Abstract
Infection of mice with murine gammaherpesvirus 68 (MHV-68) provides a valuable animal model for gamma-2 herpesvirus (rhadinovirus) infection and pathogenesis. The MHV-68 orf73 protein has been shown to be required for the establishment of viral latency in vivo. This study describes a novel transcriptional activation function of the MHV-68 orf73 protein and identifies the cellular bromodomain containing BET proteins Brd2/RING3, Brd3/ORFX, and BRD4 as interaction partners for the MHV-68 orf73 protein. BET protein members are known to interact with acetylated histones, and Brd2 and Brd4 have been implicated in fundamental cellular processes, including cell cycle regulation and transcriptional regulation. Using MHV-68 orf73 peptide array assays, we identified Brd2 and Brd4 interaction sites in the orf73 protein. Mutation of one binding site led to a loss of the interaction with Brd2/4 but not the retinoblastoma protein Rb, to impaired chromatin association, and to a decreased ability to activate the BET-responsive cyclin D1, D2, and E promoters. The results therefore pinpoint the binding site for Brd2/4 in a rhadinoviral orf73 protein and suggest that the recruitment of a member of the BET protein family allows the MHV-68 orf73 protein to activate the promoters of G1/S cyclins. These findings point to parallels between the transcriptional activator functions of rhadinoviral orf73 proteins and papillomavirus E2 proteins.
Many gammaherpesviruses promote lymphocyte proliferation and can persist in the lymphocyte compartment as well as in a number of other cell types, including endothelial and epithelial cells and fibroblasts (16, 17, 19, 56, 59). To ensure episome maintenance in these dividing cells, gammaherpesviruses express proteins that facilitate the replication of latent viral episomes and ensure the segregation of these viral genomes into progeny cells during cell division. The Epstein-Barr virus (EBV) EBNA-1 protein fulfills these essential functions (49, 62), as do the proteins encoded by open reading frame 73 (ORF73) of the gamma-2 herpesviruses (rhadinoviruses) Kaposi's sarcoma-associated herpesvirus (KSHV), herpesvirus saimiri (HVS), and murine gammaherpesvirus 68 (MHV-68) (4, 20, 28, 29, 43). The orf73 protein of KSHV, latency associated nuclear antigen 1 (LANA-1), is well characterized and has multiple functions. LANA-1 mediates replication and episome maintenance, acts as a transcriptional repressor and activator, and deregulates the cell division cycle (3, 24, 50). Some of these functions are mediated via the interaction of LANA-1 with a number of cellular proteins, including p53 (23), the retinoblastoma protein (48), MeCP2 (35), mSin3 (36), and multiple members of the BET (bromodomain and extra terminal domain) family of proteins (45, 47, 65). The MHV-68 orf73 protein, the KSHV LANA-1 homolog, is critical for the establishment and maintenance of a latent infection in mice (20, 43). MHV-68 orf73 is expressed in latently infected cells as well as during lytic infection (2, 15, 19, 51). The molecular details of how the MHV-68 protein functions are still largely unexplored.
BET proteins interact via their bromodomains with acetylated histones (13, 30, 33) and are highly conserved, with members in plants, yeast, Drosophila melanogaster, and up to mammals (18). An additional characteristic feature of BET proteins is the highly conserved extra terminal (ET) domain (see Fig. 1), which serves as a protein-protein interaction module in Brd2, Brd3, and Brd4 with KSHV LANA-1 (45, 47, 65) and between the yeast (Saccharomyces cerevisiae) BET protein Bdf1 and the TAF7 subunit of the general transcription factor TFIID (39). Mammalian BET proteins are encoded by four genes, Brd2, Brd3, Brd4, and Brd6, out of which Brd2, also called RING3, and Brd4 are the best characterized. Brd2 is a transcriptional regulator that plays a role in cell cycle regulation (11, 12, 33, 55). Brd2 overexpression in B lymphocytes in vivo in mice has been shown to induce lymphoma (26). Brd4 interacts with pTEFb (transcriptional elongation factor b) and thereby promotes RNA polymerase II (Pol II)-dependent transcriptional elongation (7, 32, 61). Brd4 overexpression and depletion both result in deregulated cell cycle progression (14, 38, 45). Recently, Brd4 has been shown to play an important role in the G1/S transition through its ability to stimulate transcription of G1/S-specific genes, among them, cyclin D1 and cyclin D2 (42). Furthermore, the C-terminal domain of the long isoform of Brd4 (Brd4L) (amino acids [aa] 1 to 1362) serves as a chromatin tether for different papillomaviruses via its interaction with their E2 proteins (1, 6, 63, 64). Brd4 is critical for papillomavirus E2 transcriptional activation (31, 40, 54) and may play a role in its transcriptional repression function (53, 60). The same C-terminal domain of Brd4L has recently been shown to interact with the transcriptional elongation factor pTEF and to inhibit human immunodeficiency virus type 1 (HIV-1) Tat-mediated, pTEF-dependent transcription (7). We have shown previously that KSHV LANA-1 interacts with Brd2 and Brd4 via their conserved ET domains (45, 57, 58). This interaction contributes to the association of LANA-1 with cellular heterochromatin and modulates the transcriptional activator role of the short isoform of Brd4 (Brd4S) (aa 1 to 722), which lacks the pTEFb interaction domain and by itself activates the cyclin E promoter (7, 45, 57, 58).
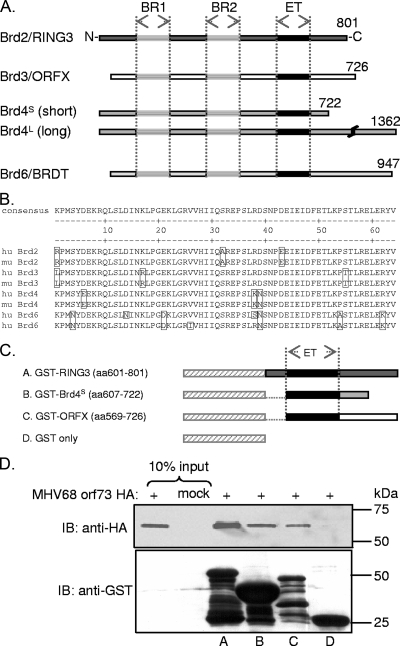
FIG. 1.
The MHV-68 orf73 protein interacts with the carboxy termini of the cellular BET proteins Brd2/RING3, Brd4S, and Brd3/ORFX. (A) Schematic depiction of the human BET proteins. BR1, bromodomain 1; BR2, bromodomain 2. (B) High degree of sequence conservation among human and murine ET domains. The human BET proteins are encoded by four genes, Brd2, Brd3, Brd4, and Brd6, and are characterized by two bromodomains and an ET domain. The BET protein sizes range from 722 to 1,362 aa for Brd4S and Brd4L, respectively. (B) ClustalW protein sequence alignment of human (hu) and murine (mu) BET protein 64-aa-long ET domains. ET domains of hu Brd2 (aa 640 to 703), mu Brd2 (aa 638 to 701), hu Brd3 (aa 570 to 633), mu Brd3 (aa 571 to 634), hu Brd4 (aa 608 to 671), mu Brd4 (aa 609 to 672), hu Brd6 (aa 508 to 571), and mu Brd6 (aa 504 to 567) were aligned. Boxed residues differ from the consensus sequence. The alignment shows 100% identity of the human and murine orthologous Brd2 ET domains, of human and murine Brd3 ET domains, and of human and murine Brd4 ET domains. The Brd6 ET domains of mice and humans are 95.3% identical. ET domain identity between paralogous BET proteins is also high, ranging from 84.4% to 92.2%. (C) Schematic of the GST-BET fusion proteins. Labels A, B, C, and D correspond to lane labeling in panel D. (D) The full-length MHV-68 orf73-HA protein interacted with GST-BET fusion proteins containing the ET domains of Brd2/RING3, Brd4, and Brd3/ORFX in GST pull-down assays. Bacterially expressed GST fusion proteins were bound to glutathione Sepharose resin and subsequently incubated with lysates from 293T cells transfected with MHV-68 orf73-HA expression plasmid. Ten percent of the input lysates was loaded for comparison. Top, anti-HA immunoblot (IB) used to detect the orf73 protein; bottom, anti-GST immunoblot used to verify expression of the GST-BET fusion proteins.
In this study we show that Brd2, Brd3, and Brd4 also interact with the MHV-68 orf73 protein. By identifying and mutating a binding site for Brd4 and Brd2 in the MHV-68 orf73 protein, we show that the orf73/BET interaction is crucial for the ability to activate the cyclin D1, D2, and E promoters. The results pinpoint the binding site for two BET proteins in a rhadinoviral orf73 protein and indicate that, similarly to papillomavirus E2, a rhadinoviral orf73 protein utilizes a member of the BET protein family to exert its transcriptional activation function.
MATERIALS AND METHODS
Cell culture methods.
Murine 3T3 fibroblasts as well as the human embryonic kidney epithelial cell line HEK 293T were cultured at subconfluence in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum, 50 IU/ml penicillin and 50 μg/ml streptomycin, and 100 μg/ml glutamine. The KSHV- and EBV-negative human Burkitt's lymphoma cell line BJAB was maintained in RPMI 1640 with 10% bovine growth serum plus 50 IU/ml penicillin and 50 μg/ml streptomycin. Spodoptera frugiperda SF9 insect cells were cultured in spinner flask cultures as previously described (45).
DNA constructs and baculoviruses.
The full-length MHV-68 orf73 clone was generated by PCR using DNA from MHV-68-infected cells and the forward primer ORF73 START (ATGCGGCCGCCGCCGCCACCATGCCCACATCCCCACCGAC, NotI site and orf73 start codon underlined), which has an optimized Kozak sequence and binds at genomic bp 104868 (GenBank accession number U97553), and the reverse primer ORF73 STOP (GCGGATCCTTAAGCGTAGTCTGGAACGTCGTATGGGTAAGCGTAGTCTGGAACGTCGTATGGGTATGTCTGAGACCCTTGTCCCTG, BamH1 site underlined), which introduces a double hemagglutinin (HA) tag and binds at genomic bp 103927. The PCR product was inserted into pVR1255 to obtain an MHV-68 orf73 full-length expression construct with a double HA tag at the 3′ end/C terminus (pVR1255 orf73). This and all other generated constructs were sequenced. All MHV-68 orf73 constructs were derived from this plasmid.
The MHV-68 orf73 constructs with internal mutations were generated using a site-directed mutagenesis kit (Stratagene), pVR1255 orf73 as the template, and appropriate primers. To introduce larger mutations, e.g., the 225-7A mutation (described below), up to three successive rounds of site-directed mutagenesis were performed using a template with intermediate mutations. Details can be obtained from the authors upon request. Constructs were sequence verified. The vector EGFPC1 was purchased from Clontech. The enhanced green fluorescent protein (EGFP)-Brd2/RING3 full-length (aa 2 to 801) and EGFP-Brd4S full-length (aa 2 to 722) vectors were described previously (45, 58). The vector myc-Rb was a kind gift from S. Mittnacht, ICR, London, United Kingdom.
For prokaryotic expression, the constructs glutathione S-transferase (GST)-Brd2/RING3 (aa 601 to 801), GST-Brd4S (aa 607 to 722), and GST-Brd3/ORFX (aa 569 to 726) (see Fig. 1C and D) were described previously (45).
All luciferase-based reporter constructs were in the pGL2basic (pGL2b; Promega) backbone. The murine cyclin D1 promoter region, encompassing 7.9 kbp (44), and the murine cyclin D2 promoter region, encompassing 2.3 kbp (8), were kindly provided by M. Eilers. Further, the human cyclin E promoter was used as the reporter plasmid (25).
The Brd2/RING3 baculovirus was described previously (47). The baculovirus for the expression of the Brd4S full-length protein (aa 2 to 722) with an amino-terminal myc epitope and a carboxy-terminal hexahistidine tag was generated from pENTR1A-Brd4 by in vitro recombination with BaculoDirect C-terminal linear DNA (Invitrogen). pENTR1A-Brd4 was generated by PCR with the Brd4S full-length template (aa 2 to 722) in pcDNA3 (45) and oligonucleotides Brd4S BAC F (AGAGGATCCATTATGGAGCAGAAGCTGATCTCCGAGGAGGACCTGTCTGCGGAGAGCGGC) and Brd4S BAC R (AGACTCGAGGCAGGACCTGTTTCGG).
Transient transfections and luciferase-based reporter assays.
For transfections, cells were grown to subconfluence in six-well plates (Greiner). HEK 293T and 3T3 cells were transfected with FuGENE6 (Roche) (FuGENE-DNA ratio of 3 μl:1 μg). For luciferase reporter assays, cells were transfected with 50 ng luciferase reporter plasmid per well together with 500, 1,000, or 2,500 ng of orf73 expression plasmid or empty vector. At 48 h posttransfection, cells were washed once in cold phosphate-buffered saline (PBS) and then lysed and scraped on ice in reporter lysis buffer (Promega). Lysates were spun at high speed for 1 min to pellet the debris. Lysates were analyzed for luciferase activity using a luciferase assay system (Promega).
Electroporation of B cells.
BJAB cells were electroporated following published procedures, with some modifications (45). Briefly, 1.3 × 107 BJAB cells were resuspended in 400 μl medium without antibiotics. Endotoxin-free DNA (20 μg) was added to the cells, and cells were electroporated as previously described (45).
Protein analyses by immunoblotting.
Cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membranes, and immunodetection of HA-tagged MHV-68 orf73 proteins was performed with a rat anti-HA monoclonal antibody (12CA5; Roche). For immunoprecipitation experiments, HA-tagged orf73 proteins were detected with a rat monoclonal anti-HA antibody (3F10; Roche). KSHV LANA-1 was detected with serum from KSHV-positive patients. Brd2/RING3 was detected with a polyclonal rabbit anti-RING3 antibody (47). Anti-green fluorescent protein (GFP) immunoblotting was performed with a monoclonal anti-GFP antibody (JL-8; BD Biosciences). myc-tagged Rb was detected with a monoclonal anti-c-myc antibody (9E10; Biomol). Cellular actin was detected with a panactin mouse monoclonal antibody (MAB 1501; Chemicon/Millipore). Endogenous Brd4 was detected with an affinity-purified rabbit polyclonal antiserum raised against the amino-terminal aa 1 to 470 of Brd4 (54). Species-specific horseradish peroxidase-conjugated secondary antibodies were used.
Expression and purification of recombinant proteins from Escherichia coli and insect cells.
GST and GST fusion proteins were expressed as previously described (47). For the purification of GST and GST fusion proteins, glutathione G Sepharose resin was used as previously established (45). Brd4S and Brd2/RING3 were expressed in SF9 insect cells and purified using a Ni+ affinity purification protocol as previously established (45).
GST fusion protein binding assays.
GST pull-down assays using GST-RING3, GST-Brd4S, and GST-ORFX (see Fig. 1C and D) with MHV-68 orf73 protein were performed as previously described (45). Briefly, GST proteins were bound to glutathione G Sepharose, washed, and then incubated with lysates from 293T cells transfected with empty vector, pVR1255 MHV-68 orf73, or pcDNA3 KSHV LANA.
Salt extraction of MHV-68 orf73 proteins from nuclear preparations.
Nuclear preparations from 293T cells that transiently expressed MHV-68 orf73 proteins were subjected to protein extraction using increasing KCl concentrations similar to a previously published protocol for KSHV LANA (57). Transfected 293T cells were lysed for 30 min in a low-ionic-strength buffer (5% glycerol, 1% NP-40, 0.2 mM EDTA, 10 mM Tris-HCl [pH 7.9], leupeptin [50 μM], benzamidine [200 μM], aprotinin [100 U/ml], pepstatin A [1 μM], phenylmethylsulfonyl fluoride [1 mM]). Nuclear material was separated from the lysates by centrifugation. Next, the pellet was incubated with the same buffer plus 50 mM KCl, followed by centrifugation. Supernatants were collected and the pellets subjected to incubation with the same buffer with 200 mM KCl. The procedure was repeated with 300 mM KCl. Supernatants were subjected to SDS-PAGE and immunoblotting.
Co-IP assays.
Coimmunoprecipitation (Co-IP) experiments with lysates from transfected 293T cells were performed similarly to previously described procedures (58). Briefly, lysates of 293T cells coexpressing EGFP or EGFP-BET fusion proteins together with MHV-68 orf73-HA proteins were incubated with a monoclonal anti-HA antibody coupled to Sepharose A resin. After intensive washing, SDS-PAGE and immunoblotting with mouse anti-GFP and rat anti-HA antibodies were performed. For myc-Rb immunoprecipitations, an anti-myc monoclonal antibody was used. The Co-IP experiments for endogenous Brd4 with ectopically expressed HA-tagged MHV-68 orf73 proteins in BJAB cells were performed similarly to a previously published procedure (45). Briefly, 1.3 × 107 BJAB cells were electroporated with 4 μg of EGFPC1 (to assess electroporation efficiency) together with 16 μg of pVR1255HA (mock), pVR1255 orf73 wt, or pVR1255 orf73 228-4A. Seventy-two hours later, 3.5 × 107 cells per sample were harvested and lysed in 3.1 ml Tris-buffered saline (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA) plus 1% Triton X-100 (TBS-T). After centrifugation at 16,000 × g for 1 min, 1 ml of supernatants was incubated overnight at 4°C with anti-HA, anti-Brd4, and as a negative control, rabbit immunoglobulin G (IgG) coupled to Sepharose A resin. After extensive washing, proteins were separated by 4 to 12% bis-Tris PAGE, blotted, and detected with specific antibodies as indicated in the figures.
Peptide array assays.
MHV-68 orf73 peptides were chemically synthesized on cellulose membranes with the Spot technique performed according to described procedures (21, 22). The entire MHV-68 orf73 was represented as overlapping peptides of 15 aa in length each shifted by 3 aa. Hence, each peptide was identical with the previous peptide in 12 out of 15 residues. Each of the following steps was carried out with 10 ml of solution on a rocking platform at room temperature unless mentioned otherwise. Washing steps lasted 10 min. To determine unspecific binding by the first and secondary antibodies, the peptide membrane was moistened with ethanol, washed three times with Tris-buffered saline (50 mM Tris-HCl [pH 7.0], 137 mM NaCl, 2.7 mM KCl) plus 0.05% Tween 20 (TBS-T) and blocked overnight with blocking buffer (20% [vol/vol] Genosys buffer [Sigma] and 5% [wt/vol] Saccharose in TBS-T [pH 8.0]) at 4°C. The membrane was then washed once with TBS-T (pH 7.0) and incubated with anti-Brd2/RING3 polyclonal rabbit antiserum diluted 1:100 (or anti-myc monoclonal antibody [9E10; Biomol] at a 10-μg/ml final concentration to detect Brd4S) in blocking buffer for 2 h, washed three times in TBS-T, and incubated with a secondary alkaline phosphatase (AP)-conjugated antibody (goat anti-rabbit AP [Harlan Serolab] or rabbit anti-mouse AP [Dako]). The AP conjugates were diluted 1:500 in blocking buffer and incubated with the membrane for 90 min. Next, the filter was washed twice with TBS-T, PBS, and citric-acid-based saline (137 mM NaCl, 2.7 mM KCl, 50 mM citric acid-1-hydrate [pH 7.0]) each. Then, the membrane was incubated in staining solution (5 mM MgCl2, 2.4 mg BCIP [5-bromo-4-chloro-3-indolylphosphate], 3 mg methyl-thiazoletetrazoline in 10 ml citric-acid-based saline) for 30 min under slow agitation. After a wash with PBS, the image of signals was documented by scanning the membrane (HP Scanjet 5370C) and signals were quantified with Phoretix array 1.0 software. The quantification resulted in a series of dimensionless values, representing gray values of each peptide signal. These values were later used for subtraction from the values obtained from the actual protein-peptide interaction experiment. To remove bound antibodies, the membrane was next stripped as follows (all steps were carried out with 15 ml of solution). The membrane was washed twice with water, once with dimethylformamide and then with dimethylformamide in a sonication bath at room temperature until the dye signals vanished completely; three times with buffer A (8 M urea, 1% [wt/vol] SDS, 0.5% [vol/vol] β-mercaptoethanol [pH 7.0]) for 5 min on a rocking platform and 5 min in the sonication bath at 40°C; three times with buffer B (10% acetic acid, 50% ethanol, 40% water); and three times with ethanol. After the membrane was stripped, the blocking step was repeated as described above, followed by a washing step with TBS-T and incubation with SF9-expressed and purified Brd2/RING3 or Brd4S protein at a concentration of 1 μg recombinant protein per ml blocking buffer for 3.5 h. Next, the membrane was washed three times with TBS-T, incubated with the respective primary and secondary antibodies, and stained as described above.
RESULTS
The MHV-68 orf73 protein interacts with cellular BET proteins via their highly conserved ET domain.
Work done by our group has demonstrated previously that the KSHV LANA-1 protein interacts with several members of the BET protein family (45, 47, 58). Initially identified in a yeast two-hybrid screen, Brd2 interacts with KSHV LANA-1 via the carboxy-terminal domains of both proteins (47). Further, we recently demonstrated that LANA-1 interacts with two other BET family members, Brd3/ORFX and Brd4 (45).
In this study, we investigated whether the MHV-68 orf73 protein, the LANA-1 homolog, might also target cellular BET proteins. Mammalian BET proteins are encoded by four genes, Brd2, Brd3, Brd4, and Brd6 (18). Brd4 encodes two alternatively spliced isoforms, Brd4S and Brd4L (Fig. 1A) (18). Two bromodomains, histone interaction modules, and the less well-characterized ET domain, a protein-protein interaction domain, are characteristic for BET protein members and are highly conserved between the paralogous proteins. A comparison of the 64-aa-long ET domains of the human and murine orthologous proteins reveals 100% sequence identity for Brd2, Brd3, and Brd4 and 95.3% identity in the case of Brd6 (Fig. 1B). The ET domain is sufficient for the interaction with KSHV LANA-1. We hypothesized that the MHV-68 orf73 protein might also interact with BET proteins via their ET domain. To test this, GST fusion proteins encompassing the ET domains of Brd2, Brd4, and Brd3 (Fig. 1C) were bacterially expressed, bound to glutathione Sepharose resin, and then incubated with lysates from cells that expressed an HA epitope-tagged version of the MHV-68 orf73 protein. The MHV-68 orf73 protein bound to GST-Brd2, GST-Brd4, and GST-Brd3 but not to GST (Fig. 1D). Furthermore, the binding of MHV-68 orf73 to BET proteins was confirmed by Co-IP experiments. To do this, MHV-68 orf73 was coexpressed with EGFP-tagged, full-length Brd2 or Brd4 protein (see Fig. 4; also, data not shown) or EGFP-tagged BET protein fragments similar to the GST fusion proteins depicted in Fig. 1C. Immunoprecipitation with an anti-GFP antibody revealed binding of MHV-68 orf73 to these BET proteins but not to EGFP alone (data not shown).
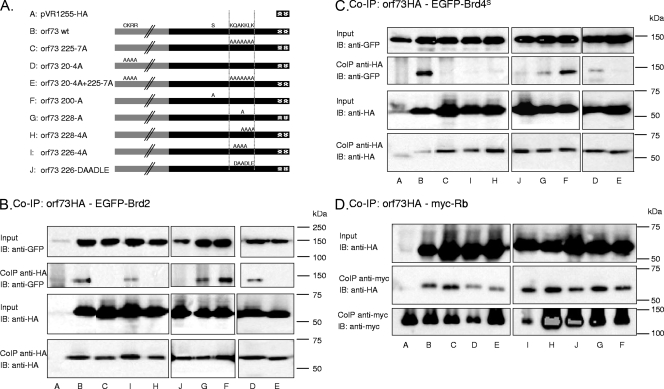
FIG. 4.
Interaction of MHV-68 orf73 protein mutants with Brd2, Brd4, and Rb. (A) Schematic depiction of MHV-68 orf73 mutants generated and used in this study. Labels A through J correspond to lane labeling in panels B, C, and D. (B) Co-IP of orf73 wt, orf73 mutants, and EGFP-Brd2. HA-tagged orf73 proteins were immunoprecipitated from lysates of 293T cells transfected with expression vectors for orf73 proteins and EGFP-Brd2 using an antibody to the HA tag. Immunoprecipitated proteins were analyzed by immunoblotting (IB) using an antibody to GFP. (C) Co-IP of orf73 wt, orf73 mutants, and EGFP-Brd4S. Experiments were performed as described for panel B. (D) Co-IP of orf73 wt, orf73 mutants, and cotransfected, myc-tagged Rb protein. Transfected, c-myc-tagged Rb was immunoprecipitated with an antibody to the c-myc epitope, and immunoprecipitates were analyzed by immunoblotting using an antibody to the HA epitope.
Mapping of the Brd2/RING3 binding site in the MHV-68 orf73 protein.
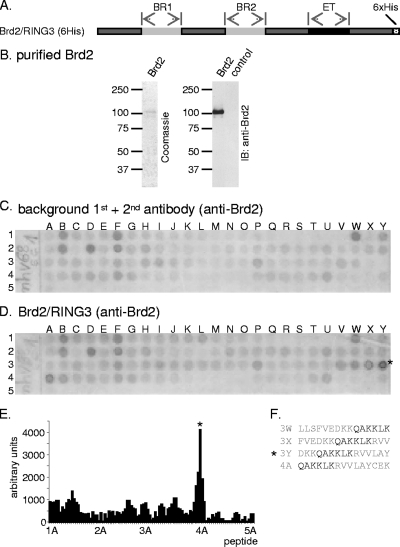
To map the linear Brd2/RING3 interaction site in the MHV-68 orf73 protein, 101 peptides representing the complete orf73 protein were synthesized, each peptide overlapping with the neighboring ones in 12 out of 15 positions. The 110-kDa protein Brd2/RING3 was expressed in SF9 insect cells, Ni+ affinity purified via its hexahistidine tag (Fig. 2A and B), and used to probe the MHV-68 orf73 peptide array (Fig. 2D). Signals for individual peptides were quantified and background signals (binding of antibodies) (Fig. 2C) subtracted from the signals obtained after BRD2 incubation (Fig. 2D). As shown in Fig. 2D to F, four peptides, 3W, 3X, 3Y, and 4A, exhibited elevated Brd2 binding. These four peptides all contained the amino acids QAKKLK, corresponding to aa 226 to 231 in the MHV-68 orf73 protein (Fig. 2F). The aa 226 to 231 (QAKKLK) were considered the footprint for direct Brd2 binding.
FIG. 2.
A peptide array assay identifies a binding site for purified Brd2 in the MHV-68 orf73 protein. (A) Schematic depiction of the baculovirus-expressed Brd2/RING3 protein that was purified and used for studies of direct interaction by peptide array assays, with results shown in panel D. BR1, bromodomain 1; BR2, bromodomain 2. (B) The purity of Brd2/RING3 was determined by SDS-PAGE followed by Coomassie staining. The identity of Brd2/RING2 was confirmed by anti-RING3 immunoblotting (IB). (C) Peptides representing the complete MHV-68 orf73 protein with each peptide overlapping with the previous one in 12 out of 15 residues were first incubated with SF9 lysate from untransfected SF9 insect cells and then probed with a primary rabbit anti-RING3 polyclonal antibody followed by incubation with a secondary AP-conjugated antibody to determine background staining. The nomenclature of peptides starts with 1A in the top left corner. (D) After the membrane was stripped, the same peptide array was incubated with affinity-purified Brd2/RING3 protein (see legend for panel B). Bound protein was detected as described for panel C. The peptide with the strongest signal intensity is marked with an asterisk. (E) Quantification of signal intensities shown in panel D minus background intensities shown in panel C, determined using Phoretix array 1.0 quantification software. (F) Sequences of the four peptides, 3W, 3X, 3Y, and 4A, that bound RING3, all containing the amino acids QAKKLK, corresponding to positions 226 to 231 in the MHV-68 orf73 protein.
Mapping of the Brd4 binding site in the MHV-68 orf73 protein.
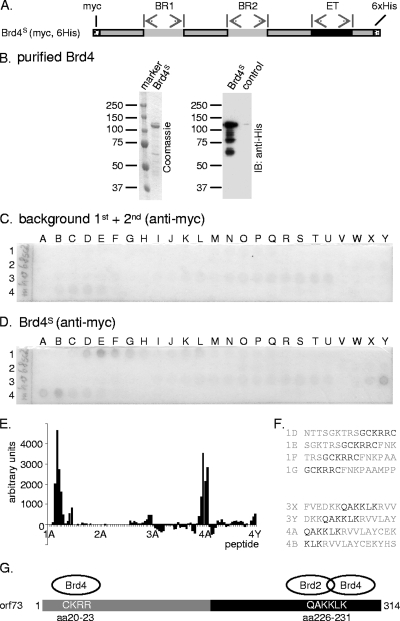
The interaction sites for Brd4 were mapped in a manner similar to that described above, using a newly synthesized MHV-68 orf73 peptide array. A Brd4 baculovirus for the expression of myc epitope- and hexahistidine-tagged Brd4S was generated, and the recombinant protein, with a molecular mass of approximately 110 kDa, was Ni+ affinity purified similarly to Brd2 (Fig. 3A and B). Probing the peptide array with the purified Brd4S protein led to the identification of two regions in the orf73 protein, represented by peptides 1D to 1G and 3X, 3Y, 4A, and 4B (Fig. 3F). Interestingly, peptides 3X, 3Y, and 4A contained the QAKKLK motif (aa 226 to 231), already identified as a Brd2 interaction site (Fig. 2F), suggesting identity or substantial overlap of the Brd2 and Brd4 binding sites in the MHV-68 orf73 protein. In contrast, the additional interaction site found for Brd4 (GCKRRC, aa 19 to 24; peptides 1D to 1G) did not react with Brd2 (Fig. 2).
FIG. 3.
A peptide array assay identifies a binding site for purified Brd4S in the MHV-68 orf73 protein. (A) Schematic depiction of the baculovirus-expressed Brd4S protein that was purified and used for studies of direct interaction by peptide array assays, with results shown in panel D. BR1, bromodomain 1; BR2, bromodomain 2. (B) The purity of Brd4S was determined by SDS-PAGE followed by Coomassie staining. The identity of Brd4S was confirmed by anti-His immunoblotting (IB). (C) A newly synthesized MHV-68 orf73 peptide array was first incubated with an anti-myc antibody followed by a secondary AP-conjugated antibody to determine background staining. (D) Next, the array was incubated with purified full-length Brd4S protein (see legend for panel B). Bound Brd4 protein was detected via its myc epitope tag. (E) Quantification of signal intensities shown in panel D minus background intensities shown in panel C. (F) Schematic depiction of the peptides that bound Brd4 (1D to 1G and 3X, 3Y, 4A, and 4B). (G) Schematic depiction of Brd2 and Brd4 binding sites in the MHV-68 orf73 protein, as determined by peptide array assays.
BET binding site mutants and their interactions with Brd2, Brd4, and pRb.
By use of site-directed mutagenesis, a number of MHV-68 orf73 mutant proteins with different mutations in their Brd2 and Brd4 binding sites and a carboxy-terminal double HA epitope tag were generated (for a schematic depiction, see Fig. 4A). In the mutant orf73 225-7A, 7 alanines replaced aa 225 to 231 (KQAKKLK in the wild-type [wt] protein), which had been identified as a site of Brd2 as well as Brd4 interaction. In the mutant orf73 20-4A, the wt amino acids CKRR (aa 20 to 23), which constituted the amino-terminal Brd4 binding footprint, were replaced by 4 alanines. These two mutations were combined in the mutant orf73 20-4A+225-7A. The mutant orf73 200-A, with serine 200 replaced with an alanine, served as a control protein, because S200 was outside any BET binding site. The mutant orf73 228-A had lysine 228 replaced with an alanine. Lysine 228 is conserved in the homologues of MHV-68 orf73, KSHV LANA-1, the rhesus rhadinovirus orf73 protein, and the HVS orf73 protein (our own analyses; 9, 52). Four alanines replaced the wt KKLK in the orf73 228-4A mutant or QAKK in the orf73 226-4A mutant. Furthermore, the mutant orf73 226-DAADLE was engineered to replace the hydrophilic, positively charged QAKKLK Brd2 and Brd4 binding site with the hydrophilic but negatively charged amino acids DAADLE.
To investigate which of these orf73 protein mutants had lost the ability to interact with Brd2 and Brd4, we carried out Co-IP assays with GFP-tagged Brd2 or Brd4 constructs and the HA-tagged MHV-68 orf73 protein mutants. As shown in Fig. 4B and C, Brd2 and Brd4S coimmunoprecipitated with the orf73 wt protein, confirming their interaction. Of the mutants described above and summarized in Fig. 4A and 6B, the mutant with the extensive 7-alanine substitution in the MHV-68 orf73 C-terminal domain (orf73 225-7A) (Fig. 4, lanes C) no longer interacted with Brd2 or Brd4. Likewise, the mutant with the 4-alanine substitution in the same region (orf73 228-4A) (Fig. 4, lanes H) and the charge reversion mutant (orf73 226-DAADLE) (Fig. 4, lanes J) failed to interact with either BET protein. In contrast, the orf73 226-4A mutant (Fig. 4, lanes I) failed to interact with Brd4S but retained some interaction with Brd2. The mutant with the single point mutation in the QAKKLK footprint, orf73 228-A (Fig. 4, lanes G), interacted with Brd2 to the same extent as the orf73 wt protein but showed reduced binding to Brd4S, while the control point mutant orf73 200-A (Fig. 4, lanes F) bound normally to both Brd4S and Brd2. Furthermore, orf73 20-4A (Fig. 4, lanes D), the 4-alanine substitution in the amino-terminal GCKRRC motif (aa 19 to 24), which on the peptide array had interacted only with Brd4S but not Brd2 (Fig. 2 and 3), showed normal binding to Brd2 and marginally reduced binding to Brd4S. As expected, the double mutant orf73 20-4A+225-7A, in which both the N-terminal and C-terminal interaction sites had been modified, lacked binding to both Brd4S and Brd2 in the Co-IP assay (Fig. 4, lanes E).
FIG. 6.
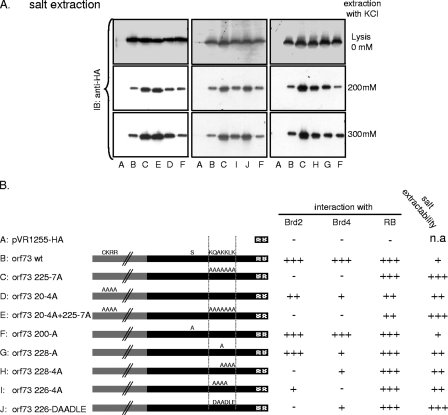
Decreased chromatin association of MHV-68 orf73 protein mutants. (A) Nuclear extracts of 293T cells with ectopically expressed MHV-68 orf73 mutants were subjected to increasing KCl concentrations to extract proteins. The supernatants of washes with 200 mM and 300 mM were analyzed by Western blotting. Expression levels of individual mutants were controlled in detergent lysates of transfected cells prior to washes with increasing KCl concentrations. IB, immunoblotting. (B) Summary of binding of orf73 wt and mutant proteins to Brd2, Brd4S, and pRB (results shown in Fig. 4), as well as extractability in high KCl concentrations. Labels A through J correspond to lane labeling in panel A. +, weak; ++, moderate; +++, strong; −, no interaction; n.a, not applicable.
To confirm that the inability of some of these mutants to interact with Brd4S and Brd2 reflected the lack of a specific binding site rather than more-extensive structural changes, we investigated the interaction of the MHV-68 orf73 protein and the mutants with the retinoblastoma protein Rb, another nuclear protein previously reported to interact with KSHV LANA (48). As shown in Fig. 4D, the orf73 wt protein and all mutants coimmunoprecipitated with myc-tagged, transfected Rb, with a possibly weaker interaction seen for mutant orf73 20-4A and the double mutant orf73 20-4A+225-7A (Fig. 4, lanes D and E). All other mutants, in particular those with the 4- and 7-alanine substitutions in the C-terminal BET interaction site (orf73 226-4A, orf73 228-4A, and orf73 225-7A) interacted with Rb to an extent comparable with that of the wt protein (Fig. 4, lanes C, H, and I). Together, these findings suggest that the orf73 KKLK motif (aa 228 to 231), identified as a BET interaction site by peptide array (Fig. 2 and 3), represents a genuine binding site for Brd2 and Brd4S in the context of the entire MHV-68 orf73 protein. Minor differences seen with these mutants (e.g., orf73 226-4A still binds to Brd2 but not to Brd4S, and orf73 228-A shows normal binding to Brd2 but reduced binding to Brd4S) suggest minor differences in the abilities of Brd2 and Brd4S to interact with the KKLK motif.
The MHV-68 orf73 protein forms complexes with endogenous Brd4 proteins in B cells, and the KKLK site in orf73 (aa 228 to 231) is critical for this interaction.
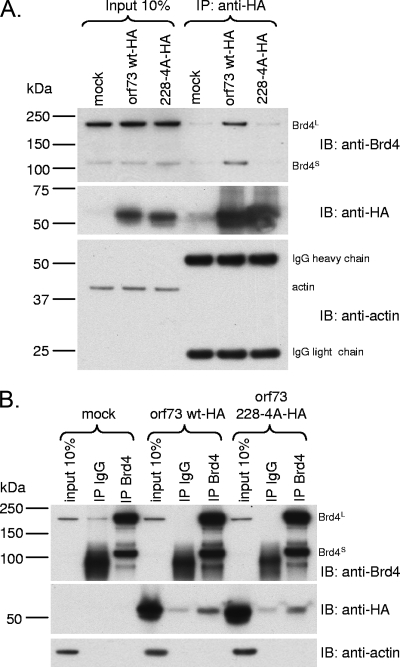
Because B cells are an important cell population of rhadinoviral latent infection, we investigated the interaction of the orf73 protein with endogenous Brd4 in B cells. To our knowledge, no antibodies to detect MHV-68 orf73 are available. Hence, human BJAB Burkitt's lymphoma cells were electroporated with expression vectors for the HA-tagged MHV-68 orf73 wt or orf73 228-4A protein to perform Co-IP experiments with anti-HA (Fig. 5A) and anti-Brd4 (Fig. 5B) resin. Both isoforms of Brd4 coimmunoprecipitated with the HA-tagged orf73 wt protein but not with the orf73 228-4A mutant protein (Fig. 5A, compare lanes 5 and 6). In the reciprocal Brd4-specific immunoprecipitation, the orf73 wt protein showed interaction with Brd4 (Fig. 5B, middle panel, lane 6). The MHV-68 orf73 228-4A mutant protein was impaired in its binding to Brd4 compared to the wt orf73 protein (Fig. 5B, compare lane 9 to lane 6) but showed binding above background levels compared to the IgG control (Fig. 5B, compare lane 9 with lane 8). This finding is in line with the observation of some residual binding of the orf73 228-4A mutant to EGFP-Brd4 (Fig. 4C; summarized in Fig. 6B).
FIG. 5.
The MHV-68 orf73 protein forms complexes with endogenous Brd4 in B cells. BJAB cells transiently expressing the HA-tagged MHV-68 orf73 or orf73 228-4A protein were used for Co-IP experiments using anti-HA resin (A) and anti-Brd4 resin (B). Immunoblotting (IB) was performed using rabbit polyclonal anti-Brd4 serum (top panels), rat monoclonal anti-HA antibody (middle panels), and mouse monoclonal antiactin antibody (bottom panels). Ten percent of the lysates used for IP were loaded as input. (A) Co-IP using anti-HA resin reveals binding of Brd4L and Brd4S to MHV-68 orf73 wt but not the orf73 228-4A mutant. (B) Reciprocal Co-IP using anti-Brd4 resin. Normal IgG served as a negative control. With longer exposure, Brd4S becomes visible in the input lanes, as seen in panel A.
Impaired Brd2 and Brd4 binding of MHV-68 orf73 mutants correlated with a weaker association to nuclear chromatin.
We and others have previously reported that the interaction of KSHV LANA-1 with Brd2/4 might contribute to the association of LANA-1 with interphase chromatin or mitotic chromosomes (58, 65). We therefore compared the associations of MHV-68 orf73 wt and mutant proteins with nuclear interphase chromatin. To do this, cells expressing HA-tagged orf73 or one of the mutants were lysed under hypotonic conditions and nuclear pellets subjected to several washes at increasing salt concentrations (50, 200, and 300 mM KCl). All orf73 constructs exhibited comparable expression levels (Fig. 6, top panel). The mutants orf73 225-7A, orf73 228-4A, orf73 20-4A+225-7A, and orf73 226-DAADLE were more readily extractable at 200 and 300 mM KCl than was the wt protein or the control orf73 200-A mutant. This can be seen in the higher signal intensity for these mutant orf73 proteins on immunoblots of the 200 mM and 300 mM KCl extractions (Fig. 6A, middle and bottom panels, lanes C, E, and J). The mutants orf73 228-A and orf73 226-4A showed intermediate phenotypes. The results of these experiments are summarized in Fig. 6B and compared to the abilities of these mutants to interact with Brd2 and Brd4S (results in Fig. 4). Overall, we noted a correlation between a decreased binding of orf73 mutants to Brd2 and Brd4S and an increased extractability in KCl buffer. These findings support a model in which the interaction of the MHV-68 orf73 proteins with Brd2 and/or Brd4 contributes to the association of orf73 with cellular chromatin.
The carboxy-terminal Brd2 and Brd4 binding site in MHV-68 orf73 is critical for the transcriptional activation of cell cycle regulatory gene promoters.
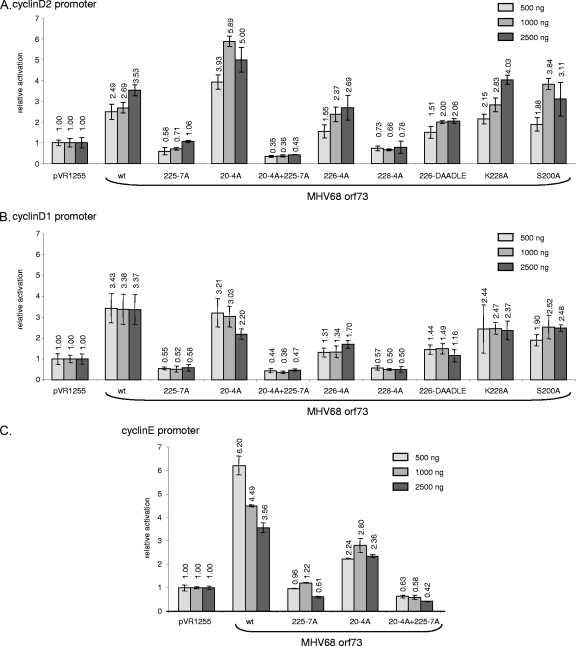
KSHV LANA-1 can act as a transcriptional repressor or activator dependent on the promoter (36, 50, 52, 57). It activates promoters of cell cycle regulatory genes, including the cyclin E and cdk2 promoters (45, 48, 65). Using luciferase reporter assays, we found that the MHV-68 orf73 wt protein also activates the promoters of the cyclins D2, D1, and E approximately three- to sixfold (Fig. 7A to C, compare pVR1255 with wt). The orf73 225-7A mutant and the orf73 20-4A+225-7A double mutant were incapable of transactivating the cyclin D2, D1, and E promoters and even moderately inhibited their activity (Fig. 7A to C). This indicated that the QAKKLK motif in the orf73 protein is critical for the ability to transactivate these promoters. The mutant orf73 228-4A also no longer transactivated (Fig. 7A and B), defining the minimal critical region to be KKLK (aa 228 to 231) in orf73. Mutating just the lysine 228 in the mutant orf73 228-A did not significantly impair the activation of the cyclin D2 promoter and only moderately reduced the transactivation of the cyclin D1 promoter.
FIG. 7.
The MHV-68 orf73 protein activates promoters of cell cycle regulatory genes, and the BET interaction site KKLK (aa 228 to 231) in the orf73 protein is critical for this function. Transient luciferase reporter assays were performed with murine 3T3 fibroblasts. Cells were cotransfected with 50 ng of promoter luciferase reporter plasmids together with 500 ng, 1,000 ng, or 2,500 ng of empty vector (pVR1255) or different MHV-68 orf73 constructs (Fig. 4A). Relative luciferase activities compared to that for the empty vector were calculated, and mean values ± standard deviations from two representative experiments in duplicate are depicted. (A) Murine cyclin D2 promoter; (B) murine cyclin D1 promoter; (C) human cyclin E promoter.
The orf73 20-4A mutant with a mutation in the GCKRRC motif identified by probing the orf73 peptide array with Brd4S (Fig. 3) showed a slightly increased activation of the cyclin D2 promoter, did not alter activation of the cyclin D1 promoter, and only moderately impaired the activation of the cyclin E promoter compared to levels for the orf73 wt protein. Hence, this motif, which is not required for efficient binding of Brd2 or Brd4S to the MHV-68 orf73 protein (Fig. 4), is also not involved in the activation of the three cell cycle-dependent promoters analyzed in this experiment. Taken together, our data demonstrate a correlation among Brd2 and Brd4 binding to the KKLK motif (aa 228 to 231) in the MHV-68 orf73 protein, the chromatin association of the orf73 protein, and the ability of the orf73 protein to act as a transcriptional activator of the promoters of the cyclin D2, D1, and E genes.
DISCUSSION
Cellular BET proteins are important interaction partners for three families of DNA viruses, papillomaviruses, gamma-2 herpesviruses, cytomegalovirus, and EBV, as well as two human retroviruses, HIV-1 and human T-lymphotropic virus type I (HTLV-1) (7, 10, 31, 34, 37, 40, 41, 45, 47, 54, 58, 63, 64). Papillomaviruses target Brd4L via their E2 proteins (63), and the gamma-2 herpesvirus KSHV has so far been shown to interact with Brd2, Brd3, and Brd4 via its LANA-1 protein (45, 65). Brd4 was also found in human cytomegalovirus immediate-early transcriptosome complexes (34), and recent evidence suggests a role for Brd4 in EBV EBNA-1 transcriptional functions (37). In the case of HIV-1, Brd4 is thought to compete with the viral Tat protein for access to the transcriptional elongation factor pTEFb, which is required for the phosphorylation of the C-terminal domain of Pol II and, together with Tat, ensures elongation of transcripts emanating from the HIV long terminal repeat (7). A peptide derived from the C-terminal domain of Brd4L can inhibit Tat-mediated transcription (7). Similarly, HTLV-I Tax and Brd4 compete for binding to pTEF (10). In this work, we demonstrated that MHV-68, the only available small-animal model for gamma-2 herpesvirus infection and pathogenesis, also interacts with the BET proteins Brd2, Brd3, and Brd4 via its orf73 protein, the LANA-1 homolog. We identified the binding site in the MHV-68 orf73 protein for Brd2 and Brd4S and showed that mutation of this binding site leads to a reduced chromatin association and failure of orf73 proteins to act as transcriptional activators of cell cycle-dependent promoters. This study is therefore the first to identify a BET protein binding site in a gamma-2 herpesvirus orf73 protein and to thereby link binding to BET proteins to the transcriptional activator function of orf73 proteins.
Similarly to KSHV LANA-1 (45, 58), the MHV-68 orf73 protein interacts with the ET domain, a 64-aa-long domain about which little is known functionally. As shown in the protein sequence alignment in Fig. 1B, the ET domains of murine and human Brd2 are 100% identical. The same is true for the ET domains of murine and human Brd3, as well as those of murine and human Brd4. In the case of Brd6, 3 of 64 aa differ between the murine and human orthologs, corresponding to 95.3% identity (Fig. 1B). Overall, this remarkably high level of ET domain conservation indicates the structural and/or functional importance of this domain.
Computational comparisons of orf73 homologs of a number of rhadinoviruses, including KSHV, HVS, rhesus rhadinovirus, retroperitoneal fibromatosis herpesvirus, and MHV-68, have revealed a conserved carboxy-terminal domain in orf73 proteins (9, 28, 52). Our work has shown previously that the integrity of this conserved domain in KSHV LANA-1 is critical for the interaction with BET proteins, because the carboxy-terminal truncation of LANA-1 beyond aa 1139 results in a loss of BET protein interaction (45, 58). Because these truncated LANA-1 mutants lose a number of functions at the same time, including the replication and transcriptional functions and the ability to form dimers, we focused this work on the delineation of the BET protein binding sites in the context of the whole MHV-68 orf73 protein. To do this, peptide array assays were performed with the orf73 protein represented by overlapping peptides of 15 aa each. These peptide arrays used incubation with purified full-length Brd2/RING3 or Brd4S proteins expressed in insect cells. This approach (i) allowed us to confirm the direct nature of the Brd2-orf73 and Brd4-orf73 interactions and (ii) allowed us to map the direct Brd2 and Brd4 interaction footprints in the MHV-68 orf73 protein (Fig. 2 and 3). Both Brd2 and Brd4 interacted with several peptides containing the amino acids QAKKLK (aa 226 to 231) in the carboxy-terminal half of orf73; Brd4, in addition, bound to several peptides containing the amino acids CGCKRR, corresponding to aa 19 to 24 in the amino-terminal part of the orf73 protein. We then confirmed the importance of the KKLK motif for the interaction with Brd2 (Fig. 4B) and Brd4S (Fig. 4C) by Co-IP assays. Furthermore, our work demonstrates that the orf73 protein engages endogenous Brd4 in B cells and confirms the KKLK motif (aa 228 to 231 in orf73) to be critical for this interaction (Fig. 5).
BET proteins are known to interact with chromatin via the interaction of their bromodomains with acetylated histones; in the case of Brd4, it has been shown that this is true for the interphase as well as mitosis (13, 33). Because of this, we tested the hypothesis whether the orf73 mutants would show impaired association with cellular chromatin. In extraction experiments with increasing KCl concentrations from nuclear preparations, the orf73 constructs with mutations in their BET protein binding sites were more readily extracted by KCl than was the orf73 wt protein or a control mutant (orf73 200-A), indicating that the mutant constructs had a weaker relative chromatin association than did the orf73 wt protein. This suggests a contribution of the orf73-BET interaction to the orf73 chromatin association. Because the association with the nuclear fraction in this assay was weakened but not completely abrogated, our data suggest that Brd2 and Brd4 are most likely not the only mechanism of chromatin association for the MHV-68 orf73 protein. In the case of KSHV LANA-1, the current model is that of a direct histone interaction via the LANA-1 amino-terminal domain (5), with the LANA-1 carboxy-terminal domain modulating the chromatin interaction to a fully functional state via its interaction with additional cellular binding partners, e.g., the BET proteins Brd2 and Brd4 or the methyl-CpG-binding protein MeCP2 or DEK (5, 27, 35, 45, 58).
KSHV LANA-1 is a promiscuous transcriptional regulator that regulates the promoters of cell cycle regulatory genes, including the cyclin E promoter, besides many other target genes (3, 45, 65). We show here that the MHV-68 orf73 protein also acts as a transcriptional activator of the cyclin D2 and cyclin E promoters, two E2F-responsive promoters, as well as the cyclin D1 promoter (Fig. 7). orf73 proteins with mutations in their carboxy-terminal Brd2 and Brd4 binding sites (e.g., the orf73 225-7A and orf73 228-4A mutants) were impaired in their ability to activate the cyclin D2, D1, and E promoters (Fig. 7). In contrast, mutating the core of the amino-terminal Brd4 binding footprint CKRR (mutant orf73 20-4A) did not have a negative effect on the transactivation of the cyclin D2 and D1 promoters and only moderately impaired the transactivation of the cyclin E promoter. Interestingly, the orf73 20-4A mutant also still interacted with Brd2 and (more weakly) with Brd4S and did not show a changed chromatin association in the salt extraction experiments at 50 mM and 200 mM KCl (Fig. 6A; also, data not shown). This suggests that the interaction of Brd4 with the site CKRR (aa 20 to 23) in the amino-terminal domain of the MHV-68 orf73 protein, which had been identified in the peptide array, is not required for the interaction with Brd2 and Brd4S and is not essential for chromatin association or the transcription activation function.
As with all site-directed mutations generated in the absence of structural information, one potential caveat could be that they affect the proper folding of the MHV-68 orf73 protein. The preserved ability of the mutants to interact with RB would argue against this. In addition, aa 225 to 231 are predicted to be part of an alpha-helical structure of orf73, and stretches of alanines, as used with the orf73 228-4A and orf73 225-7A mutants, are known to preferentially form a helical structure; therefore, our alanine mutants should fold similarly to the wt protein. Also, we found that the orf73 225-7A and orf73 228-4A mutants retained the ability of the MHV-68 orf73 wt protein to induce G2/M cell cycle arrest (not shown). Taken together, these results show that the minimal orf73 228-4A mutant has retained several functional properties of the MHV-68 orf73 proteins, while losing the ability to interact with Brd2 and Brd4S and to activate transcription from three cell cycle-dependent promoters.
Both Brd2 and Brd4 have recently been identified in protein complexes with RNA Pol II and have been shown to activate transcription (11, 32, 61). For Brd4, this occurs via the interaction of Brd4 with pTEFb, a cyclin-dependent kinase, and subsequent stimulation of RNA Pol II-dependent transcriptional elongation (32, 61). Papillomaviruses have been shown to interact with Brd4 via their E2 proteins, and this E2-Brd4 interaction is critical for the E2 transcriptional activation function (31, 40, 54). There are a number of different ways in which pTEFb can be recruited to transcription complexes, and the recruitment via the chromatin-bound activator Brd4 is one of them (46). HTLV-1 has recently been shown to modulate the Brd4/pTEFb interaction through the interaction of its Tax protein with the cyclin T1 subunit of pTEFb, thereby possibly competing with the Brd4/pTEFb interaction (10). Similarly, Brd4L has been shown to compete with HIV-1 Tat for binding to pTEFb and to thereby modulate transcription from the HIV long terminal repeat (7). The mechanism employed by HTLV-1 to target pTEFb therefore seems to be distinguishable from the mechanism employed by both papillomaviruses and gamma-2 herpesviruses, which directly bind to Brd4 (45, 54, 63, 64).
The MHV-68 orf73 protein is required for the establishment of splenic latency in mice (43). The molecular mechanisms, however, have remained elusive. Importantly, MHV-68 orf73 engages endogenous Brd4 in B cells (Fig. 5). Therefore, this study provides a first step in understanding the functions of MHV-68 orf73 on a molecular level. The MHV-68 orf73 protein is expressed throughout the viral replication cycle (19) and hence most likely plays roles during lytic as well as during latent infection. By enhancing the expression of G1/S-phase cell cycle genes (Fig. 7), orf73 may create an S-phase cellular environment favorable to viral replication. Through its interaction with the nuclear fraction (Fig. 6), possibly with host cell chromatin through its interaction with BET proteins, orf73 may play a role in latent viral replication and genome maintenance. Furthermore, the functional interaction of orf73 with BET proteins may play a role in the pathogenesis of rhadinovirus-induced B-cell lymphoma, possibly through the enhancement of BET target gene transcription, such as that of the cyclin D1, D2, and E genes (Fig. 7). Interestingly, B-cell-specific ectopic Brd2 expression in mice results in B-cell lymphoma (26). A strength of the MHV-68 system is the possibility to study aspects of rhadinoviral biology and pathogenesis in cell culture and use the findings to design in vivo experiments with infected mice. Experiments are under way to study the phenotype of orf73 proteins with mutations in their BET protein binding sites in the context of the whole virus in vivo.
In summary, this work describes a novel transcriptional activation function of the MHV-68 orf73 protein on the cyclin D1, D2, and E promoters. Furthermore, this study identifies binding sites for Brd2 and Brd4S in the MHV-68 orf73 protein and shows that the ability to activate these cell cycle promoters correlates with the interaction with BET proteins. In addition, binding to BET proteins appears to contribute to the interaction of the MHV-68 orf73 protein with cellular chromatin.
Acknowledgments
We thank Peter Valentin-Weigand for helpful discussions and suggestions early in the project and all members of the Schulz laboratory who contributed to discussions relating to this work. We thank Susanne Daenicke for expert technical assistance in the synthesis of the peptide arrays.
M.O. was funded by grants DFG GRK 745 Mucosal Host-Pathogen Interactions and DFG SPP1130. D.P. was funded by the European Union Integrated Project INCA (LSHC-CT-2005-018704). J.P.S. was funded by a Royal Society University research fellowship and Public Health Service grant CA090208 from the National Cancer Institute. Some of the later experiments were carried out by M.O. in Peter Howley's laboratory at Harvard Medical School and were supported by grant R01CA116720 (to Peter Howley) from the National Cancer Institute.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Abbate, E. A., C. Voitenleitner, and M. R. Botchan. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24877-889. [DOI] [PubMed] [Google Scholar]
- 2.Allen, R. D., III, S. Dickerson, and S. H. Speck. 2006. Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection. J. Virol. 802055-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 2803862-3874. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. [DOI] [PubMed] [Google Scholar]
- 5.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311856-861. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisgrove, D. A., T. Mahmoudi, P. Henklein, and E. Verdin. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA 10413690-13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard, C., K. Thieke, A. Maier, R. Saffrich, J. Hanley-Hyde, W. Ansorge, S. Reed, P. Sicinski, J. Bartek, and M. Eilers. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 185321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnside, K. L., J. T. Ryan, H. Bielefeldt-Ohmann, B. A. Gregory, M. E. Thouless, C. C. Tsai, and T. M. Rose. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354103-115. [DOI] [PubMed] [Google Scholar]
- 10.Cho, W. K., M. Zhou, M. K. Jang, K. Huang, S. J. Jeong, K. Ozato, and J. N. Brady. 2007. Modulation of the Brd4/P-TEFb interaction by the human T-lymphotropic virus type 1 tax protein. J. Virol. 8111179-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley, T. E., E. M. Kaine, M. Yoshida, A. Nandi, and D. J. Wolgemuth. 2002. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 161727-1737. [DOI] [PubMed] [Google Scholar]
- 12.Denis, G. V., M. E. McComb, D. V. Faller, A. Sinha, P. B. Romesser, and C. E. Costello. 2006. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 5502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 1008758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 8499-109. [DOI] [PubMed] [Google Scholar]
- 16.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 1651074-1081. [DOI] [PubMed] [Google Scholar]
- 17.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 1961363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Florence, B., and D. V. Faller. 2001. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6D1008-D1018. [DOI] [PubMed] [Google Scholar]
- 19.Forrest, J. C., C. R. Paden, R. D. Allen III, J. Collins, and S. H. Speck. 2007. ORF73-null murine gammaherpesvirus 68 reveals roles for mLANA and p53 in virus replication. J. Virol. 8111957-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 843405-3416. [DOI] [PubMed] [Google Scholar]
- 21.Frank, R. 1992. Spot synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on membrane support. Tetrahedron 489217-9232. [Google Scholar]
- 22.Frank, R., and S. Dübel. 2005. Analysis of protein interactions with immobilized peptide arrays synthesized on membrane supports, p. 591-608. In E. Golemis and P. Adams (ed.), Protein-protein interactions: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402889-894. [DOI] [PubMed] [Google Scholar]
- 24.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9300-306. [DOI] [PubMed] [Google Scholar]
- 25.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 121173-1180. [PubMed] [Google Scholar]
- 26.Greenwald, R. J., J. R. Tumang, A. Sinha, N. Currier, R. D. Cardiff, T. L. Rothstein, D. V. Faller, and G. V. Denis. 2004. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood 1031475-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths, R., and A. Whitehouse. 2007. Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J. Virol. 814021-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 772779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 7611677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, H., J. Zhang, W. Shen, X. Wang, J. Wu, J. Wu, and Y. Shi. 2007. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct. Biol. 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19523-534. [DOI] [PubMed] [Google Scholar]
- 33.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 1333-43. [DOI] [PubMed] [Google Scholar]
- 34.Kapasi, A. J., and D. H. Spector. 2008. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 7611596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 749637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, A., S. Wang, T. Nguyen, K. Shire, and L. Frappier. 2008. The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4. J. Virol. 8212009-12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruyama, T., A. Farina, A. Dey, J. Cheong, V. P. Bermudez, T. Tamura, S. Sciortino, J. Shuman, J. Hurwitz, and K. Ozato. 2002. A mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 226509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14951-962. [PMC free article] [PubMed] [Google Scholar]
- 40.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mochizuki, K., A. Nishiyama, M. K. Jang, A. Dey, A. Ghosh, T. Tamura, H. Natsume, H. Yao, and K. Ozato. 2008. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2839040-9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 7710295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller, H., J. Lukas, A. Schneider, P. Warthoe, J. Bartek, M. Eilers, and M. Strauss. 1994. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc. Natl. Acad. Sci. USA 912945-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ottinger, M., T. Christalla, K. Nathan, M. M. Brinkmann, A. Viejo-Borbolla, and T. F. Schulz. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of Brd4 and releases cells from a Brd4- and Brd2/RING3-induced G1 cell cycle arrest. J. Virol. 8010772-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 47.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 739789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 61121-1127. [DOI] [PubMed] [Google Scholar]
- 49.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42859-868. [DOI] [PubMed] [Google Scholar]
- 50.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 754955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 748532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweiger, M. R., M. Ottinger, J. You, and P. M. Howley. 2007. Brd4-independent transcriptional repression function of the papillomavirus E2 proteins. J. Virol. 819612-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinha, A., D. V. Faller, and G. V. Denis. 2005. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem. J. 387257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 1871941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 777093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viejo-Borbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. A. Sheldon, and T. F. Schulz. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 7913618-13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willer, D. O., and S. H. Speck. 2003. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 778310-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 62.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313812-815. [DOI] [PubMed] [Google Scholar]
- 63.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 64.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 7914956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You, J., V. Srinivasan, G. V. Denis, W. J. Harrington, Jr., M. E. Ballestas, K. M. Kaye, and P. M. Howley. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 808909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]