Abstract
Cytoplasmic viral RNAs with 5′ triphosphates (5′ppp) are detected by the RNA helicase RIG-I, initiating downstream signaling and alpha/beta interferon (IFN-α/β) expression that establish an antiviral state. We demonstrate here that the hepatitis C virus (HCV) 3′ untranslated region (UTR) RNA has greater activity as an immune stimulator than several flavivirus UTR RNAs. We confirmed that the HCV 3′-UTR poly(U/UC) region is the determinant for robust activation of RIG-I-mediated innate immune signaling and that its antisense sequence, poly(AG/A), is an equivalent RIG-I activator. The poly(U/UC) region of the fulminant HCV JFH-1 strain was a relatively weak activator, while the antisense JFH-1 strain poly(AG/A) RNA was very potent. Poly(U/UC) activity does not require primary nucleotide sequence adjacency to the 5′ppp, suggesting that RIG-I recognizes two independent RNA domains. Whereas poly(U) 50-nt or poly(A) 50-nt sequences were minimally active, inserting a single C or G nucleotide, respectively, into these RNAs increased IFN-β expression. Poly(U/UC) RNAs transcribed in vitro using modified uridine 2′ fluoro or pseudouridine ribonucleotides lacked signaling activity while functioning as competitive inhibitors of RIG-I binding and IFN-β expression. Nucleotide base and ribose modifications that convert activator RNAs into competitive inhibitors of RIG-I signaling may be useful as modulators of RIG-I-mediated innate immune responses and as tools to dissect the RNA binding and conformational events associated with signaling.
The initial reaction of mammalian cells to invading viral pathogens is an innate immune response mediated by host pattern recognition receptors that recognize pathogen-associated molecular patterns (PAMPs) (3). PAMPs presented during a viral infection include DNA and double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA). dsRNA PAMPs can exist in the form of the viral genome or as a replication intermediate (24, 31). Viral dsRNA is recognized in the late endosome by Toll-like receptor 3 (TLR3) (2), while uridine-rich short ssRNA is recognized by TLR7 and TLR8 in the endosomal compartments of plasmacytoid dendritic cells (8). Cytoplasmic 5′ triphosphorylated ssRNA and short dsRNA are recognized by the RNA helicase retinoic acid-inducible gene I (RIG-I) (13, 14, 22). Long cytoplasmic dsRNA, such as poly(I:C), is recognized by the related helicase melanoma differentiation-associated antigen 5 (MDA5) (14, 15). Studies with RIG-I knockout mice have shown that RIG-I is required for IFN production in response to several paramyxoviruses, the orthomyxovirus influenza virus, and the flavivirus Japanese encephalitis virus, whereas MDA5 is required for responding to picornaviruses (15, 20). RIG-I and MDA5 are individually dispensable for signaling in response to reovirus or dengue virus (DEN) infection (20). RIG-I and MDA5 cooperate to trigger an innate immune response to West Nile virus (WNV) (10).
Cytosolic 5′ triphosphates (5′ppp) generated during viral-RNA transcription or replication are required for RIG-I activation, suggesting that the 5′ppp may be a structural feature that distinguishes viral RNA from self RNA in virus-infected cells (13, 22). Capping/removing the 5′ppp or inserting modified nucleotides into the RNA abrogate RIG-I signaling (13). While capping or the absence of a 5′ppp likely prevents RNA binding to the RIG-I C terminus (7, 30), the mechanisms underlying the effects of nucleotide modifications on RIG-I activity have not been elucidated. In particular, it is not known how nucleotide modifications affect RIG-I binding.
RIG-I and MDA5 contain two N-terminal caspase activation and recruitment domains (CARDs) in addition to a helicase/ATPase domain (34). Recent data suggest that the helicase activity of RIG-I is inversely correlated with its downstream signaling activity (30), whereas RIG-I ATPase activity appears to be directly correlated with its downstream signaling activity (6, 11, 30, 34). RIG-I also contains a C-terminal domain that recognizes the 5′ppp of ssRNA and coincides with a repressor domain (RD) that regulates RIG-I signaling (7, 25, 30). After RIG-I binds to viral RNA, it undergoes conformational changes that promote self-association (25). Subsequently, RIG-I binds to the mitochondrial-membrane-associated IPS1 protein via CARD-CARD interactions, thereby activating IPS1 (16). This sets off a signaling cascade, resulting in activation of the transcription factors interferon regulatory factor 3 and NF-κB, which then induce alpha/beta interferon (IFN-α/β) production and the subsequent induction of IFN-stimulated genes (ISGs). IFN-α/β production leads to a cytotoxic response, promotes an antiviral state in neighboring uninfected cells, and helps stimulate the subsequent adaptive immune response (4).
Hepatitis C virus (HCV) is a positive-sense ssRNA virus in the family Flaviviridae. RIG-I recognizes the HCV 5′ and 3′ untranslated regions (UTRs) in TLR3-deficient human hepatoma (Huh7) cells (18, 29). RIG-I activation by 5′ppp-containing HCV ssRNAs was recently linked to sequence composition and length, specifically homopolyuridine and homopolyriboadenine motifs longer than 50 nucleotides (nt) (26). Although RIG-I signaling is activated during infection by other members of the family Flaviviridae (e.g., DEN and WNV) (6, 9, 10, 20), the relative activities of Flaviviridae RNAs as RIG-I activators have not been compared. We show here that the HCV 3′-UTR RNA is a significantly more potent RIG-I activator than 5′ppp-containing DEN, WNV, or yellow fever virus (YFV) UTR RNAs. Similar to the results of recent experiments using the HCV strain Con1 3′-UTR RNAs (26), our data confirm that the uridine-rich poly(U/UC) region of the HCV J4L6 strain 3′ UTR is the determinant for this robust immunostimulation and that its antisense sequence, poly(AG/A), is an equivalent RIG-I activator. Surprisingly, the poly(U/UC) sequence from the 3′ UTR of the fulminant HCV JFH-1 strain was a relatively weak activator, while the antisense JFH-1 strain poly(AG/A) RNA was very potent.
To gain a deeper understanding of RNA features recognized by RIG-I, we tested RNAs of various lengths and sequence compositions. Although poly(U) 50-nt or poly(A) 50-nt RNAs were inactive, the insertion of a single C or G nucleotide, respectively, increased RIG-I-mediated signaling significantly, suggesting that both sequence and length influence signaling activity. Nucleoside modifications in the base or at the 2′ ribose position have been reported to abrogate innate immune signaling through both TLR7/8 and RIG-I pathways (8, 27). To probe the mechanism of the effect on RIG-I signaling, we substituted pseudouridine for uridine or replaced uridine 2′ hydroxyls with fluoro groups in HCV 3′-UTR RNAs. The data demonstrate that, although stimulation of IFN-β is abrogated, the RIG-I-RNA binding interaction was not diminished significantly. Furthermore, the modified poly(U/UC) RNAs behave as competitive inhibitors of RIG-I binding and IFN-β induction. The data extend the range of known RNA features associated with RIG-I-mediated activation of innate immune signaling, provide a possible correlation between HCV 3′-UTR sequence identity and virulence, and describe modified RNAs that could modulate innate immune stimulation or could be used as tools to dissect specific steps in the activation process.
MATERIALS AND METHODS
Cells and viruses.
Huh7 cells were provided by P. Yang (Harvard Medical School) and S. Behrens (Fox Chase Cancer Center). Huh7 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and a 1% standard antibiotic-antimycotic solution (Invitrogen). Wild-type and RIG-I−/− murine embryonic fibroblasts (MEFs) were gifts from J. Jung (University of Southern California). The MEFs were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, a 1% standard antibiotic-antimycotic solution (Invitrogen), and 200 μg ml−1 of neomycin. Sendai virus strain Cantel was obtained from Charles River (North Franklin, CT). The vesicular stomatitis reporter virus (VSV-luc) was provided by S. Whelan (Harvard Medical School) and will be described in detail elsewhere.
DNA methods.
Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen). The pIFN-β-luc plasmid was a gift from J. Jung (University of Southern California). The pCMV-luc plasmid was purchased from Promega. T7-HCV 3′-UTR (genotype 1b, strain J4L6) and T7-HCV X in vitro transcription plasmids were gifts from R. Chung (Massachusetts General Hospital). pNS3/4A (ss1 RNA) and pEFBos FLAG-RIG-I plasmids were gifts from M. Gale (University of Washington—Seattle). The ss1 RNA corresponds to nt 3423 to 3772 of the HCV genome (29).
RNA methods.
DEN 5′- and 3′-UTR RNAs and 3′-stem-loop (SL) RNA were derived from serotype 4 virus strain 814669 (AF326573.1). The WNV 3′-SL RNA was derived from strain NY99 (FJ411043.1). The YFV 3′-SL RNA was derived from vaccine strain 17D (X03700.1). Poly(U/UC) RNA was transcribed from a T7 PCR product generated using T7-HCV 3′-UTR plasmid, a forward primer with a flanking T7 promoter sequence, and a reverse primer (21). Poly(U/UC)-ss1 and ss1-poly(U/UC) chimeric RNAs were generated by PCR using pNS3/4A and T7-HCV 3′-UTR plasmids as templates, one pair of external primers (with a forward primer containing T7 promoter sequence), and one pair of internal primers (both chimeric oligonucleotides). The recombinant PCR products were cloned in T vector (Promega) and sequenced. Correct clones were amplified in Escherichia coli, and plasmid DNA was harvested using a QiaFilter Plasmid Maxi kit (Qiagen). The plasmids were digested to release their inserts, the insert fragments were gel purified, and the purified inserts were then used as templates for transcription using the T7 Megashortscript kit (Ambion). J4L6 poly(U/UC) 80-nt, 70-nt, and 60-nt deletion RNAs were generated using the Milligan transcription method (21), where a T7 promoter primer was annealed to an oligonucleotide with reverse complementary sequence to generate a partially double-stranded template. The Milligan transcription method was also used to generate J4L6 poly(AG/A) 100-nt, J4L6 poly(AG/A) 60-nt, poly(G/GC) 60-nt, poly(U) 50-nt, poly(U) 35-nt, poly(A) 50-nt, U33CU16, A33GA16, JFH-1 poly(U/UC), and JFH-1 poly(AG/A) RNAs. All of these in vitro-transcribed RNAs contain three guanines at the extreme 5′ end to facilitate transcription by the T7 polymerase. All other unmodified RNAs were transcribed from linearized plasmid DNA templates using the T7 Megashortscript kit (Ambion). Biotinylated RNAs were transcribed using the T7 Megashortscript kit and biotin-11-cytidine-5′ppp (Roche Diagnostics). Transcripts with 2′fluoro (F)-uridine and 2′F-cytidine substitutions were made using the DuraScribe T7 transcription kit (Epicenter) according to the manufacturer's protocol. RNAs with pseudouridine substitutions were made using the T7 Megashortscript kit and pseudouridine-5′ppp (Trilink). After transcription, the DNA template was hydrolyzed with DNase, proteins were separated from the transcribed RNA by phenol-chloroform extraction, nucleotides were removed by quick-spin gel filtration column chromatography, and then the RNA was by precipitated with ethanol and ammonium acetate or sodium acetate. The RNA was sedimented by centrifugation, washed with 70% ethanol, dried briefly, and resuspended in RNase-free water. The RNA concentration was determined by absorbance in a spectrophotometer.
Transfection and luciferase reporter assays.
Huh7 cells (3 × 104 per well) were plated on a 24-well plate. After 24 h, the cells were transfected with 100 ng of pIFN-β-luc (firefly luciferase) and 1 ng of pCMV-luc (Renilla luciferase) using Lipofectamine 2000 (Invitrogen). After a 24-hour incubation, equal numbers of moles of each viral-RNA fragment were denatured for 3 min at 90°C and then renatured by slow cooling in renaturation buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA) and transfected using Lipofectamine 2000. Twenty-four hours after RNA transfection, the cells were lysed in 100 μl of passive lysis buffer (Promega), and an aliquot was analyzed using the dual-luciferase reporter assay system (Promega).
ISG56 immunoblotting.
Twenty-eight hours after Huh7 cells were transfected with the viral RNAs, the cells were lysed (10 mM Tris, pH 8, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol, 1× phosphatase inhibitor cocktail 1, 1× phosphatase inhibitor cocktail 2, 1× protease inhibitor cocktail; cocktails were purchased from Sigma Chemical Company). Cell debris was removed by centrifugation (200 × g for 3 min), and a Bradford assay was used to determine the protein concentration. Fifteen micrograms of total protein was electrophoresed into a 9% Tris-glycine sodium dodecyl sulfate (SDS)-polyacrylamide gel. Immunoblot analysis was performed using polyclonal anti-IFN-stimulated gene 56 (ISG56) antibody (provided by G. Sen, Cleveland Clinic) and monoclonal anti-beta-actin (AC-15) antibody (AbCam). Proteins were detected with a horseradish peroxidase-conjugated secondary antibody and were visualized by chemiluminescence.
Enzyme-linked immunosorbent assay.
Wild-type or RIG-I−/− MEFs were mock transfected or transfected with equal numbers of moles of renatured in vitro-transcribed HCV 3′-UTR, poly(U/UC), or X RNA. After 24 h, the cell culture supernatants were collected and analyzed for IFN-β production using an enzyme-linked immunosorbent assay (PBL Biomedical Laboratories). The levels of IFN-β expression were determined by comparison to the linear portion of a standard curve.
Preparation of RIG-I cell extract.
Cell extracts were prepared essentially as described by Chang et al., Gee et al., Takahasi et al., and Yoneyama et al. (6, 11, 30, 34). Briefly, subconfluent Huh7 cells were transfected with pEFBos FLAG-RIG-I plasmid and then incubated for about 48 h. The cells were scraped into cold phosphate-buffered saline, sedimented by centrifugation, and then resuspended in 2 volumes of hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1× protease cocktail inhibitor [cocktail purchased from Sigma Chemical Company], 1 mM dithiothreitol). The cells were allowed to swell for 15 min on ice and were then broken using a Dounce homogenizer. The extract was clarified by centrifugation (15 min at 14,000 rpm and 4°C), and the protein concentrations of the supernatants were determined using a Bradford assay. Aliquots were stored at −80°C.
Competitive RIG-I pull-down assay.
We standardized our RIG-I binding analyses across the experiments by using competitive pull-down assays with biotinylated poly(U/UC) RNAs. This was important in order to rule out the possibility that biotinylated RNAs with other sequences may not have interacted equivalently with the streptavidin particles, thereby preventing binding comparisons. One microgram of biotinylated HCV strain J4L6 poly(U/UC) RNA was incubated for 1 h at 25°C with or without excess nonbiotinylated competitor RNA and 30 μg of FLAG-RIG-I cell extract. Following the incubation, the mixture was transferred into 400 μl of wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40) containing 50 μl streptavidin MagneSphere paramagnetic particles (Promega) and rocked at 4°C for 2 h. The RNA-protein complexes were collected by magnetic separation, washed three times with wash buffer, resuspended in SDS-polyacrylamide gel electrophoresis sample buffer, boiled for 5 min, and electrophoresed into a Tris-glycine 7.5% SDS-polyacrylamide gel. FLAG-tagged protein within the pull-down fraction was analyzed by immunoblotting using M2 anti-FLAG antibody (M2 antibody purchased from Sigma Chemical Company).
RESULTS
Comparative analysis of activation of TLR3-independent signaling by viral UTR RNAs.
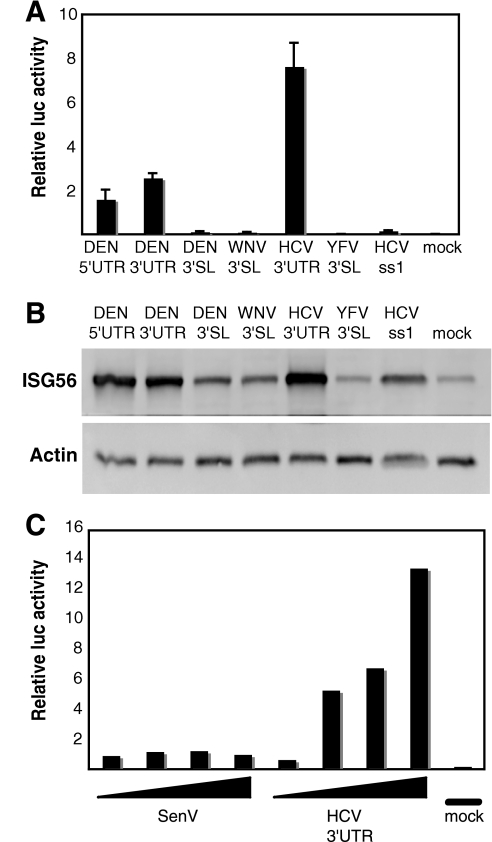
To determine if flavivirus UTR RNAs activate TLR3-independent signaling, we transfected equal numbers of moles of renatured 5′-triphosphorylated full-length DEN 5′- or 3′-UTR RNA or the conserved 3′-terminal SL RNAs from DEN, WNV, or YFV into TLR3-deficient Huh7 cells (18) and measured expression from an IFN-β promoter-luciferase reporter as an indication of early signaling events. The HCV 3′-UTR RNA served as a positive control, while the HCV ss1 RNA and no RNA (mock) were used as negative controls (29). The full-length DEN 5′- and 3′-UTR RNAs (105 nt and 383 nt, respectively) activated IFN-β to moderate levels compared to the negative controls, but the DEN, WNV, and YFV SL RNAs (110 nt) were minimally active (Fig. 1A). As expected, the HCV 3′-UTR RNA was a strong activator (Fig. 1A) (29). To confirm the luciferase reporter data using a direct assay for IFN production in response to the viral RNAs, we measured downstream ISG56 expression by immunoblotting. The ISG56 expression pattern mirrored that of the IFN reporter assay (Fig. 1B). These data suggest that endogenous IFN-β was produced and released in response to the transfected RNAs, leading to downstream ISG expression (1). Activation of IFN expression by the HCV 3′ UTR was also compared with that induced by Sendai virus infection. As shown in Fig. 1C, the HCV 3′ UTR was significantly better as an activator of IFN-β than Sendai virus infection. These results demonstrate that the HCV 3′-UTR RNA is a considerably more potent activator of TLR3-independent innate immune signaling than flavivirus UTR RNAs and Sendai virus. We assessed the role of the 5′ppp in activation by treating the in vitro-transcribed HCV 3′-UTR, DEN 5′-UTR, and DEN 3′-UTR RNAs with a phosphatase or by adding an m7GpppG cap (13). Both phosphatase treatment and capping the 5′ end reduced activation of the IFN-β reporter to background levels (data not shown). These results confirm that the 5′ppp has a critical role in activation of IFN-β expression. We note, however, that despite the fact that all of the viral RNAs tested (Fig. 1) contained a 5′ppp, we observed significant variability in the extent of IFN-β activation. We conclude that the 5′ppp is necessary but not sufficient for IFN-β activation.
FIG. 1.
IFN-β induction potentials of HCV and flavivirus UTR RNAs. (A) Twenty-four hours after being plated, Huh7 cells were cotransfected with plasmids encoding firefly or Renilla luciferase under the control of the IFN-β promoter or constitutive cytomegalovirus promoter, respectively. Following a 24-hour further incubation period, the cells were mock transfected or transfected in triplicate with equal numbers of moles of renatured in vitro-transcribed viral 5′- or 3′-UTR or 3′-SL RNAs. Twenty-four hours later, the cells were lysed, and aliquots of the extracts were analyzed using a dual-luciferase assay. The firefly luciferase light unit values were divided by the Renilla light units (transfection efficiency control) to generate the relative luciferase (luc) value. The bars show average relative luciferase values plus standard deviations. (B) Twenty-eight hours after viral-RNA transfection, the cells were lysed and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting for ISG56 and actin. (C) Huh7 cells were transfected or infected with increasing amounts of HCV 3′-UTR RNA (50 ng, 250 ng, 650 ng, and 1 μg) or Sendai virus (SenV) (50, 100, 250, or 500 hemagglutinin units), and IFN-β reporter activation was measured 24 h later as described in the legend to Fig. 1A.
The poly(U/UC) sequence is responsible for the potent activation of IFN-β by the HCV 3′ UTR.
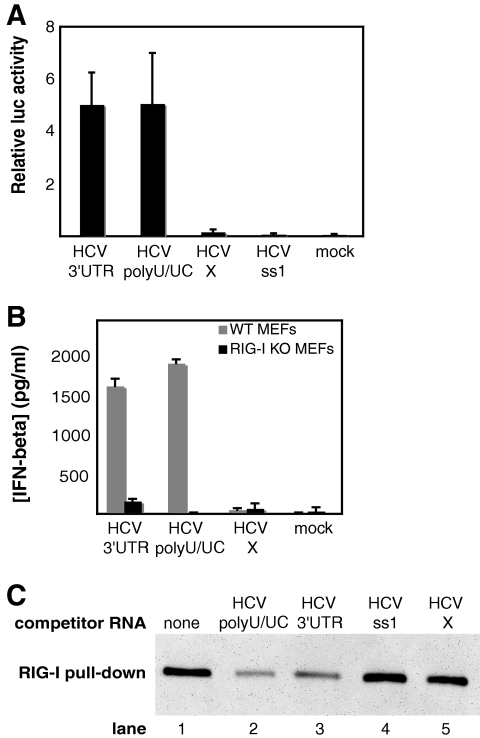
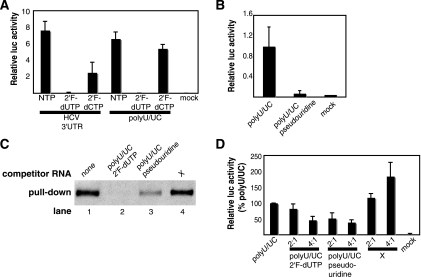
To understand why the HCV 3′-UTR RNA is significantly more potent than the other viral RNAs tested, we tested each half of the 3′-UTR RNA independently. While this work was in preparation for publication (D. Uzri and L. Gehrke, presented at the 26th Annual Meeting of the American Society for Virology, Corvallis, OR, 14 to 18 July 2007), Saito et al. reported a similar approach for analyzing the RNAs of HCV type 1b strain Con1 (26). The 5′ half of the 3′ UTR is a poly(U/UC) sequence that is unlikely to form stable secondary structure because it is very pyrimidine rich and forms few canonical Watson-Crick base pairs. The 3′ half of the 3′ UTR has the potential to fold into three SLs and is referred to as the HCV X RNA (32). When transfected into Huh7 cells, the poly(U/UC) region activated the IFN-β reporter to levels comparable to those of the full-length 3′ UTR; however, the HCV X RNA was minimally active (Fig. 2A). To verify that the poly(U/UC) RNA activates innate immune signaling through RIG-I, we compared the ability of the poly(U/UC) RNA to induce IFN-β in wild-type and RIG-I knockout MEFs. The poly(U/UC) RNA was a potent inducer of IFN-β in the wild-type MEFs, but this activation was completely abrogated in RIG-I knockout MEFs, demonstrating that RIG-I is necessary for signaling in response to the poly(U/UC) RNA (Fig. 2B). We also performed a competitive RIG-I pull-down assay using extract from Huh7 cells overexpressing FLAG-RIG-I (25) to determine if poly(U/UC) RNA can bind to RIG-I (Fig. 2C). Biotinylated poly(U/UC) RNA pulled down RIG-I from the extract (lane 1), and nonbiotinylated poly(U/UC) and HCV 3′-UTR RNAs were able to compete for 83% and 73% of this binding, respectively (Fig. 2C, lanes 2 and 3). Nonbiotinylated ss1 and X RNAs were less effective competitors of RIG-I binding (0% and 27% reduction, respectively) (Fig. 2C, lanes 4 and 5). To verify that the X RNA was not less stable than the HCV 3′-UTR or poly(U/UC) RNAs, we measured the stability of radiolabeled HCV 3′-UTR, poly(U/UC), and X RNAs over time in RIG-I cell extract and found that the X RNA was not less stable than the HCV 3′-UTR or poly(U/UC) RNA (data not shown).
FIG. 2.
Identification of the poly(U/UC) region of the HCV 3′ UTR as the determinant of RIG-I activation. (A) HCV poly(U/UC) and X RNAs were transcribed in vitro, and equal numbers of moles of the RNAs were transfected into Huh7 cells. Their potencies in activating the IFN-β reporter were determined as for Fig. 1A. The bars show average relative luciferase (luc) values plus standard deviations. (B) Wild-type (WT) or RIG-I knockout (KO) MEFs were mock transfected or transfected in triplicate with equal numbers of moles of in vitro-transcribed HCV 3′-UTR, poly(U/UC), or X RNA. After a 24-hour incubation period, an enzyme-linked immunosorbent assay was used to measure IFN-β protein levels from cell culture media. The bars show average amounts of mouse IFN-β protein levels plus standard deviations. (C) One microgram of biotinylated poly(U/UC) RNA was incubated with or without a 2.5-fold molar excess of nonbiotinylated competitor RNA and 30 μg of FLAG-RIG-I-containing Huh7 cell extract. RNA-protein complexes were recovered by pull-down assay using streptavidin magnetic particles. FLAG-tagged RIG-I protein within the pull-down fraction was analyzed by immunoblotting using M2 anti-FLAG antibody.
Activation does not require the 5′ppp and activating RNA domain to be immediately adjacent.
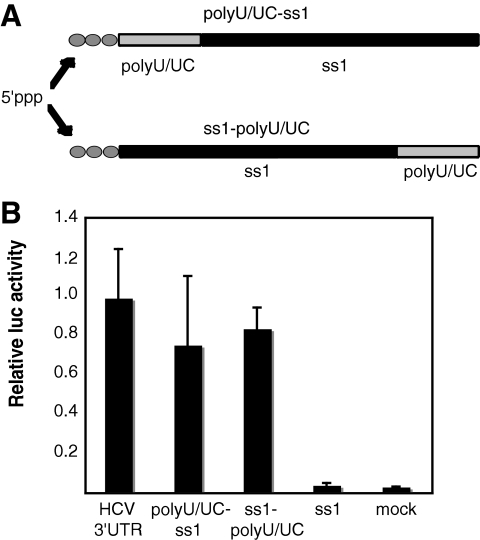
To determine if potent induction of IFN-β expression required that the 5′ppp be immediately adjacent to the activating nucleotide sequence, we created chimeric RNAs containing the poly(U/UC) sequence fused to the 5′ or 3′ terminus of the 350-nt HCV ss1 nonactivating sequence. Schematic representations of these constructs are presented in Fig. 3A. If the 5′ppp needed to be immediately adjacent to the poly(U/UC) sequence for potent activation, then the ss1-poly(U/UC) RNA would be expected to have less activation potential than the poly(U/UC)-ss1 RNA. RNAs were transcribed and tested for activation of IFN-β reporter expression. The results (Fig. 3B) suggest that both chimeric RNAs are signaling activators. These data are evidence that the poly(U/UC) region is a functional motif that can be separated from the 5′ppp by over 300 nt while retaining potent activity.
FIG. 3.
Separating the 5′ppp and the poly(U/UC) region does not disrupt signaling. (A) Schematic representations of the chimeric RNAs, showing the activating poly(U/UC) region positioned upstream of ss1 and immediately adjacent to the 5′ppp or downstream of ss1 and distant from the 5′ppp. (B) The chimeric RNAs were in vitro transcribed, and their abilities to activate the IFN-β reporter were tested as for Fig. 1A. The bars show average relative luciferase (luc) values plus standard deviations.
Interrupted short homopolymeric uridine and adenine RNAs activate signaling.
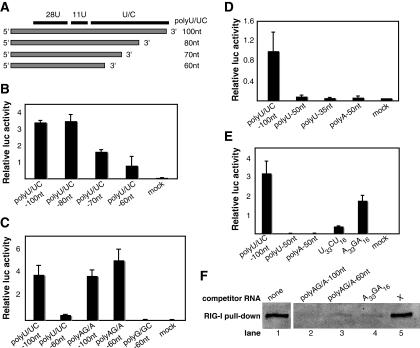
To determine if there is a minimal length or motif required for potent activation by the poly(U/UC) RNA, we transcribed RNAs with 3′-terminal deletions (Fig. 4A) and, following transfection into Huh7 cells, measured luciferase expression from the IFN-β-luc plasmid. As shown in Fig. 4B, poly(U/UC) 80 nt retained full activity compared to full-length poly(U/UC) 100 nt. However, further 3′ deletions resulted in gradual reductions in activity (Fig. 4B). 5′-terminal deletions of poly(U/UC) were also tested, and deletion of 30 nt or more from the 5′ end (which shortened or deleted the 28U tract) completely abrogated activity (data not shown). These data suggest that the intact 28U region is critical for activation. Since the poly(U/UC) RNA is 70% uridine, we decided to test whether potent RIG-I stimulation is limited to uridine-rich sequences. We tested the activity of the complement of poly(U/UC), the poly(AG/A) RNA, which is relevant to HCV infection because it is generated during minus-strand synthesis. Interestingly, the poly(AG/A) RNA had activity comparable to that of the full-length poly(U/UC) RNA (Fig. 4C), suggesting that potent RIG-I activation is not limited to uridine-rich sequences. The poly(AG/A) 60-nt deletion RNA (containing 28A and 11A tracts) also exhibited full activity, while the corresponding poly(U/UC) 60-nt RNA (containing 28U and 11U tracts) showed reduced activity and the poly(G/GC) 60-nt RNA (containing 28G and 11G tracts) showed minimal activity (Fig. 4C). These data suggest that length requirements for potent RIG-I activation vary with the nucleotide sequence. Since uridine- and adenine-rich sequences were both potent RIG-I activators, we determined if 50-nt RNAs containing a homopolymeric tract of uridine or adenine of 50 nt are also strong immunostimulators. Both poly(U) 50 nt and poly(A) 50 nt (26) activated IFN-β expression poorly (Fig. 4D). To determine whether their lack of activity was due to insufficient length (50 nt) or because homopolymeric composition is not adequate for potent RIG-I signaling, we tested the activities of 50-nt poly(U) and poly(A) RNAs with a cytidine or guanine nucleotide inserted after the 33rd U or A, respectively. The rationale for this approach is based on the observation that in the J4L6 poly(U/UC) RNA, runs of uridine are not continuous but are interrupted by cytidine (Fig. 4A). Interestingly, both U33CU16 and A33GA16 RNAs exhibited higher activity than homopolymeric poly(U) 50-nt and poly(A) 50-nt RNAs (15- and 52-fold higher, respectively) (Fig. 4E), suggesting that interrupting the homopolymeric sequence with a single C or G nucleotide enhances activation potential. To confirm that the activities of the most immunostimulatory adenine-rich RNAs correlated with RIG-I-RNA binding, competitive pull-down experiments were performed. The data (Fig. 4F) demonstrate that the poly(AG/A) 100-nt (lane 2; 86% reduction), poly(AG/A) 60-nt (lane 3; 88% reduction), and A33GA16 (lane 4; 89% reduction) RNAs were effective competitors of RIG-I binding to the biotinylated poly(U/UC) RNA. X RNA (lane 5; 15% increase) did not compete for binding. Importantly, these results suggest that activation of RIG-I is a function of both RNA sequence and length.
FIG. 4.
Examining the roles of RNA sequence composition and length in RIG-I activation. (A) Schematic representations of the different HCV strain J4L6 poly(U/UC) 3′ deletion RNAs tested. The black bars at the top indicate (left to right) 28 contiguous U residues followed by 11 contiguous U residues followed by a downstream U/C region. The gaps between the bars correspond to single cytosine residues. (B to E) Activation of the IFN-β reporter by in vitro-transcribed RNAs. The bars show average relative luciferase (luc) values plus standard deviations. (B) Activation by poly(U/UC) 3′ deletion RNAs. (C) Activation by poly(AG/A) 100-nt (full-length), poly(AG/A) 60-nt, and poly(G/GC) 60-nt RNAs. (D) Activation by 50-nt and 35-nt homopolymeric uridine RNAs and 50-nt homopolymeric adenine RNA. (E) Activation by 50-nt homopolymeric uridine and adenine RNAs interrupted with a single C or G nucleotide, respectively. (F) One microgram of biotinylated poly(U/UC) RNA was incubated with or without threefold molar excess of nonbiotinylated competitor RNA and 30 μg of FLAG-RIG-I cell extract. The competitive-binding assays were carried out as described in the legend to Fig. 2C.
Modified RNAs are competitive inhibitors of signaling activity and RIG-I binding.
The immunostimulatory activity of 5′ppp RNA was shown to be inhibited by capping or removing the 5′ppp or by introducing nucleoside base and ribose modifications (13). We tested the effects of ribose 2′ hydroxyl modification on the immunostimulatory potential of the HCV 3′-UTR and poly(U/UC) RNAs. Consistent with previous reports (13), replacement of uridine 2′ hydroxyls with fluoro groups in the HCV 3′-UTR and poly(U/UC) RNAs reduced IFN-β activity to background levels (Fig. 5A). Less dramatic reductions in activity were observed when cytidine 2′ hydroxyls were replaced with fluoro groups (Fig. 5A), likely because there are fewer cytidines than uridines in both RNAs. Next, we tested how incorporation of a modified base affected IFN-β activation; 100% replacement of uridine with pseudouridine, which has the same chemical formula as uridine but a shifted glycosidic bond, also abrogated activity (Fig. 5B). To determine whether the inhibition of IFN-β induction by incorporation of 2′F-dUTP or pseudouridine was due to reduction in binding to RIG-I, we performed a competitive RIG-I pull-down assay. In the presence of threefold molar excess of 2′F-dUTP- or pseudouridine-modified poly(U/UC) RNA, the amounts of RIG-I pulled down by biotinylated poly(U/UC) RNA were reduced 99% and 74%, respectively (Fig. 5C, lanes 2 and 3). X RNA was less effective as a competitor (Fig. 5C, lane 4; 18% reduction). These results suggest that RIG-I signaling is diminished by 2′ hydroxyl and base modifications but that the modifications do not affect RIG-I binding to the RNA. To determine if these modified RNAs function as competitive inhibitors of IFN-β induction, we cotransfected unmodified poly(U/UC) RNA with equal or excess molar quantities of 2′F-dUTP- or pseudouridine-modified poly(U/UC) RNA (Fig. 5D). Both 2′F-dUTP- and pseudouridine-modified poly(U/UC) RNAs functioned as competitive inhibitors of IFN-β induction, while the X RNA was not inhibitory (Fig. 5D). These data suggest that the modified RNAs trap RIG-I in an inactive complex, thereby decreasing innate immune signaling. To begin to understand why the modified RNAs failed to activate signaling, we asked whether base and ribose modifications affected the conformation of the poly(U/UC) RNA as measured by native gel electrophoresis. When pseudouridine- and 2′F-dUTP-modified poly(U/UC) RNAs were electrophoresed into a native polyacrylamide gel, we found that the pseudouridine-modified poly(U/UC) RNA displayed slower migration than unmodified RNA, suggesting a larger Stokes radius. Conversely, the 2′ hydroxyl substitutions did not affect RNA mobility under these conditions (data not shown).
FIG. 5.
Ribose and base modifications affect RNA innate immune stimulation potential. (A and B) Activation of the IFN-β reporter by in vitro-transcribed RNAs. The bars show average relative luciferase (luc) values plus standard deviations. (A) Activation by HCV 3′-UTR and poly(U/UC) RNAs transcribed with 2′F-dUTP in place of UTP or 2′F-dCTP in place of CTP. (B) Activation by poly(U/UC) RNA transcribed with pseudouridine-5′ppp in place of UTP. (C) One microgram of biotinylated poly(U/UC) RNA was incubated with or without a threefold molar excess of nonbiotinylated competitor RNA and 30 μg of FLAG-RIG-I cell extract. The competitive-binding assays were carried out as described in the legend to Fig. 2C. (D) Unmodified poly(U/UC) RNA was transfected alone or with 2′F-dUTP- or pseudouridine-modified poly(U/UC) RNA at 2:1 or 4:1 (modified/unmodified) molar excesses. IFN-β reporter activity was measured 24 h posttransfection as described in the legend to Fig. 1A. The data are presented as percentages of the control unmodified poly(U/UC) RNA activity (100%). The bars show average relative luciferase values plus standard deviations.
JFH-1 poly(AG/A) RNA, but not JFH-1 poly(U/UC) RNA, is highly immunostimulatory.
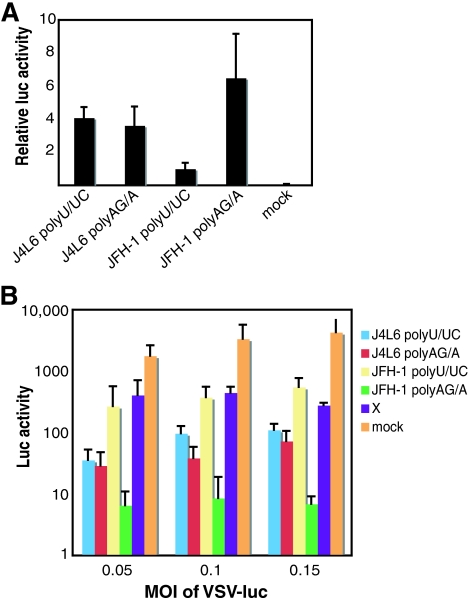
The poly(U/UC) and poly(AG/A) sequences tested thus far in this study were from the HCV genotype 1b J4L6 strain. The length and composition of the poly(U/UC) region vary somewhat among different HCV strains (17), and we considered the possibility that these variations could result in strain-specific innate immune stimulation. Saito et al. reported that full-length genomic HCV RNA (strain Con1) lacking the 3′ UTR had much lower immunostimulatory potential than full-length genomic RNA, suggesting that the poly(U/UC) sequence is a critical determinant for RIG-I-mediated signaling in response to the full-length viral genomic RNA (26). The HCV genotype 2a JFH-1 strain, which was isolated from a patient with fulminant hepatitis in Japan, is unique in that it replicates efficiently and produces infectious particles in cell culture (19, 33, 36). We reasoned that the enhanced replicative properties of the JFH-1 virus might correlate with diminished activation of innate immune signaling in comparison with other HCV strains. Therefore, we compared the immunostimulatory activities of the JFH-1 poly(U/UC) and poly(AG/A) RNAs to those of the J4L6 RNAs (Fig. 6A). Interestingly, the JFH-1 poly(U/UC) RNA had half the immunostimulatory activity of the J4L6 poly(U/UC) and poly(AG/A) RNAs (Fig. 6A), a finding that correlates with the higher replicative potential of the JFH-1 strain. In contrast, the JFH-1 poly(AG/A) RNA was more stimulatory than the J4L6 poly(U/UC) and poly(AG/A) RNAs (Fig. 6A). To determine if the observed activity in the IFN-β reporter assay (Fig. 6A) correlated with a virus infection model, we transfected Huh7 cells with J4L6 or JFH-1 poly(U/UC) or poly(AG/A) RNA, waited 24 h, and then infected them with VSV expressing firefly luciferase (VSV-luc) at increasing MOIs. Activation of innate immune signaling by the RNAs was expected to establish an antiviral state that would limit VSV replication. The results (Fig. 6B) demonstrate that inhibition of VSV-luc replication correlates well with IFN-β reporter induction by the different RNAs. Taken together, the results shown in Fig. 6 strongly suggest that innate immune signaling potential is defined in part by specific sequences present in the HCV 3′-UTR poly(U/UC) and poly(AG/A) domains, which vary among the different strains.
FIG. 6.
Comparison of the HCV type 1b J4L6 and HCV type 2a JFH-1 strain poly(U/UC) and poly(AG/A) RNAs as activators of innate immune stimulation. (A) HCV J4L6 and JFH-1 RNAs were transcribed in vitro and transfected into Huh7 cells as described in the legend to Fig. 1A. The bars show average relative luciferase (luc) values plus standard deviations. (B) Twenty-four hours after Huh7 cells were transfected with equal numbers of moles of J4L6 or JFH-1 poly(U/UC), poly(AG/A), or X RNA, the cells were infected with VSV-luc at MOIs of 0.05, 0.1, and 0.15. Four hours postinfection, the cells were lysed and luciferase activity was assayed as a measure of VSV replication. The bars show average luciferase values plus standard deviations.
DISCUSSION
In this paper, we have examined sequence and structural elements required for potent RIG-I-mediated innate immune signaling that manifests as IFN-β expression and the establishment of an antiviral state. In addition to confirming 5′ triphosphorylated uridine- and adenine-rich ssRNAs as potent activators of RIG-I signaling (26), the results presented here suggest that (i) the 3′-UTR RNAs or the structurally conserved 3′-terminal SL RNAs from several flaviviruses are comparatively weak signaling activators when evaluated side by side with HCV 3′-UTR RNA, (ii) immediate adjacency of the 5′ppp and the HCV poly(U/UC) sequence is not essential (i.e., separating the two determinants by over 300 nt did not reduce signaling significantly), (iii) RIG-I-RNA binding and signaling activity are determined by both the RNA length and nucleotide sequence (e.g., inactive short homopolymeric uridine and adenine RNA sequences can be converted to signaling activators by interrupting the homopolymer with single C or G nucleotides, respectively), (iv) nucleoside base and ribose 2′ hydroxyl modifications that block signaling activity do not inhibit the binding interaction between RNA and RIG-I, and (v) poly(U/UC) and poly(AG/A) sequences from two different HCV strains have distinct immunostimulatory potentials that may be factors in defining their different replicative capacities.
Following the report that the 3′ UTR of the HCV RNA activates TLR3-independent innate immune signaling (29), we carried out experiments to determine if corresponding regions from other Flaviviridae members had similar potentials. The results (Fig. 1) demonstrate that, although the 5′ and 3′ UTRs of DEN (type 4) elicited measurable stimulation of innate immune signaling, the smaller, highly structurally conserved 3′-terminal SL RNAs (5, 12) of DEN (type 4), YFV (the 17D vaccine strain), and WNV (strain NY99) viruses were minimally active. The increased activity of the DEN 5′ and 3′ UTRs did not correlate with elevated uridine or adenine compositions compared to the SL RNAs (DEN 5′ UTR, 29% U, 29% A; DEN 3′ UTR, 17% U, 29% A; DEN 3′ SL, 23% U, 25% A; WNV 3′ SL, 19% U, 24% A; YFV 3′ SL, 21% U, 28% A; HCV 3′ UTR, 49% U, 14% A; ss1, 19% U, 20% A). Removing the 5′ppp from the HCV 3′ UTR, DEN 5′ UTR, and DEN 3′ UTR completely abrogated IFN-β stimulation, suggesting that activation was mediated through RIG-I and not MDA5 (data not shown) (7). After observing the large relative differences in RNA immunostimulation potentials (Fig. 1), we subsequently localized the robust activation of IFN-β expression to the HCV poly(U/UC) region (Fig. 2). While this work was in preparation for publication (D. Uzri and L. Gehrke, presented at the 26th Annual Meeting of the American Society for Virology, Corvallis, OR, 14 to 18 July 2007), Saito et al. reported similar results using 3′-UTR sequences from a related HCV strain, Con1 (26). The precise sequences in the DEN 5′- and 3′-UTR RNAs that activate signaling have not been mapped. However, these RNAs lack uridine or adenine repeats that are found in the HCV 3′-UTR sense and antisense RNAs, suggesting that sequences other than poly(U/UC) or poly(AG/A) may have weak activating potential.
We considered the physiological relevance of testing activation potentials of short (60- to 350-nt) RNA sequences compared to the full-length RNAs. It has been reported that both HCV replicon RNA (29) and full-length genomic RNA (26) activate signaling. HCV genomic RNA lacking the 3′ UTR has much less immunostimulatory potential than full-length genomic RNA (26), suggesting that both the 5′ppp and the poly(U/UC) sequence can be sensed by RIG-I, despite the length of the primary nucleotide sequence separating them. Here, we generated chimeric RNAs that allowed us to test the dependence of activation on immediate adjacency of the 5′ppp and the poly(U/UC) RNA sequence. The results (Fig. 3) revealed that potent activation does not require the immediate adjacency of the 5′ppp and the activating poly(U/UC) sequence. Mechanistic models suggest that the 5′ppp is bound by the RIG-I C-terminal domain (7, 30) and that the poly(U/UC) nucleotide sequence is bound elsewhere, presumably by the RNA helicase (DECH) domain. The fact that RIG-I can sense the 5′ppp and RNA nucleotide activation motifs, despite their separation by 350 nt (Fig. 3) or by over 9,000 nt of primary sequence in the case of the HCV replicon and genomic RNAs (26, 29), suggests that RIG-I may have significant structural flexibility or may undergo cofolding events with the RNA in order to bridge the 5′ppp and nucleotide activation domain. It is possible that long-range RNA-RNA or protein-RNA interactions bridge the 5′ and 3′ ends of the HCV RNA, thereby enhancing RIG-I sensing of both the 5′ppp and downstream nucleotide activation domains.
The data described here provide new insights into the definition of the RNA determinants required for innate immune signaling. Our data (Fig. 4) and those of others (26) suggest that activation of IFN-β reporter expression is proportional to the length of the poly(U/UC) region, with little activity observed using poly(U/UC) RNAs with fewer than 60 nt. Although homopolymeric poly(U) 50-nt and poly(A) 50-nt RNAs were inactive in our assays (Fig. 4D) (26), interrupting these homopolymers by inserting a single C or G nucleotide, respectively, increased IFN-β reporter expression significantly (Fig. 4E). These experiments followed from the observation that the uridine tracts in the J4L6 poly(U/UC) sequence are interrupted by intermittent cytidines (Fig. 4A). We considered the possibility that introducing an additional interruption into the poly(U/UC) or poly(AG/A) RNA might further enhance activity. The results (data not shown) indicate that inserting a single C or G nucleotide to interrupt the 43U or 43A tracts of the JFH-1 poly(U/UC) or poly(AG/A) RNA, creating 14U and 28U or 14A and 28A tracts, respectively, did not increase IFN-β reporter expression. We speculate that when starting with a homopolymer, nucleotide interruptions may increase RIG-I activation until a plateau is reached, after which further interruptions have no effect or may even reduce activation of RIG-I. Others have noted that RNA-protein interactions are enhanced by discontinuities in RNA structure (e.g., bulged nucleotides and noncanonical base pairs) (28); however, defining the molecular basis for the enhanced activity of these short (50-nt) interrupted homopolymer RNAs will require further experimentation. RIG-I is activated by both the poly(U/UC) RNA and its antisense sequence, poly(AG/A), but not by poly(G/GC) (Fig. 4) (26). The poly(U/UC) and poly(AG/A) binding surfaces on RIG-I have not been defined, and it will be interesting to determine how RIG-I can bind specifically and functionally to both polypyrimidine- and polypurine-rich RNA sequences.
It was reported previously that RNAs with ribose 2′ hydroxyl substitutions (e.g., 2′-O-methyl and 2′F) exhibit reduced signaling through TLR7 and RIG-I (8, 13, 27). Replacing uridine with pseudouridine, which has a shifted glycosidic bond, is also known to abrogate RIG-I signaling (13). Correspondingly, HCV 3′-UTR and poly(U/UC) RNAs transcribed with 2′F-dUTP and 2′F-dCTP showed diminished stimulatory activities (Fig. 5A), as did poly(U/UC) RNA transcribed with pseudouridine (Fig. 5B). We demonstrate here that, despite these activity losses, the binding interactions between RIG-I and 2′F-dUTP- or pseudouridine-modified poly(U/UC) RNAs were retained (Fig. 5C). The mechanisms underlying the activity losses are not understood; however, the results reported here strongly indicate that the disruption of RIG-I-RNA binding can be excluded. The in vitro binding data (Fig. 5C) were extended in a functional assay in which modified RNAs were shown to diminish IFN-β induction when cotransfected with unmodified activator poly(U/UC) RNA (Fig. 5D), likely because of competitive RIG-I binding. These results suggest that RIG-I molecules that are bound to RNAs containing modified nucleotides are trapped in an inactive intermediate form (Fig. 7). A recent report of single-molecule reconstructions of RIG-I bound to phosphorothioated oligodeoxynucleotides is consistent with this hypothesis (23).
FIG. 7.
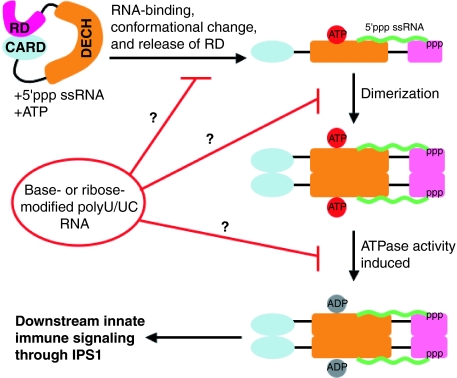
Model defining steps of RIG-I activation and where modified RNAs could block signaling. The actual order of steps may be different than what is shown here. Ribose 2′ hydroxyl and base modifications do not affect binding to RIG-I but may inhibit RIG-I activation at one of the downstream steps. Additional details can be found in the text.
The lengths and nucleotide sequences of the poly(U/UC) regions vary somewhat among different HCV isolates (17), and we considered the possibility that these variations might be associated with differences in innate immune responses. The HCV genotype 2a JFH-1 strain, which was isolated from a patient with fulminant hepatitis in Japan, is unusual in that it replicates efficiently and produces infectious particles in cell culture (19, 33, 36). We reasoned that the enhanced replicative properties of the JFH-1 virus might correlate with diminished activation of innate immune signaling in comparison with other HCV strains. Saito et al. reported that genomic HCV RNA (strain Con1) lacking the 3′ UTR was less active in innate immune signaling than full-length genomic RNA, suggesting that the poly(U/UC) sequence is a critical determinant for RIG-I-mediated signaling in response to full-length genomic RNA (26). The J4L6 poly(U/UC) sequence contains an uninterrupted 28-uridine tract, while the comparable JFH-1 sequence has a longer uninterrupted 43-uridine tract. Interestingly, the isolated JFH-1 poly(U/UC) RNA had half the immunostimulatory activity of the J4L6 poly(U/UC) and poly(AG/A) RNAs (Fig. 6). Alternatively, the JFH-1 poly(AG/A) RNA was nearly twice as stimulatory as the J4L6 poly(U/UC) and poly(AG/A) RNAs. Assuming that the activities of the isolated poly(U/UC) RNAs correlate with their behavior in the context of full-length genomic RNA (26), then the full-length JFH-1 strain genomic RNA would be predicted to have poor immunostimulatory activity. The corresponding poly(AG/A)-containing antisense RNA may not elicit a strong antiviral response because (i) for most positive-strand RNA viruses, antisense strands are present in much smaller amounts than sense strands; (ii) the poly(AG/A) RNA may be shielded by RNA binding proteins; and/or (iii) the accumulation of antisense strands may be accompanied by a parallel increase in translated viral proteins, including NS3/4A, which is known to block RIG-I signaling by cleaving IPS1 from mitochondrial membranes. The presence of the poly(U/UC) region is highly conserved in HCV isolates and is essential for viral-RNA replication (35); therefore, its recognition by the RIG-I signaling pathway may illustrate the evolution of an elegant host antiviral response mechanism.
A model that extends previous summaries (25) and also indicates how modified RNAs may be used to dissect the steps in the signaling pathway is presented in Fig. 7. In the absence of RNA ligand, RIG-I is a latent molecule whose activity is downregulated by a C-terminal RD (25). A positively charged groove, and specifically lysine 858, in the RD is likely the 5′ppp-binding site of RIG-I (7). However, a maximum of 3 nt can fit in this groove, suggesting that nucleotide-activating determinants, such as the poly(U/UC) sequence, bind to the RIG-I DECH box domain (7). The RIG-I RD activates the RIG-I ATPase by RNA-dependent dimerization (i.e., ATPase activity is stimulated by dimer formation) (7). Upon substitution of the 2′ ribose position or replacement of uridine with pseudouridine, RIG-I binding to the RNA is unaffected compared to RNA with unmodified nucleotides; however, downstream signaling is abrogated. It is possible that the modified poly(U/UC) RNAs inhibit the subsequent RIG-I conformational changes, dimerization, or ATPase activation. Further experimentation will be required to address these questions. The competitive activity of the modified RNAs (Fig. 5) suggests that they may be useful as modulators of RIG-I-mediated innate immune responses.
Acknowledgments
We thank J. Jung (Keck School of Medicine, University of Southern California, Los Angeles), P. Yang (Harvard Medical School, Boston, MA), S. Whelan (Harvard Medical School, Boston, MA), R. Chung (Massachusetts General Hospital, Boston, MA), G. Sen (Cleveland Clinic Foundation, Cleveland, OH), S. Behrens (Fox Chase Cancer Center), and M. Gale (University of Washington, Seattle) for providing reagents. We thank G. W. Martin (Massachusetts Institute of Technology, Cambridge), R. Gomila (Tufts Medical School, Boston, MA), S. Whelan (Harvard Medical School, Boston, MA), and R. Chung (Massachusetts General Hospital) for helpful discussions and comments.
This work was supported by U.S. Public Health Service awards GM42504 and P30 DK034854 through the Harvard Digestive Diseases Center.
Footnotes
Published ahead of print on 18 February 2009.
REFERENCES
- 1.Aaronson, D. S., and C. M. Horvath. 2002. A road map for those who don't know JAK-STAT. Science 2961653-1655. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413732-738. [DOI] [PubMed] [Google Scholar]
- 3.Beg, A. A. 2002. Endogenous ligands of Toll-like receptors: implications for regulating inflammatory and immune responses. Trends Immunol. 23509-512. [DOI] [PubMed] [Google Scholar]
- 4.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430257-263. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153113-121. [DOI] [PubMed] [Google Scholar]
- 6.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8157-171. [DOI] [PubMed] [Google Scholar]
- 7.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29169-179. [DOI] [PubMed] [Google Scholar]
- 8.Diebold, S. S., C. Massacrier, S. Akira, C. Paturel, Y. Morel, and C. Reis e Sousa. 2006. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 363256-3267. [DOI] [PubMed] [Google Scholar]
- 9.Fredericksen, B. L., and M. Gale, Jr. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 802913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredericksen, B. L., B. C. Keller, J. Fornek, M. G. Katze, and M. Gale, Jr. 2008. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee, P., P. K. Chua, J. Gevorkyan, K. Klumpp, I. Najera, D. C. Swinney, and J. Deval. 2008. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 2839488-9496. [DOI] [PubMed] [Google Scholar]
- 12.Grange, T., M. Bouloy, and M. Girard. 1985. Stable secondary structures at the 3′-end of the genome of yellow fever virus (17 D vaccine strain). FEBS Lett. 188159-163. [DOI] [PubMed] [Google Scholar]
- 13.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 14.Kato, H., O. Takeuchi, E. Mikamo-Satoh, R. Hirai, T. Kawai, K. Matsushita, A. Hiiragi, T. S. Dermody, T. Fujita, and S. Akira. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2051601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 16.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 17.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 703363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 1022992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 20.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 158783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 23.Ranjith-Kumar, C. T., A. Murali, W. Dong, D. Srisathiyanarayanan, R. Vaughan, J. Ortiz-Alacantara, K. Bhardwaj, X. Li, P. Li, and C. C. Kao. 2008. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J. Biol. Chem. 2841155-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito, T., and M. Gale, Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 1917-23. [DOI] [PubMed] [Google Scholar]
- 25.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito, T., D. M. Owen, F. Jiang, J. Marcotrigiano, and M. Gale. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sioud, M. 2006. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hydroxyl uridines in immune responses. Eur. J. Immunol. 361222-1230. [DOI] [PubMed] [Google Scholar]
- 28.Steitz, T. A. 1993. Similarities and differences between RNA and DNA recognition by proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 792689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahasi, K., M. Yoneyama, T. Nishihori, R. Hirai, H. Kumeta, R. Narita, M. Gale, Jr., F. Inagaki, and T. Fujita. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29428-440. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi, O., and S. Akira. 2007. Recognition of viruses by innate immunity. Immunol. Rev. 220214-224. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 703307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 35.You, S., and C. M. Rice. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J. Virol. 82184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]