Abstract
A newly discovered virally encoded deubiquitinating enzyme (DUB) is strictly conserved across the Herpesviridae. Epstein-Barr virus (EBV) BPLF1 encodes a tegument protein (3,149 amino acids) that exhibits deubiquitinating (DUB) activity that is lost upon mutation of the active-site cysteine. However, targets for the herpesviral DUBs have remained elusive. To investigate a predicted interaction between EBV BPLF1 and EBV ribonucleotide reductase (RR), a functional clone of the first 246 N-terminal amino acids of BPLF1 (BPLF1 1-246) was constructed. Immunoprecipitation verified an interaction between the small subunit of the viral RR2 and BPLF1 proteins. In addition, the large subunit (RR1) of the RR appeared to be ubiquitinated both in vivo and in vitro; however, ubiquitinated forms of the small subunit, RR2, were not detected. Ubiquitination of RR1 requires the expression of both subunits of the RR complex. Furthermore, coexpression of RR1 and RR2 with BPLF1 1-246 abolishes ubiquitination of RR1. EBV RR1, RR2, and BPLF1 1-246 colocalized to the cytoplasm in HEK 293T cells. Finally, expression of enzymatically active BPLF1 1-246 decreased RR activity, whereas a nonfunctional active-site mutant (BPLF1 C61S) had no effect. These results indicate that the EBV deubiquitinating enzyme interacts with, deubiquitinates, and influences the activity of the EBV RR. This is the first verified protein target of the EBV deubiquitinating enzyme.
Ubiquitination of proteins plays an indispensable role in many cellular and viral processes (22). Ubiquitination proceeds in steps through the activity of three types of proteins: E1, ubiquitin (Ub)-activating enzymes; E2, Ub-conjugating proteins; and E3 enzymes, which are Ub ligases (29). Wild-type Ub is ∼8 kDa in length and is attached to its substrate via its C-terminal glycine 76 to lysine residues in the target protein (32). Ub modification of proteins occurs on lysine residues and may consist of a single Ub (monoubiquitination) or multiple Ubs in which a chain is linked together on the same substrate site (polyubiquitination). Ub has seven potential lysines (K6, K11, K27, K29, K33, K48, and K63) that can be used to form polyubiquitin chains (29). Polyubiquitin linkages through K48 or K63 have been well studied. K48-linked Ub chains typically, but not always, target proteins to the proteasome for degradation (26, 33). In contrast, K63-linked Ub chains are typically involved in the signaling, trafficking, and regulatory responses of their protein substrates (1, 9, 19, 28, 39). More recently appreciated is that protein ubiquitination can be reversed through the action of deubiquitinating enzymes (DUBs) (43, 44).
All members of the Herpesviridae family encode a large tegument protein containing deubiquitinating activity located within their N-terminal regions (34). Members of this unique class of deubiquitinating enzymes function as cysteine proteases and have a strictly conserved catalytic triad composed of a cysteine, histidine, and aspartic acid residue. Sequence homology over the N-terminal region is only about 15% among members of the Herpesviridae (34). Herpesvirus DUBs do not share sequence homology or conserved domains with known eukaryotic DUBs (25). However, the catalytic triad is strictly conserved and is essential for DUB activity among the herpesvirus family as well as for cellular DUBs (25, 34). Mutation of the active-site cysteine or histidine of the human cytomegalovirus (CMV) counterpart pUL48 resulted in a complete loss of DUB activity (25, 42). Such mutations clearly reduced production of infectious human CMV and pseudorabies virus in cell culture (5, 42), demonstrating an important role for virally encoded DUB activity in the virus life cycle.
BPLF1, a late lytic cycle gene of Epstein-Barr virus (EBV), produces a full-length transcript of 9.5 kb and an apparent smaller product approximately 3 kb in length (35). BPLF1 is 3,149 amino acids in length and contains the deubiquitinating activity within its first 205 amino acids (34). The N-terminal fragment of the herpes simplex virus (HSV) homolog (UL36), which encompasses DUB and cleaves polyubiquitin chains, appears to be specific for K48 linkages; cleavage of K63-Ub chains was not detected (25). In addition, DUBs from the alphaherpesviruses and gammaherpesviruses are Ub specific and show little or no preference for other Ub-like molecules, such as Nedd8, SUMO, and ISG15 (21, 25).
Recently, an EBV interactome map was constructed with the use of rapid yeast two-hybrid screening (6). This map predicted many possible cellular partners but only one EBV-interacting partner with BPLF1, BaRF1; BPLF1 was also predicted to interact with itself. BaRF1 is the small subunit (RR2) of the EBV ribonucleotide reductase, and it interacted with a protein fragment containing BPLF1's first 1,000 amino acids (6).
EBV RR2 is a lytic cycle gene product 302 amino acids in length. EBV RR2, which has been studied relatively little, is similar to cellular RR2 in that it must interact with the large RR subunit (RR1) to be functionally active (24). Mg2+ and ATP are required for cellular RR activity, but both are dispensable for HSV RR activity (23). Cellular RR activity is controlled by regulating RR2 stability; RR2 levels are reported as undetectable during G1 phase, but high levels are detected during S phase (4), which may be clues to cellular function. Cellular RR1 protein levels remain constant throughout the cell cycle (14). In addition, the cellular small subunit, RR2, can be polyubiquitinated in vitro (8). From a biological standpoint, functional HSV RR is essential for neurovirulence in mice but is not needed for virus replication in tissue culture cells (7, 20).
During initial experiments, in which BPLF1 mRNA expression was suppressed with short hairpin RNA (shRNA), decreases in viral particle production resulted, but the effects could not be attributed directly to DUB activity of BPLF1 since expression of the entire gene was suppressed. In this report, we verify a physical and functional interaction between a fragment containing the first 246 amino acids of BPLF1 and EBV RR, which results in deubiquitination of the EBV RR large subunit (RR1). Importantly, enzymatically active BPLF1 downregulates viral ribonucleotide reductase activity. The results suggest that ubiquitination of the large subunit (RR1) of the EBV RR may upregulate its activity, which is then suppressed by deubiquitination. These data point to the first protein target identified for a viral DUB and suggest a potential role of BPLF1 deubiquitinating activity in regulation of EBV replication.
MATERIALS AND METHODS
Cell lines, growth, and transfection.
HEK 293T and 293EBV+ (green fluorescent protein [GFP]-tagged recombinant B95.8 EBV) cells (11) were cultured in Dulbecco's modified Eagle medium (CellGro) supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 μg/ml), and amphotericin B (250 ng/ml). Transfections were performed with the use of Effectene transfection reagent (Qiagen) following the manufacturer's protocol, and cells were harvested after 48 h.
Construction of shRNAs against BPLF1.
shRNAs (19-mer) predicted to knock down BPLF1 were designed, constructed, and ligated into pSuperRetro.puro (OligoEngine) at the BglII and XhoI sites. shRNAs were named according to the base number they map to in the BPLF1 sequence. 893 was constructed using the forward primer 5′-GATCCCCGGCGAATAATACCGTATAATTCAAGAGATTATACGGTATTATTCGCCTTTTTC-3′ and the reverse primer 5′-TCGAGAAAAAGGCGAATAATACCGTATAATCTCTTGAATTATACGGTATTA TTCGCCGGG-3′. The corresponding forward and reverse scrambled primers (893S) were 5′-GATCCCCGCGAACATAAGATAAGTGCTTTCAAGAGAAGCACTTATCTTATGTTCGTTTTTC-3′ and 5′-TCGAGAAAAACGAACATAAGATAAGTGCTTCTCTTGAAAGCACTTATCTTATGTTCGCGGG -3′, respectively. The 3650 forward and reverse primers were 5′-GATCCCCGTGAAATCCATCACCTTCTATTCAAGAGATAGAAGGTGATGGATTTCATTTTTC-3′ and 5′-TCGAGAAAAATCAAATCCATCACCTTCTATCTCTTGAATAGAAGGTGATGGATTTCACGGG-3′, respectively. The corresponding forward and reverse scrambled primers (3650S) were 5′-GATCCCCGTATCTTATCATACCGCAACTTCAAGAGAGTTGCGGTATGATAAGATATTTTTC-3′ and 5′-TCGAGAAAAATATCTTATCATACCGCAACTCTCTTGAAGTTGCGGTATGATAAGATACGGG-3′, respectively. Primers were placed in annealing buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 100 mM NaCl), brought to a boil, and allowed to slowly cool to room temperature. Annealed primers were ligated to BglII and XhoI cut pSuperRetro.puro vector with T4 DNA ligase (NEB). Correct shRNA sequences were verified after transformation.
Reverse transcriptase PCR (RT-PCR).
293EBV+ cells were transfected with shRNA constructs and the BZLF1-expressing construct (to induce the lytic cycle) (3). After 72 h, the supernatant fluid and cells were harvested. RNA was extracted from the cell pellet with use of the Zymogen mini RNA isolation II kit (Orange, CA) and subjected to DNase treatment with the Zymogen DNA-Free RNA kit. RNA was quantified, and 4 μg was used with the high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) to generate cDNA. Samples were then treated with RNase H. PCR analysis was performed with primers mapping to either the N-terminal or C-terminal region of BPLF1. PCR conditions were 95°C for 2 min followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 1.5 min with a final extension at 72°C for 10 min. PCR products were then visualized after separation on 1% agarose gel.
Real-time PCR.
Supernatant fluid of shRNA-transfected, lytically induced 293EBV+ cells was harvested and filtered after 72 h. Genomic DNA was extracted from 88 μl of supernatant fluid with the DNeasy kit (Qiagen) and analyzed by real-time PCR with the ABI 7900HT real-time PCR system. Primer and probe sequences were EBVW-1 (5′-GCAGCCGCCCAGTCTCT-3′), EBVW-2 (5′-ACAGACAGTGCACAGGAGCCT-3′), and EBVW-FAM (5′-6-carboxyfluorescein-AAAAGCTGGCGCCCTTGCCTG-6-carboxytetramethylrhodamine-3′). PCR conditions were 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. Viral DNA copy numbers were determined by dilution of a quantitated standard of EBV B95-8 DNA (Advanced Biotechnologies, MD).
Western blotting.
After transfection, immunoprecipitation (IP), or in vitro ubiquitination/deubiquitination, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, blocked with 5% milk in Tris-buffered saline-Tween 20, and incubated with appropriate primary antibody overnight at 4°C. The nitrocellulose membrane was then washed and probed with either goat or rabbit anti-mouse antibody (Santa Cruz Biotechnology) at 1:3,000 in 5% milk for 1 h at room temperature. Bands were visualized with enhanced chemiluminescence reagent from Pierce (Rockford, IL).
Ub-AMC assay.
Ub-AMC (1 μM; Biomol) was incubated with 1 μM purified N-terminal BPLF1 or 100 nM Ub C-terminal hydrolase (UCH-L1) (Boston Biochem) in reaction buffer containing 50 mM Tris, pH 7.5, 2 mM ATP, and 2 mM dithiothreitol (DTT) with and without Ub aldehyde (Ub-al; Santa Cruz Biotechnology). Reaction mixtures were incubated at 37°C for 10 min, and then samples in triplicate were excited by exposure to light at a wavelength of 340 nm and emission was measured at 460 nm.
Construction and purification of EBV clones.
The first 246 amino acids of BPLF1 (BPLF1 N-term) were amplified by PCR analysis using a forward primer (5′-ATGAGTAACGGCGACTGGGGGCAAAGCCAG-3′) and a reverse primer (5′-ATGCGGGGGTCCTCTGTCTC-3′). PCR analysis conditions were one cycle of 95°C for 2 min and then 40 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 1.5 min, followed by a 10-min extension. Purified B95-8 EBV DNA was used as template (Advanced Biotechnologies). PCR products were treated with Taq polymerase (Roche) to add adenosine overhangs and ligated into pSC-A with the use of the Strataclone PCR cloning kit (Stratagene) following the manufacturer's guidelines. After sequence verification, the clone was inserted into His-tagged pET28a (Novagen) at the EcoRI site and transformed into BL21-Gold DE3 pLysS cells (Stratagene). A large culture was grown and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cell pellet was resuspended in 40 mM HEPES, pH 7.9, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 500 mM KCl, 10% glycerol, 0.2% NP-40, and 10 mM imidazole. Cells were lysed by sonication, and cleared lysates were incubated with Ni-nitrilotriacetic acid agarose beads (Qiagen). The column was loaded and washed, and purified protein was eluted with 300 mM imidazole in resuspension buffer.
The BPLF1 N-terminal fragment was cloned into the pFLAG-CMV-2 (Sigma) vector for expression in 293T cells. To this end, the pSC-A BPLF1 N-terminal fragment vector served as template for PCR analysis in which primers provided HindIII and XbaI sites. PCR conditions were the same as those described above except the annealing temperature was 52°C. The PCR product and pFLAG-CMV-2 vector were digested with HindIII and XbaI and ligated to yield a pFLAG CMV-2 BPLF1 N-terminal fragment containing an N-terminal FLAG tag (BPLF1 1-246).
EBV RR2 was removed from previously cloned pEGFP-C1 (Clonetech) with BglII and EcoRI and was placed into pCMV-Tag-3B at the BamHI and EcoRI sites. This vector contains an N-terminal myc tag. RR1 was cloned into pcDNA3, and an N-terminal hemagglutinin (HA) tag was added by PCR.
BPLF1 C61S was constructed by utilizing site-directed mutagenesis on BPLF1 1-246. The sequences of the forward primer (C61S FWD) and reverse primer (C61S REV) were 5′-CTTTGCCGGCATCCAGAGCGTCAGCAACTG-3′ and 5′-CAGTTGCTGACGCTCTGGATGCCGCAAAG-3′, respectively. PCR conditions were 95°C for 2 min and then 30 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 8 min, followed by a 10-min extension at 72°C.
IP.
293T cells were transfected with FLAG-tagged BPLF1 1-246, myc-tagged RR2, or both. After 48 h, cells were harvested and placed in lysis buffer (30 mM NaCl, 1% NP-40, 10 mM Tris, pH 7.5, 1× protease inhibitor cocktail tablet [Roche], 5 mM DTT, and 1 mM phenylmethylsulfonyl fluoride). Cells were lysed by freeze-thawing (three times), and samples were precleared with protein A/G beads (Santa Cruz Biotechnology) at room temperature for 30 min. Supernatant fluids were collected, 1 μg/ml anti-FLAG (Sigma) was added, and samples were oscillated at 4°C for 1 h. Protein A/G beads (30 μl) were added, and samples were incubated overnight at 4°C. Beads were washed three times in lysis buffer, placed in SDS sample buffer, and separated by SDS-PAGE. Western blotting was performed as described above except mouse anti-myc (1:200; Santa Cruz Biotechnology) served as the secondary antibody.
In vitro ubiquitination and deubiquitination assays.
293EBV+ cells were transfected with BZLF1 (to activate lytic infection) along with EBV RR1 and RR2. After 48 h, cells were harvested and lysed in residual phosphate-buffered saline (PBS; from PBS rinses during harvesting) by freeze-thawing three times. Samples were placed in reaction mixture buffer (50 mM Tris, pH 7.5, 2 mM Mg-ATP [Boston Biochemical], 2 mM DTT, 25 μM MG132 [Biomol], 6 μl/sample rabbit reticulocytes [fraction II; Biomol] to provide E1 and E2 enzymes, 2 μM Ub-al [Santa Cruz Biotechnology; only for ubiquitination assay], 20 μM His6-tagged Ub [Biomol], and 1× protease inhibitor cocktail [Roche]) and were incubated at 37°C for the indicated amounts of time. For the deubiquitination assay only, Ub-al was omitted, and purified EBV BPLF1 1-246 (2 μM) was added after the initial incubation step and then incubated at 37°C for 1 h. Samples were loaded on SDS-PAGE and transferred to nitrocellulose membrane. Ubiquitinated/deubiquitinated EBV R1 and R2 were detected with anti-EaR (R1) (Santa Cruz Biotechnology), anti-HA (R1) (Santa Cruz Biotechnology), and anti-myc (R2) antibodies.
Immunofluorescence assay.
HEK 293T cells were grown and transfected on poly-lysine-treated coverslips. After 24 h, cells were washed with PBS and fixed with 4% paraformaldehyde. Cells were then permeabilized with 0.5% Triton X-100 and were blocked with 10% normal donkey serum. Primary antibody diluted in 1.5% normal donkey serum was added (anti-FLAG [BPLF1 1-247], anti-HA [EBV RR1], and anti-myc [EBV RR2]), and cells were stored at 4°C overnight. Cells were washed with PBS and exposed to secondary antibody (donkey anti-mouse Alexa Fluor 594 and donkey anti-rabbit Alexa Fluor 488; Invitrogen) at 37°C for 1 h. Cells were stained with 0.1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI), mounted, and examined with an Axiovert fluorescent microscope.
Cell compartmentalization assay.
HEK 293T cells were transfected with BPLF1 1-246, BPLF1 C61S, RR1, and RR2. Cells were harvested after 48 h, and cell pellets were processed with the Qproteome cell compartment kit (Qiagen). Fractions were then separated by SDS-PAGE, transferred to nitrocellulose membranes, and visualized with the appropriate antibodies.
In situ measurements of RR activity.
To quantify the RR activity, incorporation of [3H]cytidine into DNA was performed as described previously (40). HEK 293T cells were transfected with BPLF1 1-246, BPLF1 C61S, RR1, and RR2 in triplicate. After 24 h, [3H]cytidine (MP Biomedicals) was added to the medium and samples were incubated for an additional 24 h. Cells were harvested and treated with RNase, and DNA was purified with the use of the DNeasy kit (Qiagen) and eluted in a 200-μl volume. Incorporation of 3H into DNA was determined by scintillation counting.
RESULTS
shRNA against BPLF1 decreases mRNA levels and reduces viral genome production.
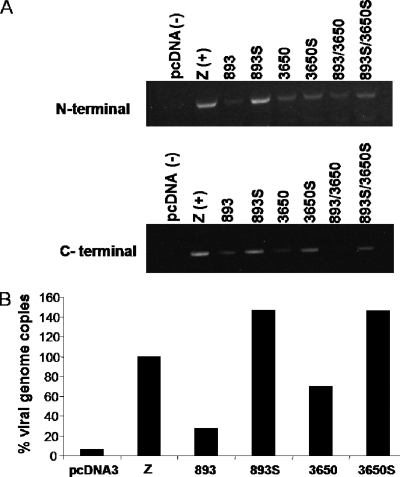
BPLF1 shRNAs were designed and transfected into 293EBV+ cells (11) along with the EBV transactivator, BZLF1 (Z), to induce viral replication. After 72 h, cells were harvested and total RNA was extracted and subjected to RT-PCR. Primers for both N-terminal and C-terminal regions were used to analyze mRNA levels. Figure 1A shows a representative experiment illustrating that shRNA 893 and 3650 effectively decreased amounts of BPLF1 mRNA. The scrambled shRNA, 3650S, does exhibit some nonspecific knockdown of the N-terminal RNA levels.
FIG. 1.
Knockdown of BPLF1 mRNA levels decreases the EBV genome copy number. 293EBV+ cells were induced by transfection with Z (BZLF1) along with shRNA against BPLF1. 893S and 3650S denote scrambled forms of 893 and 3650 shRNA constructs. Z serves as the positive control and pcDNA3 as the negative control. (A) RT-PCR of RNA from 293EBV+ cells. Seventy-two hours after transfection, cells were lysed and total RNA was extracted and subjected to RT-PCR using primers against the N-terminal or C-terminal portions of BPLF1. (B) Real-time PCR of 4-μl supernatant fluids from shRNA-transfected 293EBV+ cells at 72 h. Genomic DNA was extracted from supernatant fluids and analyzed by real-time PCR with a TaqMan probe corresponding to the BamHI sequence. The total genome copies present in the positive control (Z) were set to 100%, and samples are reported as a percentage of Z.
The effects of BPLF1 shRNAs on virus production were determined by assaying the number of viral genome copies in the supernatant fluid of the lytically induced 293EBV+ cells with the use of real-time PCR and a TaqMan primer against the BamH1W segment (17, 18). The results in Fig. 1B demonstrate that expression of 893 and 3650 shRNAs results in reduced genome copies compared with the scrambled controls. These data demonstrate that BPLF1 contributes to EBV replication. However, these effects cannot be directly attributed to DUB activity alone since BPLF1 serves viral maturation functions that may not require the deubiquitinating activity of the protein. We therefore set out to investigate the effects of BPLF1 DUB by examining its interaction with a viral lytic cycle protein.
EBV BPLF1 1-246 expresses deubiquitinating activity.
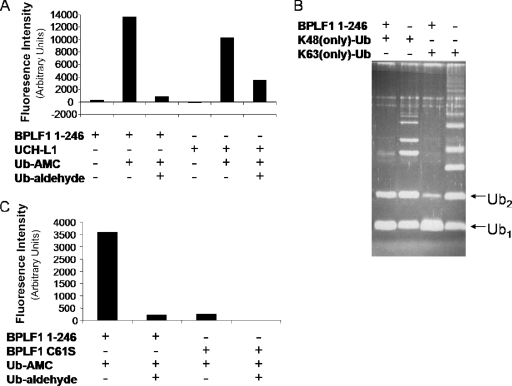
An N-terminal clone of BPLF1 containing the first 246 amino acids was constructed, expressed, and purified from Escherichia coli, and DUB activity was measured by fluorometry. BPLF1 1-246 was incubated with a fluorogenic substrate (Ub-AMC; Biomol) which, when cleaved by a deubiquitinating enzyme, releases photons, allowing the fluorescence intensity to be determined. BPLF1 1-246 caused a high level of cleavage of Ub-AMC, which was reversed by Ub-al, an irreversible inhibitor of DUBs (Fig. 2A). UCH-L1, a cellular DUB with the same conserved active site as that of BPLF1, was used to validate the assay and produced similar results.
FIG. 2.
BPLF1 1-246 exhibits deubiquitinating activity. (A and C) Functional assay of BPLF1 1-246. BPLF1 1-246 was incubated with Ub-AMC at 37°C for 10 min. Samples were excited at 340 nm, and emission was measured at 460 nm. Ub-al was added where indicated. UCH-L1 is a known cellular DUB, and BPLF1 C61S is an enzymatically inactive mutant. (B) Purified BPLF1 was incubated overnight with and without K48 and K63 Ub chains (Ub2 to Ub7), separated by SDS-PAGE, and visualized with Sypro Ruby protein stain. Mono-Ub and di-Ub are labeled Ub1 and Ub2. The bands located above the labeled mono-Ub and di-Ub bands represent the longer Ub chains (Ub3 to Ub7).
To determine the type of Ub chain cleaved by the EBV enzyme, Ub chains linked through Ub-lysine 48 or Ub-lysine 63, in which all lysines except 48 or 63 were mutated, were used as substrates for BPLF1 1-246 (Fig. 2B). Both K48 and K63-linked Ub chains were reduced mostly to mono-Ub (Ub1) and di-Ub (Ub2) forms by the DUB activity with an apparent slight (but reproducible) preference for K63-linked chains. The results indicate that both K48- and K63-linked Ub chains are substrates for EBV DUB in vitro. These observations leave open the possibilities that BPLF1 1-246 may have roles in rescuing substrate from degradation in some instances and be involved in regulating protein function in others.
Finally, an inactive DUB mutant was constructed by site-directed mutagenesis. The cysteine residue C61 in the catalytic triad also composed of a histidine and an aspartic acid residue was converted to serine (BPLF1 C61S) in the BPLF1 1-246 clone. BPLF1 C61S did not exhibit DUB activity in the Ub-AMC assay when FLAG-tagged versions of BPLF1 1-246 and BPLF1 C61S were expressed in 293T cells and purified by IP (Fig. 2C). Loss of DUB activity was observed with BPLF1 C61S compared with that of the wild-type BPLF1 1-246. This result indicates that the active-site C61 residue is essential for EBV DUB activity and that BPLF1 C61S serves as a valid inactive mutant. These data together show that BPLF1 1-246 is a functional DUB, cleaves Ub chains linked by K48 and K63, and has a strictly conserved cysteine residue which is required for its activity.
EBV RR interacts with BPLF1 1-246.
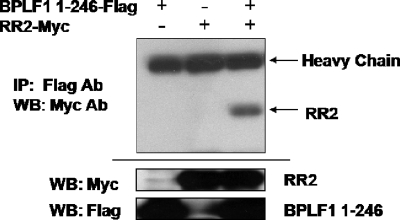
Once a functional clone was obtained, we searched for a target of the EBV DUB enzyme. Interactome mapping had suggested a possible interaction between EBV RR and BPLF1 (6). To determine whether BPLF1 1-246 could in fact interact with EBV RR, specifically RR2, FLAG-tagged BPLF1 1-246 and myc-tagged RR2 were coexpressed in 293T cells. Cells were harvested and lysed, and IPs were performed with FLAG antibody followed by immunoblotting with c-myc antibody. Figure 3 demonstrates the interaction between FLAG-tagged BPLF1 1-246 and myc-tagged RR2 proteins, either physically or through complex formation. When EBV RR2 or BPLF1 1-246 was expressed alone, no precipitating protein bands were detected. Precipitation of RR2 was detected only when both proteins were coexpressed, suggesting that they may interact physically. Additionally, it was found that the interaction of RR2 and BPLF1 was maintained in the presence of RR1.
FIG. 3.
EBV RR2 interacts with BPLF1 1-246. 293T cells were transfected with BPLF1 1-246, EBV RR2, or both as indicated. IPs were done using FLAG antibody followed by Western blotting (WB) with myc antibodies. myc-tagged RR2 is highlighted with an arrow. Input controls (20% of total cell lysate) are located at the bottom.
EBV BPLF1 1-246 localizes intracellularly with EBV RR.
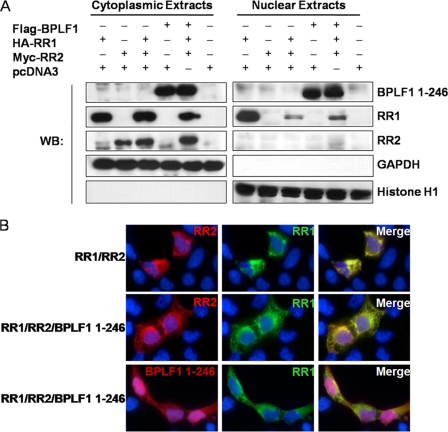
Cell fractionation was used to determine the cellular localization of RR1, RR2, and BPLF1 1-246 (Fig. 4A). 293T cells were transfected with RR1, RR2, and BPLF1 1-246, and cellular expression of these products was analyzed by cell fractionation. When RR1 was expressed alone, the product was found in both the cytoplasmic and nuclear fractions; however, when RR1 is expressed with RR2 or both RR2 and BPLF1 1-246, the localization is greatly shifted to the cytoplasmic fraction. RR2 is likely necessary for proper folding and localization of RR1 since both subunits are necessary for RR activity (10, 13, 30). RR2 is consistently located in the cytoplasm regardless of whether it is expressed alone or in conjunction with RR1. BPLF1 1-246 is found in both the nucleus and cytoplasm when expressed alone or with RR1 or RR2. Upon concurrent expression, RR1, RR2, and BPLF1 1-246 all colocalize to the cytoplasm.
FIG. 4.
EBV RR1, RR2, and BPLF1 1-246 localize in the cytoplasm of 293T cells. (A) 293T cells were transfected with EBV RR1, RR2, and BPLF1 1-246 for 48 h. Fractionated extracts were analyzed by Western blotting (WB). The notation above the blots indicates the transfecting plasmids. The notation to the right indicates the protein blotted against. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as a cytoplasmic marker, and histone H1 served as the nuclear marker. (B) 293T cells were transfected with HA-tagged RR1, c-myc-tagged RR2, and FLAG-tagged BPLF1 1-246 as indicated to the left. Immunofluorescence staining was performed with antibody against HA, myc, and FLAG. Nuclear DAPI staining is shown in blue. All panels represent ×100 magnification. Top panels depict cells transfected with RR1 and RR2 stained for RR1 and RR2. Middle and bottom panels depict cells transfected with RR1, RR2, and BPLF1 1-246 stained for RR1 and RR2 or RR1 and BPLF1 1-246, respectively. Merged images are shown to the right.
Immunofluorescence was utilized to substantiate the cellular localization of EBV RR1, RR2, and BPLF1 1-246. 293T cells were transfected with EBV RR1, RR2, and BPLF1 1-246. After 48 h, cells were washed, fixed, permeabilized, and labeled with fluorescent antibodies and examined by microscopy (Fig. 4B). When RR1 and RR2 are both expressed without BPLF1 1-246, RR1 and RR2 are in the cytoplasm (Fig. 4B, top). When RR1 and RR2 are expressed in the presence of BPLF1 1-246 (Fig. 4B, middle), localization of RR1 and RR2 remains largely cytoplasmic but seems to be in a more ordered structure. It should also be noted that a small amount of RR2 may now be associated with the nucleus, which was not observed when analyzed by cell fractionation (Fig. 4A). BPLF1 1-246 is located in both the cytoplasm and nucleus of the cell when RR1, RR2, and BPLF1 1-246 are coexpressed (Fig. 4B, bottom). BPLF1 1-246 colocalizes with R1 and R2 in the cytoplasm of 293T cells as shown in the merged pictures (Fig. 4B).
EBV RR is ubiquitinated on its large subunit and is deubiquitinated by BPLF1 1-246.
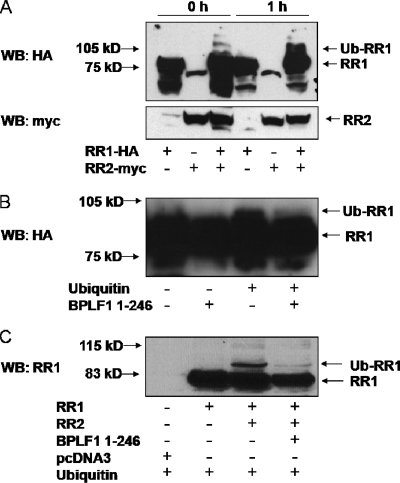
Since BPLF1 1-246 interacted with the EBV RR, we next investigated whether it was a target for the DUB activity. The first step was to determine if EBV RR is ubiquitinated. No reports of ubiquitination of EBV RR were found; however, ubiquitination of the small subunit (RR2) of mammalian RR has been reported (8). To investigate the ubiquitination of RR, we made use of an in vitro ubiquitination system. Briefly, 293EBV+ cells (11) were transfected with EBV subunits RR1, RR2, or both. After 48 h, the transfected cells were harvested and lysed and samples were placed in reaction buffer containing fraction II rabbit reticulocytes to provide E1 and E2 enzymes and His-tagged Ub, Mg-ATP, and Ub-al to inhibit cellular DUBs in reticulocytes. After a 1-h incubation at 37°C, Western blotting displayed a banding pattern consistent with the ubiquitination of RR1 (Fig. 5A). RR2 ubiquitination was not observed regardless of whether RR1 was present. The arrows in Fig. 5A indicate the RR1, RR2, and ubiquitinated forms. RR1 was almost entirely monoubiquitinated; however, fainter bands could be observed, which may relate to polyubiquitinated products. A small amount of ubiquitinated RR1 could also be observed at 0 h, which may have occurred intracellularly from endogenous sources. Faster-migrating bands below RR1 were identified but are believed to be nonspecific. These data demonstrate that ubiquitination occurs only when both subunits of EBV RR are expressed together and that only RR1 is ubiquitinated in vitro. We believe these are the first reports of EBV RR ubiquitination and that they offer a contrast with cellular RR in which only RR2 is ubiquitinated.
FIG. 5.
EBV RR1 is ubiquitinated in vitro and in vivo and is deubiquitinated by BPLF1 1-246. (A) 293EBV+ cells were transfected with RR1, RR2, or both as indicated. Cells were harvested after 48 h, and lysates were mixed with reaction buffer containing rabbit reticulocytes and exogenous Ub. Samples were then incubated at 37°C for the indicated amounts of time and subjected to Western blotting (WB). The top panel shows probing with HA antibody, and the bottom panel shows probing with c-myc antibody. The locations of RR1, RR2, and ubiquitinated RR1 (Ub-RR1) bands are indicated by arrows. (B) 293EBV+ cells were transfected with EBV RR1 and RR2. Cells were harvested after 48 h, lysates were mixed with reaction buffer, and exogenous Ub was added where indicated, followed by incubation of samples at 37°C for 1 h. Purified BPLF1 1-246 was added to the indicated samples, and the reaction was continued for another hour at 37°C. Arrows note the location of RR1 and ubiquitinated RR1. The blot was probed with anti-EBV EaR (RR1) and is overexposed to show RR1 ubiquitination. (C) 293T cells were transfected with RR1, RR2, and BPLF1 1-246 as indicated. All samples were additionally transfected with HA-tagged Ub and subjected to Western blotting against RR1. Arrows note the location of RR1 and ubiquitinated RR1.
We next tested whether DUB activity of BPLF1 1-246 could remove Ub from RR1 in vitro in a modified deubiquitination system. 293EBV+ cells were transfected with the large and small subunits of EBV RR, harvested, split into four equal fractions, and placed in reaction buffer as outlined above except that Ub-al was omitted. His-tagged Ub was added where noted (Fig. 5B). Samples were then placed at 37°C for 1 h, 5 μM of purified BPLF1 1-246 was added where noted, and incubation continued for another hour. The immunoblot was overexposed to detect ubiquitinated RR1. The small amount of ubiquitinated product detected is most likely due to cellular DUBs present in the rabbit reticulocytes. A band migrating above RR1 was detected in lane 3 (Fig. 5B), consistent with ubiquitination as all reaction mixtures were the product of one transfected sample. In lane 4 (Fig. 5B), ubiquitinated RR1 was not detected, indicating that BPLF1 1-246 is capable of removing Ub from RR1. While the additional band migrating above RR1 is approximately 8 kDa larger (the molecular mass of Ub) than RR1, it is not possible to determine if ubiquitination/deubiquitination is the result of monoubiquitin or polyubiquitin chain addition/removal, as additional ubiquitinated products may be too faint to be detected in this experiment.
We next determined if RR1 could be ubiquitinated and subsequently deubiquitinated by BPLF1 1-246 in vivo. 293T cells were transfected with RR1, RR2, BPLF1 1-246, and Ub as indicated (Fig. 5C). After 30 h, cells were harvested and proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibody against EBV RR1 (anti-EaR). Overexpression of Ub resulted in ubiquitinated RR1 but only when both RR1 and RR2 were expressed together (Fig. 5C, lane 2 versus lane 3). This is in agreement with the observation that RR1 ubiquitination occurs only when both subunits are expressed (Fig. 5A). RR2 is likely necessary for proper folding of RR1 and its subsequent ubiquitination. The addition of overexpressed BPLF1 1-246 results in the loss of the ubiquitinated RR1 forms (Fig. 5C, lanes 3 and 4). Repeated experiments resulted in the same banding pattern. These results suggest that EBV RR1 is ubiquitinated and that BPLF1 1-246 can deubiquitinate RR1 in vivo as well as in vitro. The more slowly migrating bands suggest additional ubiquitinated forms that may also be reduced in amount. We were unable to show by IP the presence of ubiquitinated RR1, perhaps due to the small amount of ubiquitinated RR1 or the inability of the antibody we used to recognize the modified RR1.
BPLF1 1-246 decreases RR activity.
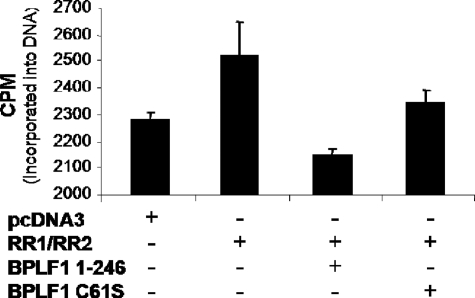
Preliminary tests of EBV RR activity in vitro resulted in enzymatic activity that was too low and variable to permit conclusive study of the effect of BPLF1 on the viral RR. Therefore, we moved to an in vivo system to measure EBV RR activity. After verifying that EBV RR could be deubiquitinated, we examined whether the enzymatic activity of BPLF1 influenced the RR activity. RR activity was assayed by adding [3H]cytidine to the medium of 293T cells transfected with EBV RR1 and RR2 alone or together with BPLF1 1-246 or BPLF1 C61S. Cytidine is converted to CDP after entering the cell and then to dCDP by cellular or EBV RR activity, and it has been shown that there is a good correlation between RR activity and incorporation of labeled cytidine into DNA (41). Cells transfected with EBV RR1 and RR2 produced an increase in incorporated 3H-labeled DNA above background levels produced by cellular RR (Fig. 6). Introduction of BPLF1 1-246 into the system significantly decreased RR activity, whereas the inactive BPLF1 C61S mutant did not result in a similar decrease in RR activity. These results indicate that EBV RR increases total RR activity above endogenous cellular levels and that the deubiquitinating activity of BPLF1 affects RR function by suppressing its activity through removal of Ub.
FIG. 6.
BPLF1 1-246 decreases RR activity. 293T cells were transfected with labeled plasmids as indicated. After 24 h, [3H]cytidine was added to the medium for an additional 24 h. Cells were harvested and treated with RNase, and DNA was extracted and purified. 3H incorporation into DNA was determined by scintillation counting. Experiments were performed in triplicate; error bars represent the standard error of the mean.
DISCUSSION
Ubiquitination and deubiquitination are important processes in both viral and cellular life cycles. However, until the surprising discovery that viruses encode deubiquitinating enzymes, only cellular DUBs were known to carry out deubiquitination of ubiquitinated proteins (34). In the case of EBV, cellular DUBs have been implicated in deubiquitination and stabilization of β-catenin in latently infected cells (36). In contrast, the newly discovered class of herpesviral DUBs is expressed during the viral lytic cycle (35). These DUBs display very little homology to known cellular DUBs, although the active site is identical to those of many cellular DUBs (25). The function of viral DUBs, whether directed at viral replicative processes or at cellular targets needed for viral replication, remains completely unknown. After determining that the full-length gene product of BPLF1 is important for viral replication, we undertook these experiments in order to explore possible functions of the EBV DUB by identifying substrates for the enzyme. We focused on the EBV RR as a likely target.
We first found that the N-terminal 246 amino acids of BPLF1 can function alone as a DUB and that one of the strictly conserved active-site residues (C61S) is essential for its function. In addition, the deubiquitinating activity of BPLF1 1-246 cleaves both K48- and K63-linked Ub chains. This result suggests that EBV DUB activity may be involved in processes that protect the substrate from proteasomal degradation, which is typically associated with K48-linked Ub chains, and that it may also play a functional or regulatory role, exemplified by K63-linked Ub protein chains. Interestingly, this result differs from that reported with the HSV homolog, UL36, which appears to be specific for K48-linked ubiquitination. This result suggests that activity of EBV DUB may be broader than that reported for HSV (25). Future studies with full-length BPLF1 protein may uncover specificity for protein targets or the type of Ub cleavage that cannot be detected with the N-terminal fragment. It is also still unknown if viral DUBs can cleave Ub precursors, which would expand further the role of the newly discovered viral enzymes.
Members of the Herpesviridae encode their own RRs. However, HSV RR differs from EBV RR in both protein size and function. Both contain RR activity, but HSV-encoded RR also contains kinase and antiapoptotic activity (27, 31, 37, 38). However, the regions that encode these activities are not represented in the EBV RR gene. The only verified function of EBV RR is its RR activity. This work adds significantly to the little that is known about EBV RR function and its regulation and should open a new area for the study of viral and possibly cellular RRs.
The localization of BPLF1 1-246 was not changed by the presence of EBV RR1 or RR2 or both. These findings seem logical since the full-length BPLF1 herpesviral homolog (UL36) plays a role in viral maturation in that it is important for release of the viral genome to the nucleus (2), and there is accumulation of nonenveloped particles in the cytoplasm when functional UL36 is absent (12). Additionally, it was found that EBV RR1 and RR2 colocalize in the cytoplasm, consistent with published literature for cellular RR in which both RR1 and RR2 are found in the cytoplasm (15, 16). EBV RR1 seems to undergo a locational shift when expressed in the presence of RR2. These findings are compatible with operation of the subunits in a complex and suggest that both subunits may be required for proper folding and function of the enzyme.
We show that EBV RR1 but not RR2 is ubiquitinated both in vitro and in vivo. This finding contrasts with the cellular reductase in which RR2 is ubiquitinated, and enzymatic activity is determined by the stability of RR2 (4, 14). Whether this difference between cellular and viral RRs also applies to other herpesvirus RRs appears not to have been reported. However, HSV RR is similar to cellular RR in that it is active only when both subunits are expressed together. We also found that EBV RR1 is ubiquitinated only when RR2 is present, which supports the idea that ubiquitination plays a regulatory role in RR function since the enzyme is functional only when the subunits can act as a complex. Additional experiments in which RR1 and RR2 were expressed in 293T cells in the presence of various proteasome inhibitors and overexpressed Ub did not result in an increase in RR1 protein levels (data not shown). This finding suggests that ubiquitination of EBV RR1 affects the function of the protein rather than targeting it for proteasomal degradation.
Deubiquitination of RR1 in the presence of BPLF1 1-246 was demonstrated both in vivo and in vitro. These data indicate not only that RR and BPLF1 1-246 interact but that EBV RR1 is also a substrate for deubiquitination by the viral enzyme. Taken along with the findings that BPLF1 1-246 preferentially cleaves K63-linked Ub chains and that no accumulation of ubiquitinated RR1 is detected in the presence of proteasome inhibitors, the data suggest that RR1 ubiquitination may upregulate RR activity and that deubiquitination of RR1 by BPLF1 DUB activity may decrease RR activity.
Finally, RR activity was measured by following the conversion of [3H]cytidine until its incorporation into DNA. Overexpression of EBV RR in 293T cells resulted in increased DNA labeling, indicating that viral RR does increase the conversion of nucleoside triphosphates to deoxynucleoside triphosphates (dNTPs) compared with the negative control in which only cellular enzymes are expressed. This is logical in the sense that EBV is typically reactivated in resting B lymphocytes which are depleted in their dNTP pools. During initial infection, EBV RR may play a critical role in increasing dNTP levels to facilitate viral replication.
Decreases in RR activity were observed when BPLF1 1-246 was expressed but not with an enzymatically inactive mutant DUB. Since herpesviral DUB activity is believed to be specific for ubiquitinated substrates, it follows that BPLF1 downregulates RR activity by the removal of Ub. The modest difference observed in the loss of activity of RR may be due to the fact that only a small percentage of RR was ubiquitinated in our experiments. Future experiments are designed to determine if EBV DUB activity is restricted to the viral RR or if it may also influence cellular RR activity.
Acknowledgments
This work was supported by National Cancer Institute grants HL64851-06 and CA09156.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Abbott, D. W., Y. Yang, J. E. Hutti, S. Madhavarapu, M. A. Kelliher, and L. C. Cantley. 2007. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell. Biol. 276012-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 361099-1104. [DOI] [PubMed] [Google Scholar]
- 4.Björklund, S., S. Skog, B. Tribukait, and L. Thelander. 1990. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 295452-5458. [DOI] [PubMed] [Google Scholar]
- 5.Böttcher, S., C. Maresch, H. Granzow, B. G. Klupp, J. P. Teifke, and T. C. Mettenleiter. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 826009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderwood, M. A., K. Venkatesan, L. Xing, M. R. Chase, A. Vazquez, A. M. Holthaus, A. E. Ewence, N. Li, T. Hirozane-Kishikawa, D. E. Hill, M. Vidal, E. Kieff, and E. Johannsen. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 1047606-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, J. M., I. McDougall, H. S. Marsden, V. G. Preston, D. M. Ryan, and J. H. Subak-Sharpe. 1988. Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J. Gen. Virol. 692607-2612. [DOI] [PubMed] [Google Scholar]
- 8.Chabes, A. L., C. M. Pfleger, M. W. Kirschner, and L. Thelander. 2003. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA 1003925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Z. J. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, E. A., P. Gaudreau, P. Brazeau, and Y. Langelier. 1986. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature 321441-443. [DOI] [PubMed] [Google Scholar]
- 11.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 958245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 7411608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutia, B. M., M. C. Frame, J. H. Subak-Sharpe, W. N. Clark, and H. S. Marsden. 1986. Specific inhibition of herpesvirus ribonucleotide reductase by synthetic peptides. Nature 321439-441. [DOI] [PubMed] [Google Scholar]
- 14.Engstrom, Y., S. Eriksson, I. Jildevik, S. Skog, L. Thelander, and B. Tribukait. 1985. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 2609114-9116. [PubMed] [Google Scholar]
- 15.Engström, Y., and B. Rozell. 1988. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J. 71615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engström, Y., B. Rozell, H. A. Hansson, S. Stemme, and L. Thelander. 1984. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 3863-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, H., and M. L. Gulley. 2001. Epstein-Barr viral load measurement as a marker of EBV-related disease. Mol. Diagn. 6279-289. [DOI] [PubMed] [Google Scholar]
- 18.Fan, H., S. C. Kim, C. O. Chima, B. F. Israel, K. M. Lawless, P. A. Eagan, S. Elmore, D. T. Moore, S. A. Schichman, L. J. Swinnen, and M. L. Gulley. 2005. Epstein-Barr viral load as a marker of lymphoma in AIDS patients. J. Med. Virol. 7559-69. [DOI] [PubMed] [Google Scholar]
- 19.Fukushima, T., S. Matsuzawa, C. L. Kress, J. M. Bruey, M. Krajewska, S. Lefebvre, J. M. Zapata, Z. Ronai, and J. C. Reed. 2007. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc. Natl. Acad. Sci. USA 1046371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, D. J., and S. K. Weller. 1988. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology 16641-51. [DOI] [PubMed] [Google Scholar]
- 21.Gredmark, S., C. Schlieker, V. Quesada, E. Spooner, and H. L. Ploegh. 2007. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 8110300-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 23.Huszar, D., and S. Bacchetti. 1981. Partial purification and characterization of the ribonucleotide reductase induced by herpes simplex virus infection of mammalian cells. J. Virol. 37580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 6771-98. [DOI] [PubMed] [Google Scholar]
- 25.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 26.Kirkpatrick, D. S., N. A. Hathaway, J. Hanna, S. Elsasser, J. Rush, D. Finley, R. W. King, and S. P. Gygi. 2006. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8700-710. [DOI] [PubMed] [Google Scholar]
- 27.Langelier, Y., S. Bergeron, S. Chabaud, J. Lippens, C. Guilbault, A. M. Sasseville, S. Denis, D. D. Mosser, and B. Massie. 2002. The R1 subunit of herpes simplex virus ribonucleotide reductase protects cells against apoptosis at, or upstream of, caspase-8 activation. J. Gen. Virol. 832779-2789. [DOI] [PubMed] [Google Scholar]
- 28.Langie, S. A., A. M. Knaapen, C. H. Ramaekers, J. Theys, J. Brun, R. W. Godschalk, F. J. van Schooten, P. Lambin, D. A. Gray, B. G. Wouters, and R. K. Chiu. 2007. Formation of lysine 63-linked poly-ubiquitin chains protects human lung cells against benzo[a]pyrene-diol-epoxide-induced mutagenicity. DNA Repair 6852-862. [DOI] [PubMed] [Google Scholar]
- 29.Li, W., and Y. Ye. 2008. Polyubiquitin chains: functions, structures, and mechanisms. Cell. Mol. Life Sci. 652397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClements, W., G. Yamanaka, V. Garsky, H. Perry, S. Bacchetti, R. Colonno, and R. B. Stein. 1988. Oligopeptides inhibit the ribonucleotide reductase of herpes simplex virus by causing subunit separation. Virology 162270-273. [DOI] [PubMed] [Google Scholar]
- 31.Perkins, D., E. F. Pereira, M. Gober, P. J. Yarowsky, and L. Aurelian. 2002. The herpes simplex virus type 2 R1 protein kinase (ICP10 PK) blocks apoptosis in hippocampal neurons, involving activation of the MEK/MAPK survival pathway. J. Virol. 761435-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petroski, M. D. 2008. The ubiquitin system, disease, and drug discovery. BMC Biochem. 9(Suppl. 1)S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 169555-72. [DOI] [PubMed] [Google Scholar]
- 34.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 7915582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmaus, S., H. Wolf, and F. Schwarzmann. 2004. The reading frame BPLF1 of Epstein-Barr virus: a homologue of herpes simplex virus protein VP16. Virus Genes 29267-277. [DOI] [PubMed] [Google Scholar]
- 36.Shackelford, J., and J. S. Pagano. 2004. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol. Cell. Biol. 245089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, C. C. 2005. The herpes simplex virus type 2 protein ICP10PK: a master of versatility. Front Biosci. 102820-2831. [DOI] [PubMed] [Google Scholar]
- 38.Smith, C. C., and L. Aurelian. 1997. The large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is associated with the virion tegument and has PK activity. Virology 234235-242. [DOI] [PubMed] [Google Scholar]
- 39.Stawiecka-Mirota, M., W. Pokrzywa, J. Morvan, T. Zoladek, R. Haguenauer-Tsapis, D. Urban-Grimal, and P. Morsomme. 2007. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic 81280-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szekeres, T., K. Gharehbaghi, M. Fritzer, M. Woody, A. Srivastava, B. van't Riet, H. N. Jayaram, and H. L. Elford. 1994. Biochemical and antitumor activity of trimidox, a new inhibitor of ribonucleotide reductase. Cancer Chemother. Pharmacol. 3463-66. [DOI] [PubMed] [Google Scholar]
- 41.Tihan, T., H. L. Elford, and J. G. Cory. 1991. Studies on the mechanisms of inhibition of L1210 cell growth by 3,4-dihydroxybenzohydroxamic acid and 3,4-dihydroxybenzamidoxime. Adv. Enzyme Regul. 3171-83. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 806003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 111245-1256. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson, K. D., K. M. Lee, S. Deshpande, P. Duerksen-Hughes, J. M. Boss, and J. Pohl. 1989. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246670-673. [DOI] [PubMed] [Google Scholar]