Abstract
Viral infection elicits the activation of numerous cellular signal transduction pathways, leading to the induction of both innate and adaptive immune responses in the host. In particular, interferon regulatory factor 3 (IRF3) has been shown to be essential for the induction of an antiviral response. Current models suggest that virus replication causes phosphorylation of C-terminal serine and threonine residues on IRF3, leading to its dimerization and translocation to the nucleus, where it activates interferon. Upon entry of replication-deficient Newcastle disease virus (NDV) particles, however, we failed to detect IRF3 dimerization or hyperphosphorylation, despite robust interferon-stimulated gene (ISG) and antiviral state induction and confirmation by small interfering RNA knockdown that IRF3 is essential for this response. To further compare the effects of various viruses and their replication status on IRF3 activation and to determine the minimal posttranslational modification required for IRF3 activation, two-dimensional gel electrophoresis and native polyacrylamide gel electrophoresis were employed. However, we failed to identify a minimal posttranslational modification of IRF3 that correlated with downstream biological activity, and the extent of posttranslational modification observed on IRF3 did not correlate with the degree of subsequent ISG induction. Thus, current techniques used to detect IRF3 activation are insufficient to infer its role in mediating downstream biological response induction and should be utilized with caution.
Mammals have evolved intricate innate and adaptive immune responses to combat pathogen infections. Following recognition of a virus by its host, a number of signal transduction pathways are activated, leading to both inflammatory and interferon (IFN)-mediated responses, which function to limit further dissemination of the virus throughout the host. Primarily, the first cell types to become infected by viruses are nonimmune cells like fibroblasts and epithelial cells. Entry of viruses into these cell types leads to the activation of type I IFN. Virus-induced activation of type I IFN genes is mediated by regulatory sequences in their promoters and requires transcription factors such as IFN regulatory factor 3 (IRF3), NF-κB, and AP-1 (17). Once type I IFN mRNAs are translated, they act in both an autocrine and paracrine manner during the innate immune response to limit virus replication by activating the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signal transduction cascade, leading to the activation of several hundred IFN-stimulated genes (ISGs) (8). Collectively, these ISGs act to limit further virus replication. IRF3 can also directly bind to the IFN-stimulated response element motif, which is located in the promoter region of ISGs, indicating that IRF3 also plays a role in direct induction of ISGs and of an antiviral response in the absence of IFN production (19).
The current model of IRF3 activation is derived predominantly from studying the binding and entry of single-stranded RNA viruses like Sendai virus (SeV) or Newcastle disease virus (NDV). Viral components are recognized by either membrane-bound (Toll-like receptor) or cytoplasmic (RIG-I, MDA5, or DAI) sensors, leading to the activation of the viral activated kinase, TBK1 (10, 28). Activated TBK1 phosphorylates IRF3 on distinct serine and threonine residues at its C terminus, leading to the release of its autoinhibitory domain and subsequent dimerization with another phosphorylated IRF3 molecule (23). This dimerized IRF3 is thought to translocate to the nucleus, where it then binds to IFN-stimulated response elements in the promoters of ISGs, including IFN-β. In the literature, hyperphosphorylation of IRF3 is defined as the addition of multiple phosphate moieties on specific serine and threonine residues in its carboxy terminus, which have previously been shown to be required for IRF3 dimerization and activation of ISGs, and results in a laddering effect on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (26, 27, 31). It is unclear, however, whether all viruses induce similar posttranslational modifications on IRF3.
In the literature, studies assessing IRF3 activation routinely involve the use of elevated levels of replication-competent virus. However, hyperphosphorylation of IRF3 was not detected following treatment of human fibroblasts with replication-deficient enveloped virus particles like herpes simplex virus, NDV, and SeV (6) even though a biologically active antiviral response was detected, suggesting that neither virus replication nor IRF3 hyperphosphorylation was required for the induction of an antiviral response following virus particle entry. Studies have shown that, in fact, hyperphosphorylation on IRF3 may be a by-product of virus replication, leading to unstable intermediates that are targets for degradation (4, 5). Thus, detectable IRF3 activation (or the lack thereof) is not suitably predictive of a biological response (5, 27, 28, 31) as there appears to be no correlation between IRF3 hyperphosphorylation and subsequent ISG induction.
While mounting evidence suggests that diverse viruses and their structural components are recognized by different intracellular sensors and that diverse viruses and their stages of infection lead to differential posttranslational modification of IRF3, all known pathways of IRF3 activation converge on TBK1 (or the related virus-activated kinase IκB kinaseɛ [IKKɛ]). Thus, it remains to be determined whether there is an observable posttranslational modification of IRF3 that is essential for its biological activity and that can be used as a predictive marker of IRF3 activation. To address this issue, we investigated the ability of both human cytomegalovirus (HCMV) and NDV, two enveloped viruses, to activate endogenous IRF3 using physiologically relevant amounts of virus in the presence and absence of virus replication. Using native gel electrophoresis and both one- and two-dimensional gel electrophoresis, we failed to detect a minimal posttranslational modification of IRF3 that correlated with the biological activity of this transcription factor. Furthermore, the extent of posttranslational modifications observed on IRF3 did not correlate with IRF3-mediated ISG induction. These data suggest that conventional techniques used to study and monitor IRF3 activation and infer subsequent biological response induction are not sufficiently predictive and should be utilized with caution.
MATERIALS AND METHODS
Cell culture, transfections, and virus infections.
Human embryonic lung (HEL) fibroblasts (ATCC) and Vero monkey kidney epithelial cells (ATCC) were grown in Dulbecco's modified essential medium supplemented with 10% or 5% fetal calf serum, respectively. Human embryonic kidney 293 cells were grown in minimal essential medium F-11 supplemented with 10% fetal calf serum. FLAG-tagged TBK1 and IRF3 plasmid constructs were a kind gift from R. Lin (McGill University). Briefly, 500 ng of plasmid DNA was transfected into 1 × 106 cells for 16 h using Lipofectamine 2000 (Invitrogen) prior to infection with any viruses. HCMV (strain AD169) (kindly provided by T. Compton) was propagated on HEL fibroblasts. NDV was kindly provided by E. Nagy. SeV was obtained from Charles River Laboratories and used at 80 hemagglutination units (HAU)/106 cells. UV inactivation of viruses was performed using a UV Stratalinker 2400 (Stratagene) for the length of time required to prevent IE1/IE2 and hemagglutinin-neuraminadase viral gene expression for HCMV and NDV, respectively, as measured by indirect immunofluorescence and reverse transcription-PCR (RT-PCR) analysis (see Fig. S1 in the supplemental material). The primers used to detect hemagglutinin-neuraminadase viral gene expression were the following: Forward, 5′-GCATACCGACTAGGATTTCC-3′; and reverse, 5′-CGAGCACAGCATATCACAACC-3′. Standard plaque reduction assays using replication-competent vesicular stomatitis virus (VSV) were performed as previously described (6). Individual virus preparations were serially diluted on HEL fibroblasts following UV inactivation to determine the multiplicity of infection (MOI) equivalent required to elicit an antiviral response. Supernatants were transferred to naïve Vero cells to test for the production of IFN using a standard plaque reduction assay.
Preparation of cellular extracts.
Whole-cell extracts were prepared as previously described (15). Cytoplasmic and nuclear extracts were prepared as previously described (3). Briefly, cells were washed twice with cold 1× phosphate-buffered saline (PBS) and then once with cold 0.2× PBS. On-plate lysis was carried out with cytoplasmic buffer (10 mM HEPES [pH 7.3], 10 mM potassium chloride, 1.5 mM magnesium chloride, 50 mM sodium fluoride, 1% Triton X-100, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and protease inhibitor cocktail [Sigma]). Lysates were spun at 12,000 × g for 3 min. Supernatants were collected and labeled, and pellets were washed once with cytoplasmic buffer to remove residual cytoplasmic proteins. Pellets were incubated with nuclear buffer (20 mM HEPES [pH 7.3], 25% glycerol, 420 mM sodium chloride, 1.5 mM magnesium chloride, 0.2 mM EDTA [pH 8.0], 50 mM sodium fluoride, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and protease inhibitor cocktail [Sigma]). Lysates were incubated on ice for 30 min with occasional vortexing and then spun at 13,000 × g for 10 min. Supernatants were desalted using a protein desalting spin column (Pierce). Protein concentrations were determined using the Bradford assay (Bio-Rad). Native or SDS sample buffer was added to an aliquot of each extract and then placed on ice or boiled for 10 min, respectively. Cytoplasmic or desalted nuclear extracts used in two-dimensional gel electrophoresis were incubated in rehydration buffer [7 M urea, 2 M thiourea, 2% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 0.5% immobilized pH gradient buffer (pH 4 to 7), 40 mM dithiothreitol, and 0.002% bromophenol blue].
Two-dimensional gel electrophoresis.
To separate proteins in the first dimension, 25 μg of either cytoplasmic or desalted nuclear extracts was spiked with 0.04 μg/μl of bovine serum albumin (BSA; Pierce) prior to equilibrating 13-cm Immobiline DryStrip gels (pH 4 to 7) (GE Healthcare) for 16 h at room temperature. After rehydration, isoelectric focusing was performed (25,000 V/h) on an Ettan IPGphor manifold system (GE Healthcare). After equilibration, the gel strips were transferred to a 7.5% SDS-PAGE gel for separation in the second dimension prior to immunoblot analysis. Following transfer of proteins to polyvinylidene difluoride, membranes were stained with Ponceau S to identify BSA, which facilitated comparative analysis between blots.
Immunoblot analysis.
Polyacrylamide gels were transferred onto either polyvinylidene difluoride (Millipore) or nitrocellulose (GE Healthcare) membranes for qualitative or quantitative immunoblot analysis, respectively. Antibodies used to recognize denatured IRF3 (Santa Cruz SC-9082) or native IRF3 (Immuno-Biological Laboratories Co. Ltd.) were used at a dilution of 1:1,000 in 5% BSA in Tris-buffered saline-Tween (0.1%). The phosphospecific IRF3 (IRF3 P396) (Cell Signaling) was used at a dilution of 1:1,000. The ISG-56 antibody (provided by G. Sen) was used at a dilution of 1:5,000, and the alpha-actin (Santa Cruz SC-1616) antibody was used at a dilution of 1:1,000. The monoclonal FLAG antibody (Sigma) was used at a dilution of 1:1,000. Secondary antibodies conjugated to horseradish peroxidase were used at a dilution of 1:5,000. Quantitative Western blots were developed using ECL Advanced Technology (GE Healthcare), scanned on a Typhoon Trio imager (GE Healthcare), and analyzed using Image Quant TL software (GE Healthcare). Qualitative Western blots were developed using enhanced chemiluminescence technology and exposed to film to visualize proteins.
IRF3 siRNA treatment.
HEL cells were seeded to 30% confluence into six-well dishes the day before small interfering RNA (siRNA) transfection. The Stealth siRNA (Invitrogen) for human IRF3 (5′-CAACCGCAAAGAAGGGUUGCGUUUA-3′ and 5′-UAAACGCAACCCUUCUUUGCGGUUG-3′) was transfected into cells at a concentration of 50 nM using the Lipofectamine RNAi Max reagent (Invitrogen). Nontargeting siRNA was used as a control. Knockdown efficiency was monitored using quantitative Western blot analysis for IRF3. Maximal IRF3 reduction was seen at 72 h following siRNA transfection. Forty-eight hours following transfection, cells were treated with UV-inactivated HCMV (HCMV-UV) or NDV (NDV-UV) for 24 h. Supernatants were transferred onto naïve HEL or Vero cells to test for the presence of soluble IFN released following virus particle treatment. Standard plaque reduction assays and RT-PCR analysis for ISG-56 expression were carried out as previously described (6).
RESULTS
Input virus levels dictate the biological response output.
HCMV and NDV are DNA and RNA viruses, respectively, that have been used to study the role of IRF3 in the induction of an innate antiviral response in nonimmune cells, including fibroblast and epithelial cells (7, 14, 22, 29, 32). Typically, high MOIs are used, resulting in downstream type I IFN induction. Given the observations that (i) viruses can elicit IRF3-mediated antiviral responses in the presence and absence of detectable IFN (6, 22, 33), (ii) a higher threshold of virus entry is required to activate NF-κB and elicit IFN production as opposed to the IRF3-mediated IFN-independent response (16), and (iii) different viruses and virus preparations display different particle-to-PFU ratios, it is imperative in studies of virus-mediated IRF3 activation to consider input virus levels.
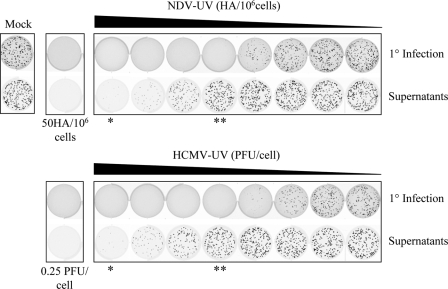
Replication-deficient HCMV or NDV virus particles were serially diluted on primary human fibroblasts (Fig. 1, top row), with corresponding supernatants transferred to naïve Vero cells (bottom row), to investigate the amount of each virus preparation required to induce an antiviral response in the presence or absence of IFN production. Vero cells were used to monitor the presence and absence of biologically active IFN since they have previously been shown to respond to, but not make, IFN due to a genetic deletion of the type I IFN locus (9). Both NDV-UV and HCMV-UV were capable of inducing an antiviral response in primary fibroblasts in the absence of detectable IFN production at low MOIs. At high MOIs, these virus particles induced an antiviral response due, in part, to the presence of IFN. It is important to note that the MOI at which a given virus preparation induced an IFN-independent antiviral state was different for individual virus preparations. For the HCMV and NDV preparations used in this study, the MOI of UV-inactivated virus capable of inducing an IFN-independent antiviral state ranged from 0.005 PFU/cell to 0.0156 PFU/cell and 0.1 HAU/106 cells to 5 HAU/106 cells, respectively. All subsequent studies (with both replicating and UV-inactivated virus) utilized 0.0156 PFU/cell and 0.25 PFU/cell (HCMV) and 5 HAU/106 cells and 50 HAU/106 cells (NDV) as low and high MOIs, respectively.
FIG. 1.
NDV-UV and HCMV-UV virus particles are capable of inducing an IFN-independent antiviral state at low MOIs. HEL fibroblasts were treated with NDV-UV at a range of 0 to 6.25 HAU/106 cells or HCMV-UV at a range of 0 to 0.0625 PFU/cell (primary infection [1°]). At 24 h postinfection supernatants were transferred onto naïve Vero cell monolayers to detect the presence of biologically active IFN. A plaque reduction assay was carried out with green fluorescent protein-tagged VSV to detect the presence of an antiviral state on each monolayer. To induce an IFN-dependent antiviral response, 50 HAU/106 cells or 0.25 PFU/cell of NDV-UV or HCMV-UV, respectively, was used in subsequent experiments. **, absence of IFN at low MOIs; *, presence of IFN at high MOIs.
IRF3 activation readouts differ between HCMV and NDV and fail to correlate with ISG induction.
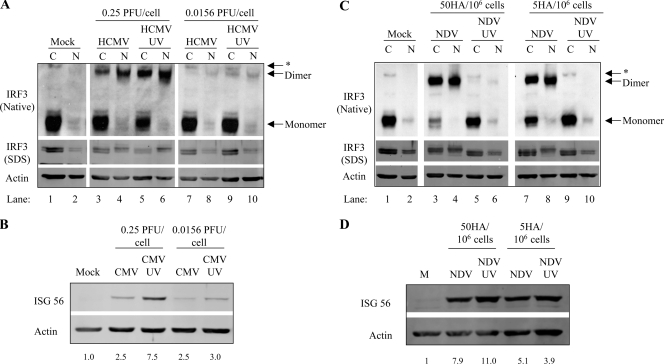
Since we along with others previously observed a disconnect between IRF3 posttranslational modification and ISG induction, a careful analysis was carried out following treatment with either replication-competent or UV-inactivated HCMV (Fig. 2A and B) or NDV (Fig. 2C and D) in primary human fibroblasts to investigate the kinetics of observable IRF3 activation following entry of two evolutionarily distinct viruses. For detection purposes, equal microgram amounts of cytoplasmic and nuclear protein extracts were analyzed, which represent an approximate sixfold enhancement of proteins in the nuclear fraction. A 10-h time course to determine the optimal time to detect IRF3 dimerization and posttranslational modification was initially carried out (see Fig. S2 in the supplemental material). Four hours and 8 h were the optimal time points to detect IRF3 activation following HCMV and NDV infection, respectively. Under resting conditions, IRF3 is a monomer that shuttles between the cytoplasm and nucleus (Fig. 2A and C, lanes 1 and 2). Following infection with HCMV at a low MOI, dimerization was minimally detected at 4 h postinfection, regardless of replication status (Fig. 2A, lanes 7 to 10), with dimers accumulating over time only in cells treated with UV-inactivated virus (see Fig. S2A in the supplemental material). Following infection with HCMV at a high MOI, dimers were detected as early as 2 h postinfection, with maximal accumulation detected at 4 h (HCMV-UV) and 6 h (replication-competent virus) (see Fig. S2A in the supplemental material). At both low and high MOIs, IRF3 posttranslational modification was detected in only the nuclear fractions (Fig. 2A; see also Fig. S2A in the supplemental material). Consistent with numerous reports in the literature, the observed posttranslational modification correlated with the appearance of Ser396-phosphorylated IRF3, detected using a phosphospecific antibody (data not shown). Both HCMV and HCMV-UV induced ISG-56 protein expression, with HCMV-UV activating ISG-56 more potently than HCMV at both high and low MOIs (Fig. 2B; see also Fig. S2B in the supplemental material).
FIG. 2.
Replication-deficient HCMV and NDV differentially activate IRF3 at high and low MOIs. (A) Primary fibroblasts treated at high and low MOIs of HCMV and HCMV-UV were harvested 4 h postinfection and subjected to native and denaturing gel electrophoresis. IRF3 is more potently activated following HCMV-UV treatment, leading to an increase in ISG-56 protein expression at 8 h (B). (C) Detection of IRF3 dimers and hyperphosphorylation are absent following NDV-UV but not NDV infection at 8 h postinfection even though ISG-56 protein can be detected in extracts from both treatments (D). ISG-56 protein levels relative to actin controls were determined by quantitative Western blot analysis and are listed below each sample (B and D). *, nonspecific band; C, cytoplasmic; N, nuclear; M, mock.
A different profile of IRF3 dimerization was detected following infection with NDV (Fig. 2C; see also Fig. S2C in the supplemental material). Following infection at both high and low MOIs, IRF3 dimers were reproducibly visualized following infection with replication-competent virus only, despite similar levels of ISG-56 protein induction (Fig. 2D; see also Fig. S2D in the supplemental material). As with HCMV infection, IRF3 hyperphosphorylation was predominantly detected in the nucleus (Fig. 2C, lanes 4 and 8) even under conditions where a substantial percentage of cytoplasmic IRF3 is present as a dimer (Fig. 2C, lanes 3 and 7). These data demonstrate a disconnect between ISG induction and detectable hallmarks of IRF3 activation and indicate that entry of diverse enveloped virus particles differentially activates IRF3 to elicit an antiviral response at high and low MOIs. Two possibilities exist to explain the absence of detectable IRF3 activation despite the induction of ISG-56 and the antiviral state. First, the sensitivity of our assays in detecting IRF3 hyperphosphorylation and dimerization could be too low. Second, it is possible that IRF3 is not essential in the induction of ISG-56 in response to entry of a subset of enveloped virus particles, including NDV, in primary human fibroblasts.
IRF3 is required for the induction of an antiviral state following entry of replication-deficient HCMV and NDV.
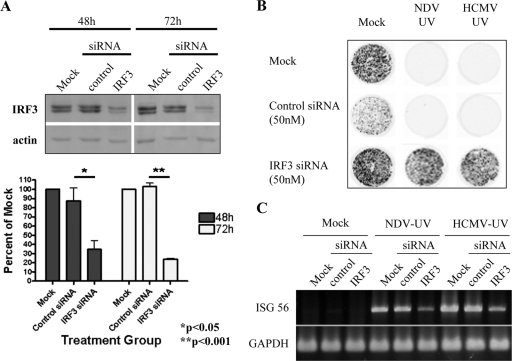
To confirm that IRF3 is essential for the induction of ISGs and an antiviral state following treatment with NDV-UV despite the absence of detectable hyperphosphorylation and dimerization, knockdown of IRF3 was performed. Following transfection of siRNA oligonucleotides specific for human IRF3, the level of IRF3 protein was significantly decreased at 72 h compared to that of cells transfected with mock or control siRNA (Fig. 3A). siRNA-mediated knockdown of IRF3 was able to completely prevent the induction of an antiviral state following treatment with NDV-UV or HCMV-UV (Fig. 3B). Although RT-PCR analysis following siRNA-mediated knockdown of IRF3 resulted in a decrease in the level of ISG-56 accumulation, low levels of this ISG were still detected, consistent with the failure to knock down IRF3 to undetectable levels (Fig. 3C). It is important to note, however, that this level of ISG-56 was insufficient to induce an antiviral state in these cells. Collectively, these data suggest that IRF3 is essential for the induction of an antiviral state in human fibroblasts following treatment with NDV-UV or HCMV-UV.
FIG. 3.
Knockdown of IRF3 prevents the induction of an antiviral state following treatment with NDV-UV or HCMV-UV. (A) Subconfluent HEL fibroblasts were pretreated with a 50 nM concentration of either siRNA specific for human IRF3 or a scrambled control siRNA. At 48 h and 72 h posttransfection, whole-cell extracts were harvested, and SDS-PAGE was performed, followed by Western blotting for IRF3. The blot was subsequently probed for actin. Blots were developed using an ECL Advanced kit and scanned using the Typhoon scanner. A graphical representation of the relative intensity of IRF3 protein compared to actin in three independent experiments is shown in the bottom panel. Statistical analysis was performed by a one-way analysis of variance with the Tukey test posthoc. (B and C) HEL fibroblasts were mock treated or treated with a control or IRF3-specific siRNA oligonucleotide for 48 h. Fibroblasts were then treated with 5 HAU/106 cells (IFN independent) of NDV-UV or 0.01 PFU/cell (IFN independent) of HCMV-UV for 24 h. A plaque reduction assay with green fluorescent protein-tagged VSV (B) or RT-PCR analysis (C) for the presence of ISG-56 expression was performed. Plates treated with green fluorescent protein-tagged VSV were then imaged using the Typhoon scanner 24 h later.
The two-dimensional profile of endogenous IRF3 differs between viral stimuli and does not correlate with ISG induction.
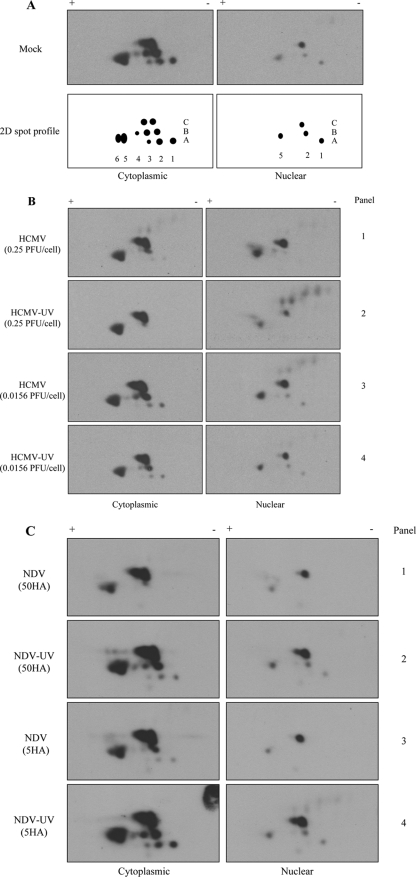
Since one-dimensional gel electrophoresis was insufficient to detect reproducible hallmarks of IRF3 activation, two-dimensional gel electrophoresis was used to assess the activation status of IRF3. Two-dimensional gel electrophoresis has previously been used to look at posttranslational modifications on IRF3 and IRF7 (20, 25). To investigate differences in IRF3 modification following infection with replicating or UV-inactivated HCMV and NDV using low or high MOIs, extracts from the experiment shown in Fig. 2 were additionally separated by two-dimensional gel electrophoresis. Mock-infected cytoplasmic and nuclear extracts produced similar, though slightly different, spot profiles over multiple experiments (compare Fig. 4A and 5A). Following infection at a high MOI with HCMV or HCMV-UV (Fig. 4B, panels 1 and 2), no new spots were detected in the cytoplasmic extracts compared to mock-infected extracts even though IRF3 dimerization was detected (Fig. 2A). Instead, a loss of spots A1 to A3 and B4 (spots are designated according to the coordinates shown in Fig. 4A) was observed. Accumulation of spots C2 and B5 in HCMV-infected nuclear extracts was observed, suggesting that these spots might be involved in IRF3 hyperphosphorylation since hyperphosphorylation was observed by one-dimensional PAGE (Fig. 2A, lane 4). With infection at a low MOI, the two-dimensional profile of IRF3 within the cytoplasm and nucleus did not change significantly compared to mock-infected extracts (Fig. 4B, panels 3 and 4) even though ISG-56 protein was detected in these extracts (Fig. 2B). Detection of nonspecific spots could sometimes be seen in the two-dimensional profile of IRF3 (Fig. 4B, panels 2 and 3, nuclear). These spots were also detected in mock-infected extracts of some experiments and are likely an artifact of individual extract preparation.
FIG. 4.
The two-dimensional profile of endogenous IRF3 is different following HCMV or NDV treatment. Human fibroblasts were treated with replication-competent or deficient HCMV (0.25 PFU/cell for IFN-dependent and 0.0156 PFU/cell for IFN-independent responses) or NDV (50 HAU/106 cells for IFN-dependent and 5 HAU/106 cells for IFN-independent responses) for 4 h or 8 h, respectively. Cytoplasmic and nuclear fractions were harvested, and individual extracts were run on a two-dimensional (2D) PAGE gel. (A) A representative two-dimensional pattern of endogenous IRF3 in mock-infected cytoplasmic and nuclear extracts from more than 10 independent experiments. The representative two-dimensional spot profile is illustrated. (B) Two-dimensional profile of HCMV- and HCMV-UV-treated samples that elicit both an IFN-dependent and -independent response. (C) Two-dimensional profile of NDV- and NDV-UV-treated samples that elicit both IFN-dependent and -independent responses. All two-dimensional blots were aligned using exogenously added BSA prior to separation of the samples in the first dimension. The numbers in the two-dimensional spot profile for cytoplasmic IRF3 correspond to the numbers in the profile for nuclear IRF3. +, pH 4; −, pH 7.
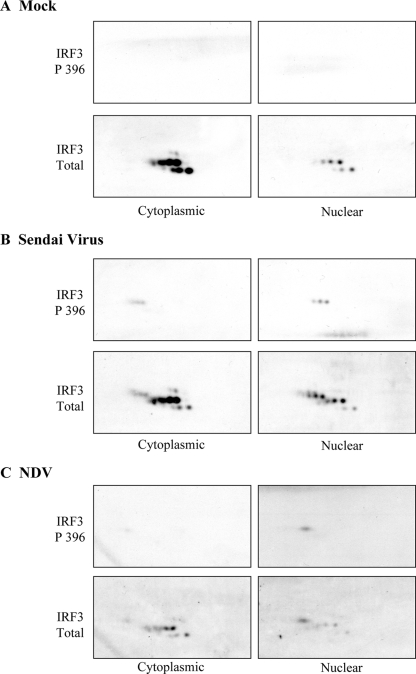
FIG. 5.
The two-dimensional profile of phosphorylated IRF3 (Ser396) is different following either SeV or NDV infection. HEL fibroblasts were either mock infected (A) or infected with Sendai virus (B) or NDV (C) for 8 h. Cytoplasmic and nuclear extracts were harvested, and two-dimensional gel electrophoresis followed by immunoblot analysis for phosphorylated IRF3 (IRF3 P396) was performed. Blots were subsequently probed for total IRF3. Exogenously added BSA was used to align spots between immobilized pH gradient strips. Amounts of SeV and NDV were enough to induce an IFN-dependent antiviral response (i.e., 80 HAU or 50 HAU per 106 cells for SeV or NDV, respectively).
Treatment of fibroblasts with NDV or NDV-UV led to a different two-dimensional IRF3 profile compared to what was seen with HCMV or HCMV-UV. Following infection with replicating NDV, the disappearance of spots A1 to A3 along with the accumulation of spot B5 was seen in cytoplasmic extracts (Fig. 4C, panels 1 and 3). Interestingly, the two-dimensional profile of IRF3 following NDV-UV treatment was virtually identical to that of mock-infected cytoplasmic extracts, regardless of the MOI (Fig. 4C, panels 2 and 4). Moreover, in NDV-infected nuclear extracts where dimers and hyperphosphorylated IRF3 were readily detected by one-dimensional gel electrophoresis, no new spots were formed relative to the mock-infected nuclear IRF3 profile. Instead, the disappearance of two spots was observed relative to the mock-infected nuclear IRF3. However, with NDV-UV treatment, where we failed to see detectable IRF3 dimerization and hyperphosphorylation, the appearance of a new spot beside C2 was detected in nuclear extracts relative to mock-infected extracts, suggesting that there is no correlation whatsoever between the various profiles of IRF3 and activation of a biological response.
The inability to readily detect novel IRF3 species fails to address whether IRF3 becomes posttranslationally modified since a newly modified species of IRF3 may overlap on a two-dimensional profile with an existing species of IRF3. Thus, to investigate whether IRF3 phosphorylation can be detected following two-dimensional gel electrophoresis, a phosphospecific IRF3 antibody was used. Extracts from fibroblasts infected with NDV and the related paramyxovirus SeV were compared since we routinely observe enhanced IRF3 phosphorylation following SeV infection of fibroblasts relative to NDV infection, despite induction of similar antiviral responses (6). Ser396 phosphorylation was not detected in mock-infected cellular extracts (Fig. 5A). Interestingly, following SeV treatment three new species of IRF3 were readily detected in both the cytoplasmic and nuclear fractions using the phosphospecific IRF3 antibody that were not detected in the mock-infected extracts (Fig. 5B). In contrast to findings in Fig. 4, a new IRF3 species, consistent with phosphorylation on Ser396, was detected in the nuclear fraction of extracts harvested from NDV-infected fibroblasts (Fig. 5C). Collectively, these data demonstrate that despite the ability of either diverse (NDV and HCMV) or related (NDV and SeV) viruses to elicit a similar robust ISG induction, current methodologies are insufficient to utilize IRF3 posttranslational modification as a readout for IRF3 biological activity. Furthermore, these data indicate that although diverse and related viruses may elicit a similar biological response, their recognition by cellular components may be distinct, leading to differential activation of downstream mediators, such as IRF3.
Dimerization of IRF3 is required for the induction of ISG-56 following treatment with NDV-UV.
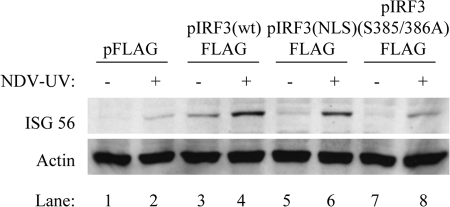
The current model for IRF3 activation suggests that phosphorylation and dimerization of IRF3 are essential for its subsequent activation of ISGs and an antiviral state following virus stimulation. There is literature showing that IRF3 dimerization is not detected following treatment of human macrophages with poly(I:C), despite robust ISG induction (24). Since dimerization of IRF3 was undetectable following NDV-UV treatment, we investigated whether IRF3 dimerization was required for ISG-56 induction following NDV-UV treatment in primary fibroblast cells. Previous work has shown that phosphorylation of Ser385 and Ser386 on IRF3 is required for dimerization (13, 18). To test whether dimerization of IRF3 was required for activation of ISGs following NDV-UV treatment, 293 cells were transfected with an IRF3 construct containing substitutions of Ala residues for Ser385 and Ser386 (pIRF3-FLAG S385/386A). As previously observed, overexpression of wild-type IRF3 activated ISG-56 to low levels in the absence of other stimuli (Fig. 6, compare lanes 1 and 3). Following NDV-UV treatment, however, the level of ISG-56 increased relative to mock infections (lanes 3 and 4). Cells overexpressing the nuclear localization signal mutant demonstrated an intermediate level of ISG induction, consistent with intermediate levels of nuclear IRF3 (data not shown). When cells were transfected with the dimerization mutant, however, the level of ISG-56 was significantly decreased relative to cells treated with wild-type IRF3 and NDV-UV (Fig. 6, compare lanes 4 and 8). These data suggest that although endogenous IRF3 dimerization is not detectable following NDV-UV treatment (Fig. 2C), this event is required for activation of the antiviral response.
FIG. 6.
IRF3 dimerization is required for the activation of ISG-56 following NDV-UV particle treatment. HEK 293 cells were transfected with the following vectors for 16 h: pFLAG, pIRF3 (wt)-FLAG (where wt is wild type), pIRF3 (NLS)-FLAG (where NLS is nuclear localization signal), and pIRF3(S385/386A)-FLAG. Monolayers were subsequently infected with NDV-UV for 8 h prior to the harvest of cytoplasmic extracts. Immunoblot analysis for the presence of ISG-56 was carried out. Actin was used as a cell loading control.
DISCUSSION
The current model of virus-mediated IRF3 activation involves the activation of the kinases TBK1/IKKɛ through both Toll-like receptor-dependent and -independent mechanisms, resulting in phosphorylation of key serine and threonine residues located in the C-terminal regulatory domain of IRF3 (11, 13, 26-28, 32). In the literature, hyperphosphorylation of IRF3 has been used as a readout for IRF3 activation, with the assumption that IRF3 hyperphosphorylation correlates with subsequent ISG induction (11, 13, 26, 28, 31). However, we along with others have shown that ISG induction does not always correlate with IRF3 hyperphosphorylation and degradation (5, 6). Furthermore, ISG induction has been observed in response to virus infection in an IRF3-independent fashion (21).
It is important both to understand the limitations of current assays and to identify biologically relevant hallmarks of IRF3 activation to properly characterize the role of this transcription factor in the innate immune response to virus infection. To fully understand the nature of the innate cellular response, it is also important to consider input virus levels since activation of different transcription factors, like IRF3 and NF-κB, requires different thresholds of virus input (16). Also, since replication-defective virus particles can activate IRF3 and NF-κB (albeit with different thresholds), it is important to consider the particle/PFU ratio of each individual virus preparation. Recently, we proposed a model in which exposure to increasing numbers of virus particles increases the complexity of the cellular response from an intracellular, IFN-independent response to one involving the secretion of cytokines and activation of immune cells, including dendritic cells and macrophages (16).
Despite a similar biological outcome (ISG-56 production and the induction of an antiviral state), HCMV and NDV modify IRF3 differently. With NDV, observable IRF3 modification is primarily dependent on replication competency and not MOI, while with HCMV, observable IRF3 modification is primarily dependent on MOI and not replication competency. The reduction in ISG-56 protein levels in HCMV-infected cells compared to HCMV-UV-treated cells may be due to production of viral proteins such as IE86 and/or pp65, which have been shown to dampen virus-mediated cytokine induction (1, 30). Despite differences in IRF3 modification between NDV and HCMV, ISG induction did not correlate with the characteristic hallmarks of IRF3 activation following virus infections, consistent with previous observations (5). The lack of observable IRF3 dimer formation following NDV-UV treatment at both high and low MOIs did not correlate with its ability to induce ISG-56. Additionally, the two-dimensional profile of endogenous cytoplasmic IRF3 is very similar in fibroblasts treated with either high MOIs of HCMV and HCMV-UV or high and low MOIs of replicating NDV. Strikingly, very little, if any, IRF3 hyperphosphorylation is detected in cytoplasmic extracts following these virus treatments even though significant IRF3 dimerization is observed following native PAGE. Moreover, the two-dimensional profiles of nuclear IRF3 following these virus treatments failed to consistently demonstrate the creation of new spots that were not already identified in the cytoplasmic fractions. Instead, IRF3 isoforms present in the cytoplasmic fractions appeared to translocate to the nucleus, which does not account for the observable hyperphosphorylation of IRF3 by one-dimensional gel electrophoresis. Further analysis of the two-dimensional profiles of IRF3 following treatment with the related paramyxoviruses SeV and NDV, which induce similar antiviral states and ISG accumulation in cells (2, 6, 12), uncovered different two-dimensional patterns using both total IRF3 and phospho-IRF3-specific antibodies. Interestingly, Reimer et al. (24) recently demonstrated that poly(I:C) treatment of human macrophages led to robust ISG induction in the absence of detectable IRF3 dimerization. In contrast, the authors observed IRF3 dimers and ISG induction in response to lipopolysaccharide, suggesting that IRF3 can be differentially activated depending on the stimulus used and illustrating the importance of using the correct assays and model systems to study IRF3 activation.
Our data also suggest that the lack of observable modification of IRF3 does not necessarily imply that either IRF3 or the modification is dispensable. Prescott et al. (21) recently identified a novel pathway that did not require viral entry, any of the characterized pattern recognition receptors, or IRF3 following Sin Nombre infection. Even though neither dimerization nor hyperphosphorylation of IRF3 was observed following NDV-UV treatment, IRF3 was found to be essential for the induction of an antiviral response, along with phosphorylation of Ser385/386. A simple interpretation would then be that, for some forms of virus stimulation (e.g., treatment of fibroblasts with NDV-UV), neither dimerization nor hyperphosphorylation of IRF3 is required. However, overexpression of a dimerization mutant of IRF3 prevented the induction of ISG-56 following NDV-UV treatment. Similarly, while the current model of IRF3 activation involves phosphorylation by TBK1/IKKɛ in the cytoplasm and then translocation to the nucleus, one- and two-dimensional profiles of IRF3 suggest that IRF3 is posttranslationally modified in the nucleus, not in the cytoplasm. Further experiments are required to address this apparent discrepancy.
There is increasing evidence in the literature to support a two-step model of IRF3 activation. First, Sarkar et al. observed that following poly(I:C)-mediated phosphorylation, dimerization, and nuclear translocation of IRF3, a second posttranslational modification on IRF3 via phosphoinositide-3 (PI3) kinase was required for full activation of IRF3 and subsequent ISG induction (25). In addition, we previously reported that antiviral response induction mediated by virus particle entry requires a novel phosphoinositide-3 kinase family member to activate IRF3 following its dimerization and nuclear translocation (15). In vitro biochemical analyses using purified IRF3 and TBK1 revealed that TBK1 was solely responsible for the phosphorylation of serine and threonine residues in site 2 (amino acids 396 to 405) and subsequent release of autoinhibition on IRF3 and interaction with CBP in the absence of dimerization, suggesting that a second kinase may be responsible for dimerization and full activation of IRF3 (18). Furthermore, Clement et al. described a mechanism in which phosphorylation of Ser396 on IRF3 enhances the antiviral response by exposing Ser339 for subsequent phosphorylation by an unidentified proline-directed kinase, resulting in hyperphosphorylation, dimerization and association with CBP (5). Taken together, these data strongly suggest the existence of other protein kinases that function to further activate the IRF3-mediated antiviral response after initial phosphorylation by the virus-activated kinases, TBK1/IKKɛ. Regardless of the likelihood of these alternative pathways, however, current techniques are insufficient to monitor and characterize the subsequent modification profile of IRF3 and infer biological activity.
Taken together, these data emphasize that despite the similarities and cross talk in innate viral recognition pathways (e.g., all known pattern recognition receptor pathways converge on TBK1), the resulting modification of IRF3 can be diverse (or undetectable), and depends on the source of virus, its replication status, and the MOI. More importantly, however, these diverse alterations of IRF3 (or lack thereof) fail to reflect downstream biological events, particularly when the activity of endogenous IRF3 is assessed in the context of infection at a low MOI. Thus, caution should be taken in the use of conventional assays to infer the requirement of IRF3 or a particular modification for induction of an innate antiviral response. Furthermore, it should be appreciated that despite similarities in the induced biological outcomes, diverse and related viruses differentially activate innate immune modifiers such as IRF3, and thus “virus activation” of these modifiers must be studied in a virus-specific fashion.
Supplementary Material
Acknowledgments
We thank R. Lin, E. Nagy, G. Sen, and T. Compton for reagents used in this study. We also acknowledge the helpful discussions with member of the Sen laboratory at the Cleveland Clinic Foundation and member of the Macri laboratory at McMaster University regarding two-dimensional gel electrophoresis.
This work was supported by a grant from the Canadian Institutes of Health Research. R.S.N. was supported by a Natural Sciences and Engineering Research Council postgraduate scholarship. K.L.M. was supported by an Rx & D Health Research Foundation career award.
Footnotes
Published ahead of print on 11 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 7810995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J., S. VanScoy, T. F. Cheng, D. Gomez, and N. C. Reich. 2008. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9168-175. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 192499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibeau-Poirier, A., S. P. Gravel, J. F. Clément, S. Rolland, G. Rodier, P. Coulombe, J. Hiscott, N. Grandvaux, S. Meloche, and M. J. Servant. 2006. Involvement of the IκB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 1775059-5067. [DOI] [PubMed] [Google Scholar]
- 5.Clement, J. F., A. Bibeau-Poirier, S. P. Gravel, N. Grandvaux, E. Bonneil, P. Thibault, S. Meloche, and M. J. Servant. 2008. Phosphorylation of IRF-3 on Ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J. Virol. 823984-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 781706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFilippis, V. R., B. Robinson, T. M. Keck, S. G. Hansen, J. A. Nelson, and K. J. Früh. 2006. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 801032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S.-M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 11.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 182986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 2799698-9702. [DOI] [PubMed] [Google Scholar]
- 14.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 183796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noyce, R. S., S. E. Collins, and K. L. Mossman. 2006. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J. Virol. 80226-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paladino, P., D. T. Cummings, R. S. Noyce, and K. L. Mossman. 2006. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 1778008-8016. [DOI] [PubMed] [Google Scholar]
- 17.Panne, D., T. Maniatis, and S. C. Harrison. 2007. An atomic model of the interferon-beta enhanceosome. Cell 1291111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panne, D., S. M. McWhirter, T. Maniatis, and S. C. Harrison. 2007. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J. Biol. Chem. 28222816-22822. [DOI] [PubMed] [Google Scholar]
- 19.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFκB- and JNK-independent activation of the 561 and IFN-β genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 996322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prakash, A., and D. E. Levy. 2006. Regulation of IRF7 through cell type-specific protein stability. Biochem. Biophys. Res. Commun. 34250-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott, J. B., P. R. Hall, V. S. Bondu-Hawkins, C. Ye, and B. Hjelle. 2007. Early innate immune responses to Sin Nombre hantavirus occur independently of IFN regulatory factor 3, characterized pattern recognition receptors, and viral entry. J. Immunol. 1791796-1802. [DOI] [PubMed] [Google Scholar]
- 22.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 758909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, B. Y., C. Liu, S. S. Lam, H. Srinath, R. Delston, J. J. Correia, R. Derynck, and K. Lin. 2003. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat. Struct. Biol. 10913-921. [DOI] [PubMed] [Google Scholar]
- 24.Reimer, T., M. Brcic, M. Schweizer, and T. W. Jungi. 2008. poly(I:C) and LPS induce distinct IRF3 and NF-κB signaling during type-I IFN and TNF responses in human macrophages. J. Leukoc. Biol. 831249-1257. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 111060-1067. [DOI] [PubMed] [Google Scholar]
- 26.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 2789441-9447. [DOI] [PubMed] [Google Scholar]
- 27.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276355-363. [DOI] [PubMed] [Google Scholar]
- 28.Sharma, S., B. R. tenOever, N. Grandvaux, G.-P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 3001148-1151. [DOI] [PubMed] [Google Scholar]
- 29.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 2768951-8957. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.tenOever, B. R., S. Sharma, W. Zou, Q. Sun, N. Grandvaux, I. Julkunen, H. Hemmi, M. Yamamoto, S. Akira, W. C. Yeh, R. Lin, and J. Hiscott. 2004. Activation of TBK1 and IKKɛ kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 7810636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 171087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 9413985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.