the 1918/rec versus the 1930/rec virus. However, no conclusions were drawn based on these observations since only three animals were kept past 7 dpi, and we were not able to determine the factors involved in the pathological changes beyond 7 dpi in the 1918/rec virus-infected pigs. Interestingly, the original reports commented on the presence of pulmonary lesions in pigs slaughtered three weeks or more after clinical recovery from the hog flu (27).
Overall, the clinical disease and virus replication observed in piglets infected with the 1918/rec influenza virus were consistent with a typical swine influenza virus infection (12, 22, 23) and not significantly different from the infection with the 1930/rec virus, representing classical swine H1N1 viruses. The clinical signs observed in our 1930/rec virus-infected pigs corresponded with the description of the disease given by Shope for pigs inoculated with the 1930 filtered samples of swine influenza virus (27, 28). In his experimental inoculations, the respiratory disease was mild compared to the field infections in swine. Shope concluded that coinfection with Haemophilus influenzae suis was necessary to induce severe disease in swine (18, 27, 28). It is generally accepted that the clinical outcome of experimental and field infections with influenza virus in swine can differ as additional factors (e.g., stress or crowding) and agents (e.g., mycoplasma, respiratory bacteria, or other viruses) are usually involved in the field, worsening the disease signs caused by the virus. Based on historical records, the swine influenza in 1918 was not a severe disease although, due to almost 100% morbidity, the concerns about classical swine fever (where mortality at that time was reaching 75% in the affected herds) brought it strongly into focus (reference 4 and references therein; see also references 27 and 30). Thus, our observation of a mild clinical disease in piglets infected experimentally with the 1918/rec virus strengthens the hypothesis that the influenza in pigs during the pandemic was caused by the same (or very close) virus as the human influenza (5, 7, 15, 30, 35, 41).
The question of the role of swine in the 1918 influenza pandemic is very interesting. Our experimental infections demonstrated that the 1918 human influenza virus causes an apparent disease in swine, suggesting that this virus was not present unnoticed in pigs prior to the reported outbreaks. The historical records indicate that the disease in swine was observed only later during the pandemic (4), with the first notable occurrence in October 1918 (30). Consequently, one could speculate that the initial interspecies transmission of influenza virus during the 1918 pandemic occurred from people to pigs and only later appeared to occasionally transmit back to people (reference 4 and references therein), likely contributing at least regionally to the maintenance and spread of the disease. The virus spread throughout the swine population, adapted to the swine host, and subsequently resulted in the current lineage of the classical H1N1 swine influenza viruses (7, 26).
In summary, our study demonstrates that swine are susceptible to infection with the 1918 pandemic influenza virus; however, this species does not appear to be as sensitive to the virus as mice, ferrets, and nonhuman primates. It supports the hypothesis based on genetic analysis that the 1918 pandemic influenza virus is likely an ancestor of both the human H1N1 viruses and the classical swine H1N1 influenza viruses, both still present in humans and pigs.
Although the acute 1918 influenza virus infection was similar to the 1930 influenza virus infection and to other influenza virus infections reported in swine, there was an interesting difference in the convalescent phase. In this study lesions were detected as late as 20 dpi, suggesting that the 1918 virus may have unique virulence determinants in comparison to other swine influenza viruses.
Acknowledgments
Funding for this work was provided from the Canadian Food Inspection Agency and ARS-USDA technical development funds.
We thank John Copps, Jason Gren, and Greg Smith for technical support.
Footnotes
Published ahead of print on 18 February 2009.
REFERENCES
- 1.Bikour, M. H., E. H. Frost, S. Deslandes, B. Talbot, and Y. Elazhary. 1995. Persistence of a 1930 swine influenza A (H1N1) virus in Quebec. J. Gen. Virol. 762539-2547. [DOI] [PubMed] [Google Scholar]
- 2.Brown, I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 7429-46. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. K., T. D. Nguyen, H. Ozaki, R. J. Webby, P. Puthavathana, C. Buranathal, A. Chaisingh, P. Auewarakul, N. T. Hanh, S. K. Ma, P. Y. Hui, Y. Guan, J. S. Peiris, and R. G. Webster. 2005. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J. Virol. 7910821-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Easterday, B. C. 2003. Swine influenza: historical perspective, p. 241-244. In P. Martelli, S. Caviarani, and A. Lavazza (ed.), Proceedings of the 4th International Symposium on Emerging and Re-emerging Pig Diseases, Rome, 29 to 2 July 2003. University of Parma, Parma, Italy.
- 5.Francis, T., and T. P. Magill. 1936. The incidence of neutralizing antibodies for human influenza virus in the serum of human individuals of different ages. J. Exp. Med. 63655-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser, L., J. Stevens, D. Zamarin, I. A. Wilson, A. García-Sastre, T. M. Tumpey, C. F. Basler, J. K. Taubenberger, and P. Palese. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 7911533-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman, O. T., W. J. Bean, Y. Kawaoka, I. Donatelli, I. Y. Guo, and R. G. Webster. 1991. Evolution of influenza A virus nucleoprotein genes: Implications for the origin of H1N1 human and classical swine viruses. J. Virol. 653704-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinen, P. P., E. A. de Boer-Luijtze, and A. T. J. Bianchi. 2001. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 82:2697-2707. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, E., S. Krauss, D. Perez, R. Webby, and R. G. Webster. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 203165-3170. [DOI] [PubMed] [Google Scholar]
- 10.Jung, K., Y. Ha, and C. Chae. 2005. Pathogenesis of swine influenza virus subtype H1N2 infection in pigs. J. Comp. Pathol. 132179-184. [DOI] [PubMed] [Google Scholar]
- 11.Jung, K., and C. Chae. 2006. Expression of Mx protein and interferon-α in pigs experimentally infected with swine influenza virus. Vet. Pathol. 43161-167. [DOI] [PubMed] [Google Scholar]
- 12.Jung, T., C. Choi, and C. Chae. 2002. Localization of swine influenza virus in naturally infected pigs. Vet. Pathol. 3910-16. [DOI] [PubMed] [Google Scholar]
- 13.Karasin, A. I., K. West, S. Carman, and C. W. Olsen. 2004. Characterization of avian H3N2 and H1N1 influenza A viruses isolated from pigs in Canada. J. Clin. Microbiol. 424349-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445319-323. [DOI] [PubMed] [Google Scholar]
- 15.Laidlaw, P. P. 1935. Epidemic influenza: a virus disease. Lancet 11118-1124. [Google Scholar]
- 16.Larsen, D. L., A. Karasin, F. Zukermann, and C. W. Olsen. 2000. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet. Microbiol. 74117-131. [DOI] [PubMed] [Google Scholar]
- 17.Lekcharoensuk, P., A. L. Vincent, K. M. Lager, A. Solórzano, A. García-Sastre, and J. A. Richt. 2005. Establishment of a pig model for the 1930 H1N1 swine influenza virus and application of reverse genetics, p.42. Abstr. XIII International Congress of Virology, 23 to 28 July 2005, San Francisco, CA.
- 18.Lewis, P. A., and R. E. Shope. 1931. Swine influenza. II. Hemophilic bacillus from the respiratory tract of infected swine. J. Exp. Med. 54361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipatov, A. S., Y. K. Kwon, L. V. Sarmento, K. M. Lager, E. Spackman, D. L. Suarez, and D. E. Swayne. 2008. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS Pathog. 4:e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich, M., A. Tuzikov, N. Bovin, A. Gambaryan, A. Klimov, M. R. Castrucci, I. Donatelli, and Y. Kawaoka. 2000. Early alterations of the receptor-binding properties of H1, H2 and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 748502-8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers, K. P., C. W. Olsen, and G. C. Gray. 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 441084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen, C. W., I. H. Brown, B. C. Easterday, and K. Van Reeth. 2006. Swine influenza, p. 469-482. In B. E. Straw, J. J. Zimmerman, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 9th ed. Iowa State Press, Ames, IA.
- 23.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 413198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richt, J. A., P. Lekcharoensuk, K. M. Lager, A. L. Vincent, C. M. Loiacono, B. H. Janke, W. H. Wu, K. J. Yoon, R. J. Webby, A. Solórzano, and A. García-Sastre. 2006. Vaccination of pigs against swine influenza viruses by using NS1-truncated modified live-virus vaccine. J. Virol. 8011009-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schickli, J. H., A. Flandorfer, T. Nakaya, L. Martinez-Sobrido, A. García-Sastre, and P. Palese. 2001. Plasmid only rescue of influenza A vaccine candidates. Philos. Trans. R. Soc. Lond. B 3561965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz, U., W. M. Fitch, S. Ludwig, J. Mandler, and C. Scholtissek. 1991. Evolution of pig influenza viruses. Virology 18361-73. [DOI] [PubMed] [Google Scholar]
- 27.Shope, R. E. 1931. Swine influenza. I. Experimental transmission and pathology. J. Exp. Med. 54349-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shope, R. E. 1931. Swine influenza. III. Filtration experiments and etiology. J. Exp. Med. 54373-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shope, R. E. 1935. The infection of mice with swine influenza virus. J. Exp. Med. 62561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shope, R. E. 1936. The incidence of neutralizing antibodies for swine influenza virus in the sera of human beings of different ages. J. Exp. Med. 63669-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spackman, E., D. A. Senne, T. J. Myers, L. L. Bulaga, L. P. Garber, M. L. Perdue, K. Lohman, L. T. Daum, and D. L. Suarez. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 403256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solórzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. García-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 797535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staeheli, P., R. Grob, E. Meier, J. G. Sutcliffe, and O. Haller. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 84518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wanf, G. Jim, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437889-893. [DOI] [PubMed] [Google Scholar]
- 35.Taubenberger, J. K., and D. M. Morens. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 1215-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tellier, R. 2006. Review of aerosol transmission of influenza A virus. Emerg. Infect. Dis. 121657-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solórzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. García-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 31077-80. [DOI] [PubMed] [Google Scholar]
- 38.Tumpey, T. M., T. R. Maines, N. van Hoeven, L. Glaser, A. Solórzano, C. Pappas, N. J. Cox, D. E. Swayne, P. Palese, J. M. Katz, and A. García-Sastre. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315655-659. [DOI] [PubMed] [Google Scholar]
- 39.Tumpey, T. M., K. J. Szretter, N. Van Hoeven, J. M. Katz, G. Kochs, O. Haller, A. García-Sastre, and P. Staeheli. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 8110818-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Reeth, K. 2007. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet. Res. 38243-260. [DOI] [PubMed] [Google Scholar]
- 41.Vana, G., and K. M. Westover. 2008. Origin of the 1918 Spanish influenza virus: a comparative genomic analysis. Mol. Phylogenet. Evol. 471100-1110. [DOI] [PubMed] [Google Scholar]
- 42.Vincent, A. L., W. Ma, K. M. Lager, B. H. Janke, and J. A. Richt. 2008. Swine influenza viruses: a North American perspective. Adv. Virus Res. 72127-154. [DOI] [PubMed] [Google Scholar]
- 43.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 748243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]