Abstract
GP64, the major envelope glycoprotein of the Autographa californica multicapsid nucleopolyhedrovirus budded virion, is important for host cell receptor binding and mediates low-pH-triggered membrane fusion during entry by endocytosis. Previous transmembrane (TM) domain replacement studies showed that the TM domain serves a critical role in GP64 function. To extend the prior studies and examine specific sequence requirements of the TM domain, we generated a variety of GP64 TM domain mutations. The mutations included 4- to 8-amino-acid deletions, as well as single and multiple point mutations. While most TM domain deletion constructs remained fusion competent, those containing deletions of eight amino acids from the C terminus did not mediate detectable fusion. The addition of a hydrophobic amino acid (A, L, or V) to the C terminus of construct C8 (a construct that contains a TM domain deletion of eight amino acids from the C terminus) restored fusion activity. These data suggest that the membrane fusion function of GP64 is dependent on a critical length of the hydrophobic TM domain. All GP64 proteins with a truncated TM domain mediated detectable virion budding with dramatically lower levels of efficiency than wild-type GP64. The effects of deletions of various lengths and positions in the TM domain were also examined for their effects on viral infectivity. Further analysis of the TM domain by single amino acid substitutions and 3-alanine scanning mutations identified important but not essential amino acid positions. These studies showed that amino acids at positions 485 to 487 and 503 to 505 are important for cell surface expression of GP64, while amino acids at positions 483 to 484 and 494 to 496 are important for virus budding. Overall, our results show that specific features and amino acid sequences, particularly the length of the hydrophobic TM domain, play critical roles in membrane anchoring, membrane fusion, virus budding, and infectivity.
In typical infections of eukaryotic cells by enveloped viruses, viral entry is mediated by the fusion of viral and cellular membranes in a process that is directed by membrane-associated viral fusion proteins. In the best models of membrane fusion, two hydrophobic domains of the viral fusion protein are critically important in fusion: the fusion peptide and the transmembrane (TM) domain. The fusion peptide is a hydrophobic domain that inserts into the target cellular membrane, thereby attaching the fusion protein to the target membrane (14). By insertion of the fusion peptide into the target membrane and anchoring of the envelope protein in the viral envelope via the TM domain, a bridge is formed between the two membranes. Subsequent structural rearrangements in the envelope fusion protein bring the viral and cellular membranes into close proximity and culminate in the merger of two bilayers and the subsequent opening of a fusion pore. The TM domain may serve several roles in this overall process. In addition to anchoring the envelope protein in the membrane, the TM domain may play a crucial role in the transition from the initial merger of the outer leaflets of the two membranes (hemifusion) to complete membrane merger and pore formation. Evidence of a more-direct role of the TM domain in this process comes from studies in which the proteinaceous TM domains of viral fusion proteins were replaced by lipid (glycosylphosphatidylinositol [GPI]) anchors or protein sequences that altered their structures. Such modifications of the TM domain may lead to partial or full arrest of fusion at the hemifusion step (22). In addition, it was recently proposed that a direct interaction between the fusion and TM peptides of hemagglutinin (HA) may be required to open the fusion pore (50).
The virus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is a member of the group I NPVs within the Alphabaculovirus genus and is the type species for the family Baculoviridae (17). Budded virions of AcMNPV appear to enter cells via a clathrin-mediated, low-pH-dependent endocytic pathway (25). During entry by endocytosis, the major envelope glycoprotein GP64 mediates low-pH-triggered membrane fusion (5). The GP64 proteins are very highly conserved in the group I NPVs and appear to be closely similar in structure and function only to GP75 proteins from thogotoviruses, a subgroup of the Orthomyxoviridae. Recent structural studies also indicate that GP64 should be classified as a class III fusion protein, along with proteins such as vesicular stomatitis virus glycoprotein (VSV G) and herpesvirus gB (21). GP64 is a type I integral membrane protein that is present on the infected cell surface and in the virion as a disulfide-linked homotrimer (38). GP64 has host cell receptor-binding activity (16), and a domain associated with receptor binding was recently mapped to an N-terminal region of the ectodomain (60). GP64 is both necessary and sufficient for mediating pH-dependent membrane fusion during viral entry (5, 55, 61). In addition to its essential role in virus entry, GP64 is also necessary for efficient budding and production of infectious virions (36, 37). The AcMNPV GP64 protein is posttranslationally modified by palmitoylation and by glycosylation at multiple sites. However, neither palmitoylation nor any single glycosylation site is necessary for GP64 synthesis, transport, budded virus (BV) production, infectivity, or membrane fusion activity (20, 59). GP64 is also posttranslationally modified by phosphorylation (30, 54), and little is known of the structural or functional implications of this modification. Two important hydrophobic regions were identified in the GP64 ectodomain and examined by mutagenesis. One region is located between amino acids (aa) 220 and 230 of the closely related Orgyia pseudotsugata (OpMNPV) GP64, and substitutions in that region disrupted normal membrane fusion activity. The second region is located between aa 327 and 335 and is important for oligomerization and cell surface expression (35). The latter region lies within a highly conserved 4-3 heptad repeat (a leucine zipper from residues 299 to 346) that is predicted to form an amphipathic alpha helix (35) and near the top of a long central helix in the postfusion structure (21). Mutations that disrupted the predicted helix or reduced the hydrophobicity along the hydrophobic face of the predicted OpMNPV GP64 leucine zipper motif were sufficient to disrupt fusion activity (35).
GP64 proteins have very short predicted cytoplasmic tail domains (CTD), ranging from approximately 3 to 8 aa in length. Earlier experimental analysis showed that the 7-aa CTD of AcMNPV GP64 is not essential for production of infectious BV, but removal of the CTD results in a measurable reduction in budding efficiency (37). Baculovirus GP64 proteins have a predicted TM domain ranging from approximately 16 to 23 amino acids. Unlike the highly conserved ectodomain amino acid sequences (which are approximately 80% identical among baculovirus GP64 proteins), sequences of the TM domains are more variable in sequence conservation (13 to 100%), and the extent of the biological function(s) of the GP64 TM domain remains unknown. In previous studies, we demonstrated that replacing the 23-aa AcMNPV GP64 TM domain with corresponding sequences from a range of viral or cellular type I membrane proteins or with a GPI addition sequence had, in many cases, severe effects on fusion activity and virus infectivity (23). This suggested that the specific amino acid sequence of the GP64 TM domain is critical for the function of GP64. In the current study, we further investigated the function of the GP64 TM domain by examining a series of GP64 proteins with modified TM domains. TM domain modifications included truncations, deletions, 3-alanine scanning mutations, and single and multiple amino acid substitutions. These studies revealed that (i) the hydrophobic length of the GP64 TM domain is critical for GP64-mediated membrane fusion activity, (ii) no specific residue of the TM domain was required for the membrane fusion activity, and (iii) several regions within the TM domain are important for cell surface localization or virus budding.
MATERIALS AND METHODS
Cells, transfections, and infections.
Spodoptera frugiperda (Sf9) cells and the cell line Sf9Op1D (that constitutively expresses the OpMNPV GP64 protein) (42) were cultured at 27°C in TNMFH medium (18) containing 10% fetal bovine serum (FBS). Transfections were carried out using CaPO4 precipitation as described earlier (4). For viral infection, BV was incubated on cells (multiplicity of infection of 5) for 1 h, and then cells were washed once in TNMFH. Times postinfection (p.i.) were calculated from the time the viral inoculum was added. As controls in transfection and infection experiments, mock transfections were performed in the absence of plasmid DNA and mock infections were performed with no virus.
Mutagenesis and construction of plasmids and bacmids.
The plasmid pGEM3ZGP64 (23), which contains the promoter region and open reading frame (ORF) of AcMNPV GP64, was used as a target for mutagenesis. The C-terminal truncation mutants were generated by PCR. The N-terminal and central region TM deletion mutations, as well as the substitution mutations and alanine scanning mutations, were generated by using site-directed, ligase-independent mutagenesis (9). The sequences of the primers used are available upon request. The modified GP64 ORFs were subcloned into the XbaI/EcoRI sites of the pBiepA vector, which contains the promoter region of AcMNPV ie1 and the poly(A) signal of AcMNPV gp64 (23). Plasmids were prepared for transfections by using a DNA Maxiprep kit (Marligen Biosciences, Inc.). In order to construct recombinant baculoviruses expressing the modified GP64 proteins, GP64 constructs in pGEM3ZGP64 were digested with KpnI and EcoRI to excise the fragment containing the promoter and GP64 ORF and the fragments were subcloned into the KpnI and EcoRI sites of the pFastBac1 plasmid (Invitrogen), resulting in removal of the AcMNPV polyhedrin promoter. The modified gp64 genes were then inserted into the polyhedrin locus of an AcMNPV gp64-null bacmid (vAc64−) by Tn7-mediated transposition as described previously (27). Constructs were confirmed by restriction enzyme digestion and DNA sequencing.
cELISA, syncytium formation, and fusion assays.
For analysis and comparisons of relative levels of cell surface-localized GP64 proteins, we used a cell surface enzyme-linked immunosorbent assay (cELISA). Sf9 cells were transfected with plasmids and incubated for 24 h to permit expression and cell surface localization, and then cells were fixed in glutaraldehyde so that cells were not permeabilized. Cell surface-localized GP64 was then detected in a cELISA assay by using monoclonal antibody (mAb) AcV5 as described previously (59). Briefly, transfected cells in 24-well plates (Corning, Inc.) were rinsed once in phosphate-buffered saline (PBS, pH 7.4) and fixed in 0.5% glutaraldehyde for 10 min at room temperature. Fixed cells were washed once with PBS and blocked by incubation for 2 h in PBS containing 1% gelatin at 27°C. Cells were then incubated in mAb AcV5 (hybridoma cell culture supernatant diluted 1:25 in PBS containing 0.5% gelatin) for 45 min at 27°C. Cells were washed three times in PBS and then incubated in a secondary goat anti-mouse antibody conjugated to beta-galactosidase (GAM-Beta-gal; diluted 1:750 in PBS containing 0.5% gelatin) for 45 min at 27°C. Cells were washed five times in PBS and then incubated at 37°C in 2 mg/ml chlorophenol red-beta-d-galactopyranoside (CPRG) in a solution of PBS containing 2 mM MgCl2 and 1% BSA. After addition of the substrate, the absorbance of the supernatant (optical density, 570 nm) was determined at various intervals using an ELISA plate reader.
For membrane fusion activity assays, Sf9 cells in 24-well plates were transfected with plasmids encoding wild-type (WT) or modified forms of GP64 as described above. At 24 h posttransfection (p.t.), TNMFH medium was removed, and cells were washed once with PBS at pH 7.4. The PBS at pH 7.4 was then replaced with PBS at pH 5.0. After a 3-min incubation period, cells were washed again with PBS at pH 7.4 and then returned to TNMFH. After 4 h of incubation at 27°C, cells were fixed with methanol for 10 min and stained using a HEMA3 stain kit (Fisher Scientific Company L.L.C.), and the number of nuclei found in syncytia was scored. The criterion for identification of a syncytial mass was the presence of at least five nuclei in a fused cell mass. Five randomly selected representative fields were evaluated for each construct. For calculations of relative levels of fusion activity, the number of nuclei in syncytia was divided by the total number of cells in a field. Those percentages were then normalized to parallel syncytium formation data from WT GP64 that was localized to the cell surface at equivalent levels.
Labeling of RBCs with R18 and calcein-AM.
Sheep red blood cells (RBCs; HemoState Laboratories) were colabeled with the lipid probe octadecyl rhodamine B chloride (R18) and the aqueous dye calcein-AM (Molecular Probes, Invitrogen) as described previously (52), with a minor modification (23). In brief, 10 μl of R18 (2 mM in ethanol) was added to RBCs (1% packed cell volume in 20 ml PBS) while gently shaking. The mixture was incubated for 30 min at room temperature in the dark, and then 20 ml of 7.5% FBS-Dulbecco's modified Eagle's medium (DMEM) was added to the suspension to absorb unbound probe. After a 20-min incubation at room temperature in the dark, the RBC suspension was washed five times in 40 ml of PBS and resuspended in 1.25 ml PBS. A 5-μl aliquot of calcein-AM (4 mM in dimethyl sulfoxide) was added to 1 ml R18-labeled RBCs, and the suspension was incubated for 1 h at 37°C in the dark. The unbound calcein-AM was also absorbed using 20 ml of 7.5% FBS-DMEM for 20 min, and the colabeled cells washed four times with PBS as described above. The double-labeled RBCs were suspended in 200 ml of PBS and used within 1 h.
Hemifusion and pore formation assay.
To analyze hemifusion and pore formation by WT GP64 and GP64 constructs containing TM domain mutations, an R18 and calcein transfer assay was performed. At 24 h p.t., transfected Sf9 cells (8 × 104 cells/well) were washed once with PBS (pH 7.4) and then incubated with R18- and calcein-labeled RBCs (0.4 ml 0.1% packed cell volume/well) for 20 min at room temperature for binding. Unbound RBCs were removed by three washes with PBS (pH 7.4), and the Sf9 cells were then incubated with PBS at pH 5.0 for 3 min at room temperature. Sf9 cells were then washed in PBS at pH 7.4 and transferred into TNMFH medium. After incubation for 20 min at 27°C, hemadsorption and the transfer of fluorescence were observed by phase-contrast and epifluorescence microscopy, respectively. Five randomly selected fields were scored for dye transfer. For each dye-labeling experiment, the efficiency of dye (R18 or calcein-AM) transfer was estimated by dividing the number of dye-containing Sf9 cells displaying mutant GP64 proteins by the number of dye-containing Sf9 cells displaying WT GP64.
Immunofluorescence analysis of cell surface GP64.
To confirm cell surface localization of GP64, Sf9 cells were plated in 24-well plates (8 × 104 cells/well) and transfected with plasmids expressing either WT or modified GP64 proteins. After 24 h p.t., cells were fixed with 4% paraformaldehyde in PBS (pH 7.4). Cells were washed with PBS and then incubated with a blocking buffer (1% gelatin in PBS, pH 7.4) at 27°C for 2 h. After being washed with PBS, the cells were incubated with a primary anti-GP64 mAb (AcV1, 1:25 dilution in PBS) at 27°C for 1 h. Cells were washed three times with PBS (pH 7.4) and incubated for 1 h at 27°C with Alexa Fluor 488-goat anti-mouse antibody (Molecular Probes, Invitrogen) diluted 1:750 in PBS. After cells were washed five times with PBS (pH 7.4), fluorescence was observed with an Olympus IX70 epifluorescence microscope.
Budding assay and measurement of GP64 incorporation into virions.
For analysis of virus budding efficiency and the relative amounts of GP64 incorporation into BV, we performed the following assays. Viruses expressing the TM-truncated and TM-deleted GP64 were amplified and their titer determined in Sf9Op1D cells. Viruses containing the alanine scanning mutants were amplified and their titers determined in Sf9 cells. The titered viruses were used to infect Sf9 cells (5 × 106) at a multiplicity of infection of 5. After inoculation and incubation for 1 h, the virus inoculum was removed and cells were washed once with TNMFH and then incubated at 27°C. At 15 h p.i., cells were washed once and starved by incubation in 2 ml methionine-free Grace's medium (Grace−met; Invitrogen) for 1 h. At 17 h p.i., the medium was replaced with 2.2 ml of Grace−met containing 200 μCi of 35S-EasyTag Express protein labeling mix (1,175.0 Ci/mmol; Perkin-Elmer Life Sciences). At 30 h p.i., 0.8 ml of TNMFH was added. The supernatants were harvested at 40 h p.i. and cleared of cell debris by brief centrifugation (10 min at 3,000 × g at 4°C) and then loaded onto a 25% sucrose cushion and centrifuged at 80,000 × g for 90 min at 4°C in an SW60 rotor. Virus pellets were resuspended in 200 μl Laemmli buffer (4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% 2-mercaptoethanol, 0.04% bromophenol blue, 0.125 M Tris, pH 6.8) containing a cocktail of protease inhibitors (complete; Roche Applied Science) and electrophoresed on 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels. Dried gels were exposed on phosphorimager screens, and screens were scanned on a Molecular Dynamics phosphorimager. Quantification of individual bands was performed by using the ImageQuant TL software package (Amersham, GE).
Western blot analysis.
Reducing and nonreducing SDS-PAGE was performed in 6% or 10% polyacrylamide gels as described previously (37). Following transfer to polyvinylidene difluoride membrane (Millipore), blots were blocked in a 4% milk Tris-buffered saline-Tween 20 solution as previously described (59). For detection of GP64, mAb AcV5 was used at a dilution of 1:1,000. Immunoreactive proteins were visualized by using alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G antibody and nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Promega) as described previously (59).
RESULTS
Expression and intracellular processing of GP64 deletion mutants.
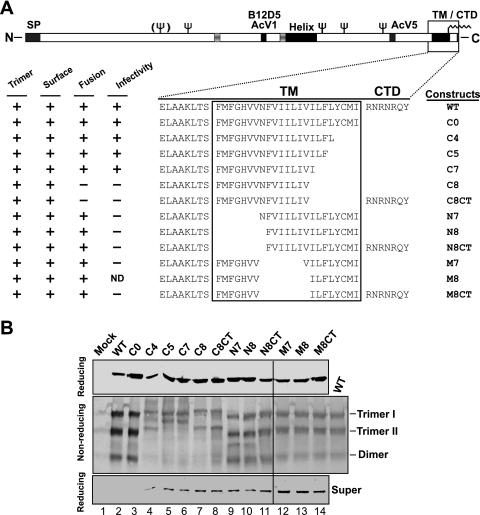
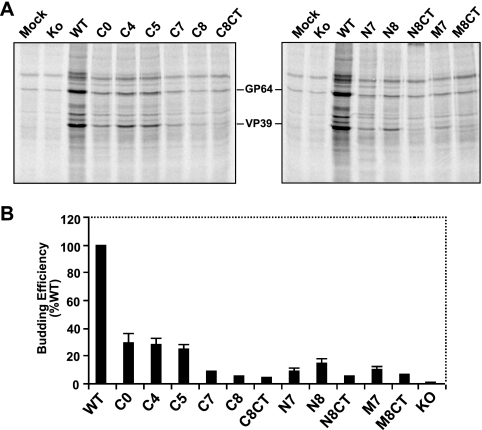
In a prior study (23), we found that substitution of heterologous TM domains from other viral and cellular membrane proteins for the GP64 TM domain had substantial negative effects on GP64 function, indicating an important role for the TM domain beyond simple anchoring of GP64 in the viral envelope. To examine the requirements of the GP64 TM domain for membrane fusion activity and viral infectivity, we generated and examined a series of truncation, deletion, and amino acid substitution mutations in the GP64 TM domain. For the initial analysis, N-terminal, central, and C-terminal portions of the hydrophobic TM domain were truncated (Fig. 1A). Although the very short (7 aa) CTD was previously shown to be nonessential, several of the larger deletion constructs were generated in both the presence and absence of the CTD (Fig. 1A). The GP64 proteins with modified TM and CTD domains were transiently expressed from plasmids in Sf9 cells. At 24 h p.t., GP64 proteins from cell lysates were examined by Western blot analysis using either reducing or nonreducing conditions for SDS-PAGE (Fig. 1B). Because GP64 trimers are associated by disulfide bonds, nonreducing conditions were used to assess GP64 oligomerization (16, 38). Oligomeric forms typical of trimer I, trimer II, and dimer were detected from most or all GP64 mutants. However, electrophoretic migration patterns of most constructs were altered from those of WT GP64. Band migration patterns were retarded for trimers of constructs with C-terminal truncations (C4, C5, C7, C8, and C8CT), while the constructs with truncations of the N-terminal and central portion of the TM domain had trimer migration patterns similar to that of WT GP64 (Fig. 1B). Examination of protein from each culture supernatant for evidence of protein shedding from the cell surface revealed that reduction of the TM domain length resulted in GP64 shedding. While the WT and control (C0) GP64 proteins were detected in the supernatant in only trace amounts, GP64 constructs containing TM domains of reduced length were detected at substantially higher levels in the supernatant (Fig. 1B, bottom). Also, inclusion of the short hydrophilic CTD at the C terminus of constructs N8, M8, and C8 did not eliminate or appear to substantially reduce the shedding of GP64 proteins with truncated TM domains (Fig. 1B, bottom, lanes 7 versus 8, 10 versus 11, and 13 versus 14, super).
FIG. 1.
Construction and expression of GP64 proteins with truncated TM domains. (A) Schematic representation of the WT GP64 protein with the truncations of the GP64 TM domain illustrated below. The schematic at the top shows the positions of several major features of the GP64 protein, including the signal peptide (SP), predicted glycosylation sites (fork symbols), mAb epitopes (AcV1, B12D5, and AcV5), an amphipathic alpha-helical domain (Helix), an acylation site (wave symbol), and the TM domain and CTD. The name of each GP64 construct is shown to the right of the sequence of the TM domain region. Results from trimerization, surface localization, membrane fusion (syncytium formation), and infectivity assays are summarized on the left: +, positive; −, negative; ND, not done. (B) Analysis of expression and oligomerization of TM-truncated GP64 constructs. Sf9 cells were transfected with plasmids encoding WT and modified GP64 constructs. Mock represents transfection with no plasmid DNA. Cells were lysed at 24 h p.t. under reducing (top) or nonreducing (middle) conditions. GP64 proteins were resolved by SDS-PAGE and detected by Western blotting using mAb AcV5. The positions of oligomeric forms of GP64 (Trimer I and II, Dimer) are identified on the right. The supernatants from transfected cells were also analyzed under reducing conditions by Western blot analysis (bottom).
Cell surface localization and conformation.
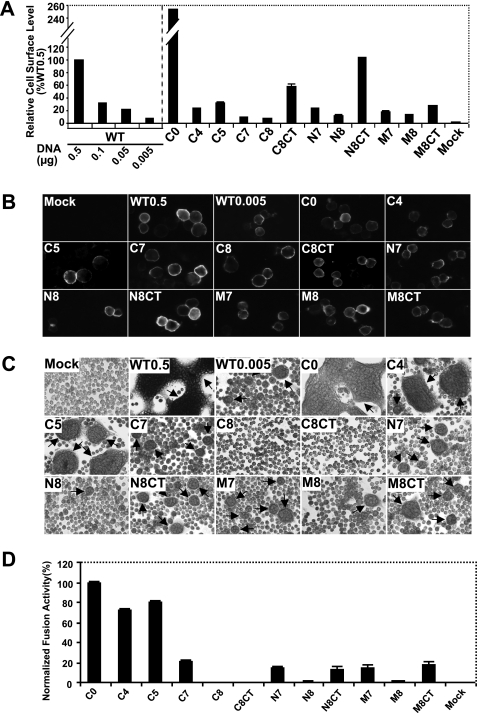
Next, we asked whether the TM-truncated GP64 proteins were transported to and localized at the cell surface. Using a cELISA protocol, we measured the cell surface level of each protein construct relative to that detected from WT GP64. Deletion of the short, 7-aa GP64 cytoplasmic tail dramatically increased the level of the GP64 C0 construct detected at the cell surface. In contrast, most constructs containing truncations of the TM domain resulted in decreased surface levels of GP64 (Fig. 2A). For example, when the TM domain was truncated by removing four or five amino acids from the C terminus, surface expression levels decreased to approximately 25 to 30% of that from WT GP64. Further truncations of seven or eight amino acids from the C terminus resulted in further reductions in cell surface levels, with constructs C7 and C8 detected at approximately 9.7% and 7.4% of the level from WT GP64. Deletion of seven or eight amino acids from the N terminus or central region of the TM domain also resulted in substantially decreased cell surface levels of the modified GP64 proteins relative to WT GP64 levels. Addition of the CTD to the C terminus of truncated protein constructs (denoted as “CT” in construct names) increased cell surface levels in most cases. Constructs C8CT and N8CT were expressed at the cell surface at substantially higher levels than C8 and N8, respectively (Fig. 2A). To confirm the presence of GP64 constructs at the cell surface and to also examine the conformation of the surface-localized GP64 constructs, we used indirect immunofluorescence with a conformation-specific mAb (AcV1) that recognizes the native, neutral pH, prefusion conformation of GP64 (19, 61). The results indicated that, like WT GP64, the TM-truncated GP64 constructs were surface localized and were in the native prefusion conformation (Fig. 2B).
FIG. 2.
Analysis of cell surface levels and membrane fusion activity of TM-truncated GP64 constructs. (A) Relative cell surface levels of GP64 constructs were measured by cELISA, using mAb AcV5. Each mutant GP64 construct was expressed by transfecting cells with 0.5 μg of the appropriate plasmid DNA. Values represent the mean results from triplicate transfections and are normalized to that of cells transfected with 0.5 μg of the plasmid DNA expressing WT GP64. Error bars represent the standard deviations from the means, and the amount of WT DNA per transfection is indicated below the graph. (B) Immunofluorescence analysis of cell surface levels of GP64 mutants using mAb AcV1. Cell surface GP64 was detected on transfected Sf9 cells fixed with 4% paraformaldehyde. (C) Analysis of membrane fusion by syncytium formation assays. Sf9 cells were transfected with plasmids expressing WT or TM-truncated GP64 proteins. At 24 h p.t., cells were exposed to a pH of 5.0 for 3 min and then incubated for 4 h and observed and photographed using phase-contrast microscopy (×200). Arrows indicate syncytial masses. WT0.5 and WT0.005 represent cells transfected with 0.5 and 0.005 μg DNA, respectively, expressing WT GP64. (D) Analysis of syncytium formation efficiency. For calculations of relative levels of fusion activity, the number of nuclei in syncytia was divided by the total number of cells in a field. Those percentages were normalized to parallel syncytium formation data from WT GP64 that was localized to the cell surface at equivalent levels. For each sample, five fields were examined. The means and standard deviations of the results of triplicate transfections are shown.
Fusion activity of GP64 truncation mutants.
We evaluated the membrane fusion activity of each TM-truncated GP64 construct by measuring fusion efficiency in a semiquantitative syncytium formation assay. Because surface levels of different GP64 constructs varied, the fusion activity of each modified GP64 construct was compared with the activity from WT GP64 that was localized to the cell surface at the same level. While surface levels of some short C-terminal truncations of the TM were reduced (Fig. 2A, C4 and C5), the normalized membrane fusion activities for those constructs were only marginally decreased. Fusion activities detected for constructs C4 and C5 were approximately 70 to 80% of that from WT GP64 (Fig. 2D). The further deletion of seven or eight amino acids from the C terminus of the TM domain resulted in substantially reduced or no detectable fusion activity (Fig. 2D, C7 and C8). We note that in a prior study (37), fusion activity was not detected from a construct equivalent to construct C7 from the current study. This difference results from differences in the pH used for triggering. We found that while no fusion activity was detected from construct C7 exposed to pH 5.2 (data not shown), as in the prior study, exposure to pH 5.0 resulted in a low level of detectable syncytium formation or fusion activity. The absence of detectable fusion activity from construct C8 may have resulted from poor anchoring in the membrane. To examine that possibility, we included the hydrophilic cytoplasmic tail in construct C8CT (Fig. 1A). While inclusion of the short hydrophilic CTD resulted in higher levels of this construct at the cell surface (Fig. 2A, C8CT versus C8), fusion activity was not restored (Fig. 2D, C8CT versus C8). Overall, GP64 constructs that contained 4- or 5-aa truncations of the C terminus of the TM domain remained highly functional for membrane fusion, but a larger TM truncation (C7) substantially reduced fusion activity, and further truncation of the TM domain (C8) completely abolished detectable fusion activity. Addition of the hydrophilic CTD to the C8 deletion resulted in greater accumulation of GP64 at the cell surface, but fusion activity was not restored.
A similar analysis was performed by generating deletions of seven or eight amino acids from the N terminus of the TM domain. Similar to the 7-aa deletion from the C terminus of the TM domain, a 7-aa deletion from the N terminus resulted in only approximately 14% of the fusion activity of WT GP64 (Fig. 2D, N7). Unlike the C-terminal deletions, a deletion of eight amino acids from the N terminus of the TM domain (N8) resulted in extremely low but detectable fusion activity (Fig. 2D, N8). And in the case of the N8 construct, addition of the hydrophilic CTD resulted in an increase in fusion activity (Fig. 1A and 2D, N8CT). Similar results were obtained when seven or eight amino acids were deleted from the central or middle portion of the TM domain (Fig. 1 and 2, M7, M8). As with the N8 construct described above, addition of the CTD to the M8 construct (M8CT) resulted in increased fusion activity (Fig. 2D, compare M8 and M8CT). In total, these results suggest that when the length of the 23-aa hydrophobic TM domain was reduced to less than 16 amino acids, the fusion activity of the mutant protein construct was either lost (C8) or very low (N8 and M8). Overall, adding the hydrophilic cytoplasmic tail increased the levels of protein at the cell surface (Fig. 2A, C8, N8, and M8 versus C8CT, N8CT, and M8CT). When fusion activity was detected, it appears that addition of the CTD resulted in an increase in the normalized membrane fusion activity (Fig. 2D, N8 and M8 versus N8CT and M8CT). However, addition of the CTD did not restore activity of the fusion-deficient C8 construct.
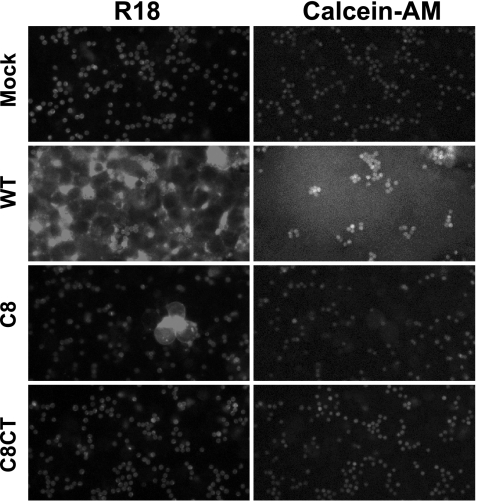
Since control comparisons with WT GP64 indicated that the fusion defects of C8 and C8CT did not result from low levels of these proteins at the cell surface, we asked whether fusion was blocked prior to the hemifusion intermediate or pore formation steps. RBCs were colabeled with a membrane dye (R18) and a soluble cytosolic dye (calcein-AM) and bound to Sf9 cells expressing various GP64 constructs. Fusion was then induced by low-pH treatment (pH 5.0 for 3 min) (see Materials and Methods). Although we observed R18 dye transfer in cells displaying construct C8 (Fig. 3), the R18 transfer efficiencies were very low (less than 1% of that from WT GP64) (data not shown). We did not observe transfer of R18 or calcein-AM from cells expressing C8CT (Fig. 3). Thus, by comparison with WT GP64, constructs containing deletions of eight amino acids from the C terminus of the TM domain appear to be severely compromised or defective in the first step in membrane fusion, the merger of the outer leaflet of the lipid bilayer.
FIG. 3.
Analysis of hemifusion and fusion pore formation by fusion-deficient GP64 proteins. RBCs that were dual labeled with membrane-restricted (R18) and cytosolic (Calcein-AM) dyes were bound to Sf9 cells transiently expressing mutant GP64 proteins. The bound cells were exposed to acidic PBS (pH 5.0) for 3 min to induce fusion and examined after a 20-min incubation period. Membrane dye transfer (R18) or cytosolic dye transfer (Calcein-AM) was monitored by fluorescence microscopy. Each mutant GP64 construct (or mock control) used for transfection is indicated at the left.
Extending the hydrophobic length of a truncated TM domain restores fusion activity of GP64.
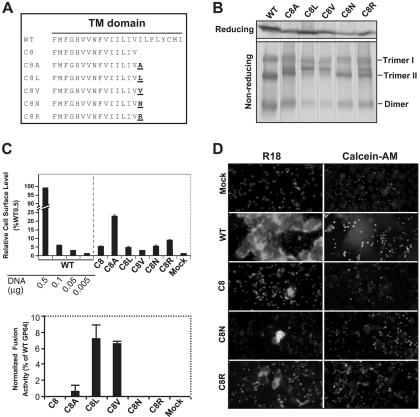
The C8 truncation of the GP64 TM domain was defective for membrane fusion, and addition of the hydrophilic cytoplasmic tail did not restore fusion activity to the resulting GP64 construct (C8CT). Therefore, we next asked whether addition of a single hydrophobic amino acid to the C-terminal end of the C8 TM domain was sufficient to restore fusion activity (Fig. 4A). For this analysis, we selected hydrophobic amino acids (A, V, and L), and as controls, we added hydrophilic amino acids (N and R) (Fig. 4A). GP64 constructs containing these TM modifications were transiently expressed in Sf9 cells and detected by Western blot analysis of cell lysates (Fig. 4B). Under both reducing and nonreducing conditions, the profiles of the modified GP64 constructs were similar but not identical to that of WT GP64. The cell surface levels of all constructs except C8A were similar and were less than 10% of that from WT GP64 (transfected and expressed under the same conditions, Fig. 4C). In contrast to the other constructs, C8A was detected at the cell surface at a higher level, corresponding to approximately 22% of that from WT GP64 (Fig. 4C, C8A, upper graph). Analysis of membrane fusion by these constructs showed that all constructs containing an additional (heterologous) hydrophobic amino acid (C8A, C8L, and C8V) induced syncytium formation. However, the fusion activity detected was low and in all cases fusion activity was less than 10% of that from WT GP64 (Fig. 4C, lower graph). No fusion activity was detected for constructs C8, C8N, and C8R by syncytium formation assay, so we examined these fusion-negative constructs for membrane merger (hemifusion) and pore formation, as described above. Constructs C8N and C8R showed detectable but low-efficiency R18 transfer that appeared similar to that of C8 and was less than 1% of that detected with WT GP64 (Fig. 4D and data not shown). No cytosolic dye transfer was observed for these same constructs. These results combined with prior data (Fig. 3) indicate that the length of the hydrophobic TM domain (≥16 aa) is critically important for fusion activity of AcMNPV GP64.
FIG. 4.
Analysis of the fusion requirements for GP64 construct C8. A single hydrophobic (A, L, or V) or hydrophilic (R or N) amino acid was added to the C terminus of the TM domain of the fusion-defective GP64 construct C8, and each resulting construct was then analyzed for restoration of membrane fusion activity. (A) Construction of GP64 constructs. The TM domain sequences of the fusion-defective C8 construct and the other constructs are shown below that of the WT GP64; amino acids added to the C terminus of the TM domain of C8 are indicated in underlined bold, and the construct name is indicated on the left. (B) Expression and trimerization of modified GP64 proteins. Sf9 cells were transfected with plasmids expressing either WT GP64 or modified GP64 construct C8A, C8L, C8V, C8N, or C8R, and expression and trimerization of GP64 were examined by Western blot analysis of cell extracts on reducing (top) and nonreducing SDS-PAGE gels (bottom). The positions of oligomeric forms (Trimer I and II and Dimer) are indicated on the right. (C) Analysis of relative cell surface levels and fusion activities of GP64 constructs. Relative cell surface levels of GP64 constructs were measured by cELISA (as described in the Fig. 2 legend). Each mutant GP64 construct was expressed by transfecting 0.5 μg plasmid DNA encoding the modified GP64 construct. Relative fusion activity was evaluated by using efficiency of syncytium formation, as described earlier (Fig. 2 legend). For each sample, five fields were examined. The means and standard deviations of the results of triplicate transfections are shown. (D) Analysis of hemifusion and fusion pore formation by GP64 protein constructs. Fluorescence micrographs show either membrane dye transfer (R18) or cytosolic dye transfer (Calcein-AM). Each mutant GP64 construct (or mock control) used for transfection is indicated at the left, and assays were performed as described in Materials and Methods and the Fig. 3 legend.
Effects of TM domain deletions on infectious BV production.
To examine the effects of GP64 TM truncations on virus infectivity, each gp64 construct (under the transcriptional control of the WT GP64 promoter) was inserted into the polyhedrin locus of a gp64-null bacmid using standard Tn7-based transposition (27, 28). In addition to the gp64 gene construct, the donor plasmid used for transposition also encoded a GUS reporter gene under the control of the AcMNPV p6.9 promoter. As a positive control, the WT AcMNPV gp64 gene was inserted into the same donor plasmid and used to rescue the gp64-null bacmid. A similar donor plasmid containing no gp64 gene was used to generate a negative control bacmid. Following the generation of a bacmid encoding each gp64 gene construct, bacmid DNA was used to transfect Sf9 cells in a transfection-infection assay (28). Transfected and infected cells were scored for GUS activity, which is (i) an indirect indicator of viral replication when detected after transfection and (ii) an indicator of infectious virion production when detected after infection. Transfection of Sf9 cells with gp64-null bacmids carrying the gp64 TM deletion constructs C0, C4, C5, and C7 resulted in rescue of gp64-null AcMNPV and production of infectious virions (Fig. 1). In contrast, the gp64-null bacmids carrying the other TM-truncated gp64 genes did not rescue viral infectivity (Fig. 1, C8, C8CT, N7, N8, N8CT, M7, and M8CT). Interestingly, our previous analysis showed that gp64 protein constructs N7, N8, N8CT, M7, and M8CT were capable of mediating membrane fusion (Fig. 2C and D) but they did not rescue viral infectivity (Fig. 1). These results suggest that the TM amino acid sequence requirements for membrane fusion and infectivity are distinct. To ensure that all the gp64 protein constructs were expressed in Sf9 cells after transfection of the bacmids, lysates of the transfected Sf9 cells were analyzed by Western blot analysis. We detected expression of all of the gp64 protein constructs in the bacmid-transfected cells (data not shown). In addition, to confirm that any defects detected were not due to other lethal mutations in the bacmid, all bacmids encoding modified gp64 genes were transfected into Sf9Op1D cells, which constitutively express the OpMNPV GP64 protein. In each case, infectious virions were produced (data not shown), indicating that the lack of virion production in Sf9 cells was due to lack of a functional GP64 protein and not to a second-site mutation.
Budding efficiency with modified GP64 constructs.
To examine possible effects of the TM domain on virion budding, we examined virion budding from viruses expressing each GP64 TM truncation construct. Each bacmid expressing a GP64 TM truncation construct was amplified as a virus, and the titer determined in Sf9Op1D cells, which express WT OpMNPV GP64. The virus produced in Sf9Op1D cells (displaying OpMNPV GP64 on the virion) was then used to infect Sf9 cells where only the modified GP64 construct was expressed. To determine if expression of only the TM-truncated GP64 affected virion budding, the infected Sf9 cells were pulse labeled with [35S]methionine and the labeled progeny virions were collected and purified. The relative quantities of virions that accumulated in cell culture supernatants were estimated by quantifying the [35S]methionine label from the major virion capsid protein, VP39, as described previously (37) (see Materials and Methods). Comparison of each virus with a control virus expressing WT GP64 revealed that removal of the cytoplasmic tail or truncation or deletion of the putative TM domain of GP64 resulted in substantially decreased virus budding (Fig. 5B). The virus budding efficiencies measured for viruses expressing GP64 constructs C0, C4, and C5 were approximately 24 to 30% of that from a virus expressing WT GP64. With the exception of construct N8, deletion of seven or eight amino acids from the TM domain appeared to result in progressive reduction in virion budding. However, because background virion budding in the absence of GP64 may range from approximately 3 to 10% of that from WT AcMNPV (37; unpublished observations) and because only the bacmid expressing the C7 construct rescued infectivity, it is unclear whether the low levels of virions detected from bacmids expressing constructs C8, C8CT, N8CT, and M8CT represent background budding or low levels mediated by the GP64 construct. In addition, comparison of larger TM deletions (with and without CTD) suggests that the addition of the cytoplasmic tail in constructs C8CT and N8CT did not detectably augment budding efficiency.
FIG. 5.
Analysis of virion budding efficiency by viruses expressing TM-truncated GP64 constructs. (A) Analysis of labeled progeny virions. Viruses expressing TM-truncated GP64 constructs were pulse labeled with [35S]methionine, and progeny BV purified as described in Materials and Methods. Labeled viral proteins were analyzed by SDS-PAGE and phosphorimager analysis. GP64 constructs are indicated above the lanes, and the positions of GP64 and VP39 proteins are indicated between the panels. Controls include an AcMNPV virus expressing WT GP64 and an AcMNPV virus containing a GP64 knockout (Ko). Each lane represents virus purified from an equivalent volume of the cell culture supernatant. (B) Budding efficiencies of TM-truncated GP64 constructs. Relative budding efficiencies were determined by comparing the quantity of [35S]methionine label in the major capsid protein (VP39) band from each virion preparation to that from a virus expressing WT GP64. Means and standard deviations are shown. KO, GP64 knockout.
Analysis of single and multiple point mutations in the GP64 TM domain.
We previously used TM domain replacements (23) and deletions to examine functional requirements of the TM domain. To further investigate the role of the TM domain, we generated and examined the effects of small, 1- to 4-aa substitution mutations within the AcMNPV GP64 TM domain. We used two strategies for these studies. First, we performed an alanine scan of the GP64 TM domain, substituting two or three alanines for amino acids of the TM domain sequence (Fig. 6A). Next, we examined a number of specific amino acid positions based on sequence conservation among the GP64 and related proteins and on current models of membrane fusion protein function.
FIG. 6.
Analysis of alanine scanning mutations in the GP64 TM domain. (A) Schematic representation of the positions of GP64 alanine scanning mutations. Sets of two or three amino acids in the TM domain were replaced with alanines as indicated (dots represent positions with no change). The name of each alanine scanning GP64 construct is indicated on the left (numbers represent positions of modified amino acids in the AcMNPV GP64 sequence). The two regions important for cell surface expression of GP64 are indicated by bars above the WT GP64 TM domain sequence, and two regions important for virus budding are similarly indicated by bars below the sequence. ED, ectodomain; CT, cytoplasmic tail. (B) Expression and trimerization of GP64 constructs with alanine scanning mutations in the TM domain. Sf9 cells were infected with viruses expressing WT GP64 or the alanine scanning constructs of GP64 shown in panel A. Expression and trimerization of GP64 were examined by Western blot analysis of cell extracts on reducing SDS-PAGE (top) or nonreducing SDS-PAGE (bottom) gel. The positions of GP64 and oligomeric forms (Trimer I and II and Dimer) are indicated at the right. (C) Relative cell surface levels and fusion activities of GP64 constructs. Relative cell surface levels of GP64 constructs were measured by cELISA of infected Sf9 cells, using mAb AcV5. Relative fusion activity of each modified GP64 construct was evaluated by measuring the efficiency of syncytium formation in infected Sf9 cells. For each sample, five fields were examined. The means and standard deviations of the results of triplicate infections are illustrated. (D) Analysis of budding efficiency and incorporation of modified GP64 constructs into budded virions. Viruses expressing modified GP64 constructs were pulse labeled with [35S]methionine, and BV purified and processed as described in Materials and Methods. The positions of GP64 and VP39 proteins are indicated at the right. Each lane represents virus purified from an equivalent volume of the cell culture supernatant. (E) Relative budding efficiencies and incorporation of GP64 into budded virions. Relative virion budding efficiency was calculated by comparison of measurements of labeled major capsid protein (VP39) in each virus. All data were normalized to the results for a virus construct expressing WT GP64 (100%). The efficiency of GP64 incorporation into virions was calculated for each virus expressing a modified GP64 protein with an alanine scanning mutation. Molar ratios of GP64:VP39 were calculated for virions of each virus based on 15 methionine residues (mutant 503-505) and 16 methionine residues (mature WT GP64 and all other mutants) or 9 methionine residues (VP39). The means and standard deviations determined from triplicate experiments are shown.
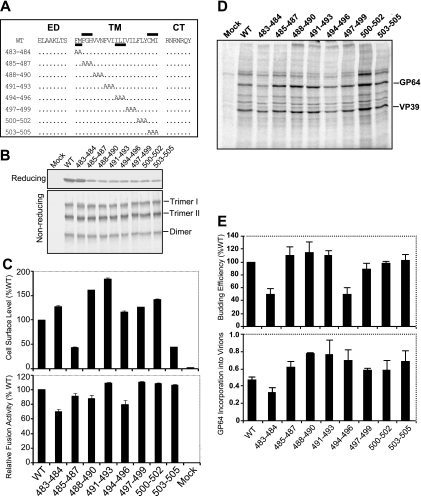
For alanine scanning mutagenesis of the GP64 TM domain, we replaced each two or three amino acids in the TM domain with alanine residues (Fig. 6A) and inserted each construct into a recombinant GP64-null virus of AcMNPV as described above. All GP64 constructs were initially examined by reducing and nonreducing gel electrophoresis and Western blot analyses to confirm that the substitution constructs were expressed and trimerized in a manner similar to that of WT GP64 in virus-infected Sf9 cells (Fig. 6B). Analysis of cell surface localization for these alanine scanning constructs showed that all constructs were expressed and localized at the cell surface, with surface levels ranging between approximately 45 and 185% of that measured from WT GP64 (Fig. 6C, upper panel). Two constructs (485-487 and 503-505) resulted in reduced surface levels of GP64, while the surface levels of all others were similar to or even greater than that of WT GP64. To determine if specific TM amino acid positions were required for the fusion function of GP64, the alanine scanning constructs of GP64 were examined in a syncytium formation assay. All alanine scanning constructs retained fusion activity, and the fusion activity mediated by these constructs was very similar to that of WT GP64, ranging from approximately 70 to 110% of that from WT GP64 (Fig. 6C, lower panel). In addition, analysis of one-step growth curves from viruses expressing the alanine scanning substitution constructions revealed that the infectious virus production from all viruses was similar to that from the control virus expressing the WT GP64 protein, with no dramatic differences from 6 h to 120 h p.i. (data not shown). To further examine possible effects of the TM domain alanine scanning substitutions on virion budding or incorporation of GP64 into virions, infected Sf9 cells were pulse labeled with [35S]methionine, progeny virions were purified, and relative levels of labeled progeny virions were estimated by measuring relative quantities of label in the major capsid protein, VP39, as described earlier. As shown in Fig. 6D, all of the GP64 constructs containing alanine scanning mutations in the TM domain were assembled into virions. The budding efficiencies of mutants 483-484 and 494-496 were decreased to around 50% of that of from WT virus, while the budding efficiencies measured for other constructs were similar to that of WT virus (Fig. 6E, top panel). Analysis of the incorporation of GP64 constructs into virions showed that GP64 incorporation was similar to (or in one case only slightly less) that observed for WT GP64. Thus, analysis of alanine scanning mutations identified no amino acid positions (or small regions) in the GP64 TM domain that were absolutely required for expression, trimerization, cell surface localization, membrane fusion, virion budding, targeting to virions, or viral infectivity. However, we identified two regions that affected cell surface localization and two regions that affected virion budding efficiency (Fig. 6C and E).
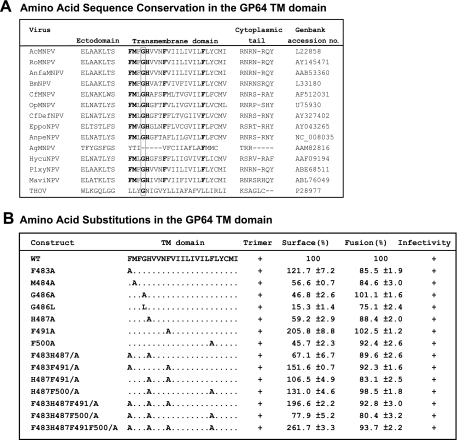
The GP64 protein is very highly conserved within the group I NPVs of the Baculoviridae, and this high level of conservation includes the TM domain (Fig. 7A). Because our initial studies showed that the GP64 TM domain could not be functionally replaced by TM domains from other (nonbaculovirus or unrelated) viral or cellular-membrane proteins, we concluded that the TM domain sequence had important functions beyond that of simply anchoring the protein in the membrane. Previous studies revealed that glycine residues within the TM domain of the VSV G protein play a critical role in membrane fusion and appear to function in the transition from hemifusion to complete membrane fusion (10). Indeed, glycine residues appear to be overrepresented in the TM domains of viral fusion proteins (2, 46). Where glycines are not present in such TM domains, it has been proposed that methionine residues may functionally substitute. In addition, more recently it was reported that phenylalanine residues in combination with glycine residues contribute to TM domain interactions (53). Because the GP64 TM domain contains only a single conserved glycine residue and multiple phenylalanine residues, we generated alanine substitution mutations in single or multiple positions to examine the potential function of the glycine or phenylalanine residues in GP64 function (Fig. 7). Substitutions for the Met residue at the N terminus of the TM domain and the single charged histidine residue were also examined either alone or in combination with other substitutions. The single and multiple substitution mutations are shown in Fig. 7B. All modified GP64 constructs were inserted into recombinant viruses (replacing native GP64) and examined for trimer formation, localization at the cell surface, fusion activity (syncytium formation efficiency), and virus infectivity (Fig. 7B). In viruses containing the GP64 TM domain substitutions of single and multiple amino acids (Fig. 7B), the GP64 proteins behaved similarly to WT GP64, with no substantial differences observed. A minor difference observed was that levels of cell surface GP64 were moderately reduced for some of the substitution mutations. Replacement of Met 484, Gly 486, His 487, or Phe 500 resulted in decreased cell surface levels (40 to 75% of the level with WT GP64) (Fig. 7B), although the fusion activity detected remained high. Thus, consistent with the alanine scanning analysis, our analysis of single and multiple amino acid substitutions at conserved residues or those identified as important in other viral membrane fusion proteins did not identify amino acids that were absolutely required for GP64 function in membrane fusion or other critical functions.
FIG. 7.
Analysis of GP64 conservation and single and multiple amino acid substitutions in the TM domain revealed no amino acid positions essential for membrane fusion or viral infectivity. (A) Sequence alignment of the TM domains of baculovirus GP64 proteins and thogotovirus GP75. Sequence alignment was performed using the program Clustal W. The position of the highly conserved glycine is boxed. Conserved methionine, histidine, and phenylalanine positions are shown in boldface. The GenBank accession number is listed for each virus. Abbreviations are as follows: RoMNPV, Rachiplusia ou MNPV; AnfaNPV, Anagrapha falcifera NPV; BmNPV, Bombyx mori NPV; CfMNPV, Choristoneura fumiferana MNPV; CfDefNPV, Choristoneura fumiferana defective NPV; EppoNPV, Epiphyas postvittana NPV; AnpeNPV, Antheraea pernyi NPV; AgNPV, Anticarsia gemmatalis NPV; HycuNPV, Hyphantria cunea NPV; PlxyMNPV, Plutella xylostella MNPV; MaviMNPV, Maruca vitrata MNPV; THOV, thogotovirus. (B) Construction and summary of effects of single and multiple site substitution mutations in the AcMNPV GP64 TM domain. The results of trimerization and infectivity assays are shown as + (positive) or − (negative). Relative cell surface levels were measured by cELISA, and relative fusion activities were evaluated using efficiency of syncytium formation from virus-infected Sf9 cells, as described in Materials and Methods. The means and standard deviations of the results of triplicate infections are shown. Infectivity was determined by transfection-infection assays as described previously (28).
DISCUSSION
For baculoviruses that encode a GP64 protein, GP64 is essential for the entry of the budded form of the virus. Compared with the highly conserved ectodomains of GP64 proteins, the TM domains are more variable in length and amino acid sequence similarity. This suggests the possibility that TM domains of GP64 proteins have conserved structural characteristics that are not apparent from an analysis of the primary amino acid sequence. The recently described structure of the postfusion form of GP64 (21) does not include the TM domain, and TM domain structures are generally rare. Recently, we found that replacement of the predicted 23-aa GP64 TM domain with corresponding TM domain sequences from a range of viral or cellular type I membrane proteins or with a GPI anchor addition sequence resulted in defects in transport, membrane fusion, virion budding, and virus infectivity (23). Only TM domains from two related viral membrane proteins (OpMNPV GP64 and thogotovirus GP75) functionally substituted for the AcMNPV GP64 TM domain sequence. These observations indicate that the TM domain is critical for the function of GP64.
To examine potential length or sequence requirements for the biological functions of the AcMNPV GP64 TM domain, we generated GP64 proteins containing truncated forms of the TM domain by removing four to eight amino acids from the N-terminal, C-terminal, or central regions of the TM domain. Although the CTD has been shown to be dispensable for fusion activity (37), we also generated GP64 constructs containing the CTD, since the addition of a cytoplasmic tail may force a truncated TM domain to span the bilayer membrane and promote membrane fusion (3). All GP64 proteins with truncated TM domains were synthesized and oligomerized in transfected cells (Fig. 1B); however, the trimer profiles of some constructs containing C-terminal deletions of the TM domain appeared to differ from that of WT GP64. The GP64 trimer is typically observed as two distinct electrophoretic forms. While the two trimer forms (I and II) have different migration rates in nonreducing SDS-PAGE, the two forms appear to have the same mass, as determined by mass spectrometry (38). It has been suggested that the two trimer forms may represent disulfide isomers (26), but it is not known whether they differ in any functional properties. The cell surface levels of TM-truncated GP64 constructs were dramatically reduced (≥67%) and appeared to decrease with successive reductions in length of the TM domain (Fig. 2). Similar results were previously reported for human immunodeficiency virus envelope glycoprotein and VSV G constructs that contained truncated TM domains (1, 39).
Recent studies of TM model peptides suggest that peptide-membrane interactions depend on peptide hydrophobicity and on the matching of hydrophobic peptide length with membrane thickness. A negative hydrophobic mismatch occurs when the hydrophobic stretch of a peptide is too short with respect to the membrane thickness (11, 13, 45, 49, 57). The observed shedding of GP64 proteins with truncated TM domains (Fig. 1B) may result from such a negative hydrophobic mismatch. Addition of the cytoplasmic tail significantly increased the cell surface levels of C8CT and N8CT and also moderately increased surface levels of M8CT (Fig. 2A), suggesting that addition of the CTD may force the truncated TM domain to adapt its structure or orientation to overcome the mismatch, as suggested for some model TM peptides (12, 15, 47). The length of the GP64 TM domain also had an effect on its fusogenicity. With successive truncations, the level of membrane fusion activity also decreased successively compared to that for WT GP64 (normalized activity). Compared with normalized WT GP64, we found that the fusion activity decreased moderately (15 to 20%) for GP64 constructs C4 and C5 and dramatically for C7 and C8 (Fig. 2D). No fusion activity was detected for C8, a construct containing only 15 amino acids of the TM domain. The N-terminal and middle regions of the TM domain were similarly sensitive, since deletion of seven or eight amino acids from those regions also resulted in dramatically reduced fusion activity (77 to 90% reduction). While addition of the CTD increased the fusion activities of N8CT and M8CT in comparison to those of N8 and M8, the fusion activities of N8CT and M8CT were still reduced by more than 90% compared with that of WT GP64. Addition of the CTD did not rescue fusion activity of C8, which was fusion negative. Because the C-terminal portion of the TM domain is somewhat more hydrophobic than the N-terminal region (as measured by hydrophobicity plots), removing the most-hydrophobic portion of the TM domain may account for both the lack of detectable fusion activity in C8 and the failure of C8CT to rescue fusion activity. Thus, these data suggested that GP64-mediated fusion requires a critical TM domain length of 15 or 16 amino acids. The critical nature of the hydrophobic length was confirmed by experiments in which a single hydrophobic amino acid (A, L, or V) was added to the C terminus of mutant C8, resulting in restored fusion activity (Fig. 4C). The addition of hydrophilic or charged amino acids did not restore activity. Interestingly, the fusion activities of the C8 mutants extended with L or V were considerably less than that of the C7 mutant that contains an I at the same position. It is of interest that in the case of the WT C7 sequence or C8L, C8V, and C8A (which restored fusion), the relative level of fusion correlates with the degree of hydrophobicity of the respective C-terminal amino acid, I, L, V, or A.
We also asked which step in membrane fusion was affected by truncations in the TM domain. We used lipid and cytosolic dyes to separately examine outer leaflet merger (the hemifusion intermediate) and pore formation, focusing on the fusion-deficient construct, C8 (a 15-aa TM domain). Using membrane and cytosolic dye labeling, we found that construct C8 induced only approximately 1% lipid dye transfer efficiency (hemifusion) compared with that of WT GP64. Addition of the 7-aa hydrophilic cytoplasmic tail did not restore fusion activity by the C8 mutant and appeared to eliminate the very low level of hemifusion. These results differ from results reported for the influenza HA protein (3). In that case, truncation of 12 amino acids from the C-terminal end of the HA TM domain (resulting in a 15-aa TM domain) did not substantially affect lipid dye transfer (hemifusion) but reduced pore formation to approximately 5% of that of the WT HA. A similar result was obtained when the deletion was from the N-terminal end of the HA TM domain. Thus, HA proteins containing TM domains of ≤15 amino acids were unable to mediate complete fusion (pore formation), but the first step (membrane merger or hemifusion) was not substantially affected. In contrast, the GP64 TM domain containing only 15 amino acids (constructs C8 and C8CT) was severely defective in the first step, membrane merger. In a recent study (23), we found that replacement of the TM domain of GP64 with either the 21-aa TM domain of OpMNPV F protein or the 23-aa TM domain of influenza virus HA resulted in chimeric GP64s that induce low-efficiency hemifusion but not pore formation. While those data suggested that the hemifusion step was largely independent of the TM domain, our current observation that TM-truncated constructs C8 and C8CT are severely compromised or defective in their ability to induce hemifusion further suggests that a critical hydrophobic length may be important for that initial hemifusion step.
Some differential effects were also observed when truncations were made from the N-terminal or middle regions of the GP64 TM domain. In those cases (N8, N8CT, M8, and M8CT), very low levels of membrane fusion were observed and fusion efficiency increased upon addition of the CTD (Fig. 2D).
Another consideration in the interpretation of results in studies such as this is the contribution or interaction of the specific lipid bilayer of the host cell membrane. In several cases, it has been demonstrated that the lipid composition of the membrane can alter peptide interactions and fusion activity (13, 40, 48, 56). The lipid composition in Sf9 cell membranes differs significantly from that of mammalian cells (29), and these differences may be associated with the lower temperatures typical of insect cell culture or insect growth. Differences in lipid composition of the membranes may explain the observed differences in fusion by the truncated GP64 and HA constructs. Another possible explanation for the observed differences is a difference in the mechanics of protein-mediated membrane fusion. Plonsky and Zimmerberg (43) found that, unlike HA-mediated fusion, AcMNPV GP64-induced fusion pores open rapidly and irreversibly and do not flicker. In addition, the initial fusion pores formed by GP64 appeared to be larger than those of HA (43). Interestingly, the minimal TM domain length for fusogenicity is in the same range for different TM peptides: 17 residues for HA (3), 16 residues for the GP64 TM domain, and between 14 and 18 residues for TM model peptides (26). Effects of the TM domain length on the fusion process were also described previously for other fusion proteins, such as those from human immunodeficiency virus (39), foamy virus (41), murine coronavirus (6), and murine leukemia virus (44), and for SNARE proteins (58).
In addition to membrane fusion, we also examined the potential roles of the TM domain in virion assembly during budding. An analysis of virus budding efficiency revealed that virus budding efficiency decreased 70 to 75% upon deletion of the cytoplasmic tail. Truncation of an additional four or five amino acids from the C-terminal end of the TM domain (constructs C4 and C5) resulted in a similarly reduced budding efficiency. Further truncation or deletion of seven or eight amino acids of the TM domain resulted in even more dramatically reduced virus budding efficiency (reductions of 86 to 96% compared to that with WT GP64) (Fig. 5B). While the reduced budding efficiency of truncated GP64 constructs appeared to correlate with reduced surface levels of the GP64 constructs, the addition of the CTD (which restored surface levels and fusion activity) (Fig. 2A and D) did not rescue the budding defects. We also examined the capacity of GP64 proteins with truncated TM domains to substitute for WT GP64, using a transfection-infection assay in Sf9 cells. Infectivity was rescued by constructs containing 16 amino acids from the N-terminal portion of the TM domain but not by constructs containing 16 amino acids from other parts of the TM domain (i.e., constructs containing deletions from the N-terminal or middle regions were not able to substitute for WT GP64) (Fig. 1, C7 versus N7 and M7). By comparing fusion assay results to results from virus infectivity studies, we can conclude that the sequence requirements for infectivity are more stringent than those for membrane fusion, which in turn are more stringent than the requirements for membrane anchoring and intracellular transport.
To further examine the GP64 TM domain and identify specific residues important for GP64 function, we generated and analyzed single and multiple amino acid substitution mutations in the TM domain. We specifically focused on conserved methionine (M484), glycine (G486), histidine (H487), and phenylalanine (F483, F487, F491, and F500) residues of the TM domain of GP64 (Fig. 7B). We also examined small regions of two to three amino acids by using an alanine scan (Fig. 6A). The glycine at 486 is conserved among almost all known baculovirus GP64 proteins and the more-distantly related thogotovirus GP75. The results of previous studies (10) suggested that at least one of the two glycine residues of VSV G protein plays a significant role in the transition from hemifusion to complete fusion. Furthermore, a study of influenza virus HA, which contains two glycines within the TM domain, demonstrated that substitution of a leucine (G250L) for the more-N-terminal glycine caused a restricted hemifusion phenotype (33). It has been proposed that these important glycine residues may function as helix breakers and thereby distort the bilayer, promoting membrane fusion. This concept is supported by the fact that a conserved proline residue (also a helix breaker) is found in the TM domains of foamy virus and murine leukemia virus envelope proteins and is essential for fusion function in both instances (41, 51). In the current study, substitution of alanine or leucine for the conserved glycine resulted in reduced cell surface levels of those GP64 constructs (Fig. 6, 485-487 and 7B), but the fusion activities of those constructs (485-487 and G486A and G486L mutants) were similar to or only slightly decreased from that of WT GP64. Similar results were found for Semliki Forest virus E1 protein. Indeed, neither of the two conserved glycine residues nor any of the five total glycine residues within the TM domain of E1 are required for fusion activity (24).
Since many or most of the Gly-less TM domains of fusion proteins possess internal methionine residues (10), we also examined the conserved methionine at position 484. Substitution of alanine for Met 484 resulted in similar or decreased levels of the GP64 construct at the cell surface but had only subtle effects on membrane fusion (Fig. 6, 483-484, and 7, M484A). We also examined the histidine (H487), based on the reported role of a charged residue within the TM domain of foamy virus envelope protein (41). Pietschmann et al. (41) found that an evolutionarily conserved positively charged amino acid, K959, within the putative TM domain of foamy virus appears to regulate fusion activity. We found no substantial effect from two substitution approaches for the GP64 TM domain histidine residue (Fig. 6, 485-487, and 7B, H487A). Recently, Unterreitmeier et al. (53) used a randomized library to biologically select TM domains that self-interact and found that higher-affinity TM domain sequences were enriched in phenylalanine. In addition, phenylalanine is frequently found associated with GxxxG motifs in TM domains of self-interacting proteins. Notably, disruption of the FxxGxxxG motif of the VSV G protein by substitution for phenylalanine or glycine residues resulted in reduced TM-TM associations (53). Substitutions for the conserved phenylalanines within the TM domain of GP64 (singly or in various combinations with H487) did not cause dramatic effects on fusion activity, although the cell surface levels were somewhat reduced for GP64 substitution construct F500A (Fig. 7B).
Using the 2- or 3-alanine scanning substitutions (Fig. 6), we found two regions, residues 485 to 487 and 503 to 505, that were important for cell surface localization of GP64. Substitution of alanines for either of these two regions resulted in cell surface levels that were decreased by more than 50% (Fig. 6C). In two additional regions, positions 483 to 484 and 494 to 496, substitution mutations resulted in virus budding that was reduced by more than 50%.
While many reports describe critical roles for the TM domains of viral fusion proteins, no uniform theme has emerged from these studies. In the case of influenza virus HA, it is clear that a TM anchor is required for full fusion activity (32, 34), but a variety of TM sequences will substitute for the native TM domain (33). In contrast, specific sequence requirements appear to be encoded within the TM domains of other fusion proteins, such as the envelope proteins from human immunodeficiency virus type I (39), murine leukemia virus (51), foamy viruses (41), coronavirus (6, 8), VSV (10), Newcastle disease virus (31), and measles virus (7). In the case of the AcMNPV GP64 protein, the results of prior studies indicated that, like the later cases, the TM domain sequence includes critical functions (23). In the present more-detailed study of the TM domain sequence, we reached two broad conclusions. First, we found that the TM domain of the GP64 protein requires a critical minimum hydrophobic length of 16 amino acids. The requirement for this length is not only related to protein anchoring but, more importantly, represents a requirement for the function of GP64 as a fusion protein. Second, our analysis of TM domain sequences by substitution mutations in conserved positions and alanine scanning show that no single amino acid in the TM domain is absolutely required for GP64 function in any of its critical roles in the virus. Combined with the results of prior studies of substitutions for the entire TM domain, the results of these studies suggest that either (i) the GP64 TM domain may include critical amino acids or sequence elements that are separate but redundant or (ii) the overall sequence forms a higher-order structure that is not disrupted by single substitutions or our alanine scanning approach, yet cannot be replaced by an unrelated heterologous TM domain. Future studies aimed at examining the higher-order structure of the GP64 TM domain may provide insight into the relationship between TM domain structure and the complex and critical process of protein-mediated membrane fusion.
Acknowledgments
We thank Gerrit Heetderks and Jian Zhou for expert technical assistance.
This work was supported by NIH grant AI33657 and BTI projects G01707-R06-1255 and B00103-R06-1255.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Adams, G. A., and J. K. Rose. 1985. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell 411007-1015. [DOI] [PubMed] [Google Scholar]
- 2.Arkin, I. T., and A. T. Brunger. 1998. Statistical analysis of predicted transmembrane alpha-helices. Biochim. Biophys. Acta 1429113-128. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 655820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 666829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, E. C., L. Heijnen, W. Luytjes, and W. J. Spaan. 1995. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology 214453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caballero, M., J. Carabana, J. Ortego, R. Fernandez-Munoz, and M. L. Celma. 1998. Measles virus fusion protein is palmitoylated on transmembrane-intracytoplasmic cysteine residues which participate in cell fusion. J. Virol. 728198-8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, K. W., Y. Sheng, and J. L. Gombold. 2000. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 269212-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, J., P. E. March, R. Lee, and D. Tillett. 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 953425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Planque, M. R., J. W. Boots, D. T. Rijkers, R. M. Liskamp, D. V. Greathouse, and J. A. Killian. 2002. The effects of hydrophobic mismatch between phosphatidylcholine bilayers and transmembrane alpha-helical peptides depend on the nature of interfacially exposed aromatic and charged residues. Biochemistry 418396-8404. [DOI] [PubMed] [Google Scholar]
- 12.de Planque, M. R., E. Goormaghtigh, D. V. Greathouse, R. E. Koeppe II, J. A. Kruijtzer, R. M. Liskamp, B. de Kruijff, and J. A. Killian. 2001. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry 405000-5010. [DOI] [PubMed] [Google Scholar]
- 13.de Planque, M. R., and J. A. Killian. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20271-284. [DOI] [PubMed] [Google Scholar]
- 14.Epand, R. M. 2003. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 1614116-121. [DOI] [PubMed] [Google Scholar]
- 15.Harzer, U., and B. Bechinger. 2000. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: a CD, 15N and 31P solid-state NMR spectroscopy investigation. Biochemistry 3913106-13114. [DOI] [PubMed] [Google Scholar]
- 16.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258455-468. [DOI] [PubMed] [Google Scholar]
- 17.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48211-234. [DOI] [PubMed] [Google Scholar]
- 18.Hink, W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226466-467. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125432-444. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis, D. L., L. Wills, G. Burow, and D. A. Bohlmeyer. 1998. Mutational analysis of the N-linked glycans on Autographa californica nucleopolyhedrovirus gp64. J. Virol. 729459-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 151024-1030. [DOI] [PubMed] [Google Scholar]
- 22.Langosch, D., M. Hofmann, and C. Ungermann. 2007. The role of transmembrane domains in membrane fusion. Cell. Mol. Life Sci. 64850-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., and G. W. Blissard. 2008. Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains. J. Virol. 823329-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao, M., and M. Kielian. 2005. The conserved glycine residues in the transmembrane domain of the Semliki Forest virus fusion protein are not required for assembly and fusion. Virology 332430-437. [DOI] [PubMed] [Google Scholar]
- 25.Long, G., X. Pan, R. Kormelink, and J. M. Vlak. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 808830-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorin, A., B. Charloteaux, Y. Fridmann-Sirkis, A. Thomas, Y. Shai, and R. Brasseur. 2007. Mode of membrane interaction and fusogenic properties of a de novo transmembrane model peptide depend on the length of the hydrophobic core. J. Biol. Chem. 28218388-18396. [DOI] [PubMed] [Google Scholar]
- 27.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site- specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 674566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 765729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marheineke, K., S. Grunewald, W. Christie, and H. Reilander. 1998. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 44149-52. [DOI] [PubMed] [Google Scholar]
- 30.Maruniak, J. W., and M. D. Summers. 1981. Autographa californica nuclear polyhedrosis virus phosphoproteins and synthesis of intracellular proteins after virus infection. Virology 10925-34. [DOI] [PubMed] [Google Scholar]
- 31.McGinnes, L., T. Sergel, and T. Morrison. 1993. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology 196101-110. [DOI] [PubMed] [Google Scholar]
- 32.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 101821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 692583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monsma, S. A., A. G. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 704607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254297-314. [DOI] [PubMed] [Google Scholar]
- 38.Oomens, A. G. P., S. A. Monsma, and G. W. Blissard. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209592-603. [DOI] [PubMed] [Google Scholar]
- 39.Owens, R. J., C. Burke, and J. K. Rose. 1994. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 68570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozdirekcan, S., D. T. Rijkers, R. M. Liskamp, and J. A. Killian. 2005. Influence of flanking residues on tilt and rotation angles of transmembrane peptides in lipid bilayers. A solid-state 2H NMR study. Biochemistry 441004-1012. [DOI] [PubMed] [Google Scholar]
- 41.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 744474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plonsky, I., M. S. Cho, A. G. P. Oomens, G. W. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 25365-76. [DOI] [PubMed] [Google Scholar]
- 43.Plonsky, I., and J. Zimmerberg. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 1351831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragheb, J. A., and W. F. Anderson. 1994. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J. Virol. 683207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren, J., S. Lew, Z. Wang, and E. London. 1997. Transmembrane orientation of hydrophobic alpha-helices is regulated both by the relationship of helix length to bilayer thickness and by the cholesterol concentration. Biochemistry 3610213-10220. [DOI] [PubMed] [Google Scholar]
- 46.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296921-936. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe, S., K. R. Barber, C. W. Grant, D. Goodyear, and M. R. Morrow. 2002. Organization of model helical peptides in lipid bilayers: insight into the behavior of single-span protein transmembrane domains. Biophys. J. 83345-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparr, E., W. L. Ash, P. V. Nazarov, D. T. Rijkers, M. A. Hemminga, D. P. Tieleman, and J. A. Killian. 2005. Self-association of transmembrane alpha-helices in model membranes: importance of helix orientation and role of hydrophobic mismatch. J. Biol. Chem. 28039324-39331. [DOI] [PubMed] [Google Scholar]
- 49.Strandberg, E., S. Ozdirekcan, D. T. Rijkers, P. C. van der Wel, R. E. Koeppe II, R. M. Liskamp, and J. A. Killian. 2004. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 863709-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamm, L. K., J. Crane, and V. Kiessling. 2003. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr. Opin. Struct. Biol. 13453-466. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, G. M., and D. A. Sanders. 1999. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Biol. Cell 102803-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ujike, M., K. Nakajima, and E. Nobusawa. 2004. Influence of acylation sites of influenza B virus hemagglutinin on fusion pore formation and dilation. J. Virol. 7811536-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unterreitmeier, S., A. Fuchs, T. Schaffler, R. G. Heym, D. Frishman, and D. Langosch. 2007. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J. Mol. Biol. 374705-718. [DOI] [PubMed] [Google Scholar]
- 54.Volkman, L. E., and P. A. Goldsmith. 1984. Budded Autographa californica NPV 64K protein: further biochemical analysis and effects of postimmunoprecipitation sample preparation conditions. Virology 139295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143185-195. [DOI] [PubMed] [Google Scholar]
- 56.Webb, R. J., J. M. East, R. P. Sharma, and A. G. Lee. 1998. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: a possible mechanism for retention in the Golgi. Biochemistry 37673-679. [DOI] [PubMed] [Google Scholar]
- 57.Weiss, T. M., P. C. van der Wel, J. A. Killian, R. E. Koeppe II, and H. W. Huang. 2003. Hydrophobic mismatch between helices and lipid bilayers. Biophys. J. 84379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y., F. Zhang, Z. Su, J. A. McNew, and Y. K. Shin. 2005. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12417-422. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, S. X., Y. Han, and G. W. Blissard. 2003. Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts. J. Virol. 776265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, J., and G. W. Blissard. 2008. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus AcMNPV. J. Virol. 824449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, J., and G. W. Blissard. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]