Abstract
Epstein-Barr nuclear antigen 1 (EBNA1) is essential for Epstein-Barr virus to immortalize naïve B cells. EBNA1 transactivates viral promoters for genes that are necessary for immortalization when it is bound to a cluster of 20 cognate binding sites, termed the family of repeats. A region of EBNA1 from amino acids (aa) 40 to 89, termed linking region 1 (LR1), has been identified previously as being sufficient for transactivation. LR1 contains two domains that are conserved in the EBNA1 orthologs of other gamma herpesviruses. The first of these, termed unique region 1 (UR1), corresponds to aa 65 to 89 of EBNA1. UR1 is necessary for transactivation and contains a conserved recognition site for cyclic AMP-dependent protein kinase (PKA), corresponding to serine 78 of EBNA1. We have pharmacologically modulated PKA activity to determine if PKA controls EBNA1's ability to transactivate. Our results indicate that PKA activators and inhibitors do not affect transactivation by EBNA1. In addition, site-directed mutagenesis demonstrates that transactivation is not influenced by the phosphorylation status of serine 78 in the UR1 domain. The second conserved domain within LR1 is a glycine-arginine repeat, corresponding to aa 40 to 54 of EBNA1. This domain, termed ATH1, functions as an AT-hook, a DNA-binding motif found in architectural transcription factors such as HMGA1a. We demonstrate that deletion of the ATH1 domain decreases EBNA1 transactivation ability, which is consistent with a transcriptional role for ATH1. Furthermore, transactivation is restored when ATH1 is replaced by equivalent AT-hook motifs from HMGA1a. Our data strongly indicate a role for AT-hooks in EBNA1's ability to transactivate, a function necessary for EBV to immortalize naïve B-cells.
Latent infection by Epstein-Barr virus (EBV) is associated with several diseases and malignancies including infectious mononucleosis, Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and lymphoproliferative diseases in immunocompromised hosts (36). Infection of naïve human B cells by EBV results in their immortalization. A subset of EBV genes is required to immortalize B cells, including the nuclear proteins EBNA1, EBNA2, EBNA3A, EBNA3C, and the membrane protein LMP1 (36). Upon binding to a set of 20 cognate binding sites, termed the family of repeats (FR), EBNA1 exerts two functions that are necessary for EBV to immortalize naïve human B cells. First, it facilitates stable replication and partitioning of EBV genomes in proliferating, latently infected cells, and, second, it activates viral promoters used to express itself and the other genes required to immortalize naïve B cells (3).
Analyses conducted using derivatives of EBNA1 have revealed a region of EBNA1 from amino acid (aa) 40 to 89, termed linking region 1 (LR1), that is sufficient for transactivation when fused to the DNA-binding domain (DBD) of EBNA1 (23). Consistent with this observation, a derivative of EBNA1 with two copies of LR1 (2×LR1) fused to the DBD, activates transcription to levels higher than wild-type EBNA1 (23). Deletion of a portion of LR1, from aa 65 to 89, substantially impairs the ability of EBNA1 to transactivate (23). Consistent with this observation, EBV containing an EBNA1 mutant in which this region, termed unique region 1 (UR1), is deleted fails to immortalize naïve B cells although it is capable of infecting transformed B-cell lines (3). UR1 contains a short sequence, KRPSCIGCKG, which is conserved in the EBNA1 orthologs of other gamma herpesviruses and includes a potential phosphorylation site for cyclic AMP (cAMP)-dependent protein kinase (PKA) at serine 78 (Ser78) of EBNA1. There is a second region within LR1, from aa 40 to 54, that is also conserved in the EBNA1 orthologs of other gamma herpesviruses. This domain, which contains a glycine-arginine repeat (GR repeat), shares sequence homology and function with a DNA-binding motif termed an AT-hook. This motif is present in architectural transcription factors such as HMGA1a (37, 38). HMGA1a, formerly known as HMG-I(Y) (9), transactivates a number of cellular and viral promoters by bending DNA to form a transcription enhanceosome (7, 24, 45) or by looping DNA to bring a distal enhancer proximal to promoter sequences (5). Given the role of HMGA1a in transactivation, it is paradoxical that a chimeric HMGA1a-DBD protein, in which the first 450 aa of EBNA1 were replaced by HMGA1a, supported the stable replication of EBV-derived plasmids when bound to the FR but not transactivation (21, 37). This paradox was clarified by the observation that a derivative of HMGA1a-DBD containing four copies of UR1 supported both transactivation and stable replication when bound to the FR (3). These findings indicate either that EBNA1's AT-hook regions are not necessary for transactivation or that transactivation requires both UR1 and AT-hook(s), assuming that the AT-hooks of HMGA1a can substitute for those of EBNA1.
In this report we have studied the contributions of a conserved potential PKA phosphorylation site within UR1, corresponding to serine 78 (Ser78), and AT-hooks toward EBNA1's ability to transactivate. Phosphorylation by PKA modulates the activity of many transcription factors including the cAMP response element binding protein (CREB), class II transactivator, Fos, and NF-κB (16, 28, 33, 41, 47). Because the potential PKA recognition site in UR1 is conserved in EBNA1 orthologs, we sought to determine whether pharmacologic modulators of PKA activity influence the ability of EBNA1 to activate transcription. Our results indicate that PKA activators, agonists, inhibitors, or antagonists do not affect EBNA1's ability to activate transcription. We have confirmed these results by site-directed mutation of Ser78. Replacements of the serine with alanine, aspartic acid, or threonine result in EBNA1 derivatives whose activity is statistically indistinguishable from wild-type EBNA1. The combination of these results indicates that Ser78 is not regulated by PKA or any other cellular kinases.
To evaluate the contributions of the AT-hook motif within LR1 (ATH1) toward EBNA1's transactivation ability, we have examined the function of an EBNA1 derivative in which ATH1 has been deleted. The resulting protein, ΔATH1, is impaired in its ability to transactivate relative to wild-type EBNA1. The AT-hook region within LR1 is three times as long as the canonical AT-hooks from HMGA1a, predicting its potential association with longer stretches of AT-rich DNA than each of HMGA1a's AT-hooks. Consistent with this, we demonstrate that addition of a single AT-hook from HMGA1a to ΔATH1 cannot restore transactivation to wild-type levels. In contrast, addition of three copies of the HMGA1a AT-hook to ΔATH1 does restore wild-type transactivation levels. Thus, our results indicate that in addition to UR1, EBNA1 requires ATH1 for optimal transactivation. We discuss the implications of this finding in light of the mechanism by which AT-hook proteins activate transcription.
MATERIALS AND METHODS
Effector and reporter plasmids.
Plasmids 1553, 1160, 1891, and 1893 used to express wild-type EBNA, the EBNA1 DBD, LR1-DBD, and 2×LR1 have been described previously (1, 25, 30). Plasmid 438 expresses a derivative of EBNA1, termed UR1-DBD, in which aa 59 to 89 of EBNA1 is fused in frame to aa 379 to 641. Plasmids 524, 525, and 531 express derivatives of EBNA1 in which Ser78 of EBNA1 has been altered to aspartic acid (S78D), alanine (S78A), and threonine (S78T), respectively. Plasmid 526 expresses a derivative of EBNA1 in which aa 65 to 89 are duplicated (2×UR1). Plasmid 254 expresses a derivative of EBNA1 in which aa 71 to 88 are deleted (ΔUR1). Plasmid 533 expresses a derivative of EBNA1 in which aa 40 to 54 are deleted (ΔATH1). Plasmids 548 and 549 express derivatives of ΔATH1 in which the deleted regions were replaced by a single AT-hook from HMGA1a [1×Hook(ΔATH1)] or three AT-hooks from HMGA1a [3×Hook(ΔATH1)]. All plasmids were constructed by site-directed mutagenesis using overlap extension PCR (2), and the desired alteration was confirmed by automated sequencing. Plasmid 53 was used as the FR-TKp-luciferase (where TKp is the thymidine kinase promoter) reporter (20), and plasmid 1033 was used as the oriP-BamHI-Cp-luciferase reporter (25). Plasmid 2145 expresses enhanced green fluorescent protein (EGFP) and was used to normalize for transfection efficiency. All plasmids were propagated in Escherichia coli strain DH5α and purified using isopycnic CsCl gradients.
Cell culture.
Experiments were performed in C33a (herpesvirus [HPV] negative) cervical cancer cells (44), BJAB (EBV-negative) Burkitt's lymphoma cells (39), and BJAB/FR-TK-luciferase cells (23). Cells were propagated as described earlier in serum and antibiotic-containing medium (20, 23). C33a cells were transfected using calcium phosphate precipitates. A total of 5 × 106 BJAB cells were electroporated in a volume of 0.5 ml of RPMI medium in 0.4-cm cuvettes with 310 V, 975 μF, and no resistance.
Immunoblotting.
Extracts from 5 × 105 live-transfected cells were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels under reducing conditions, electroblotted on to a polyvinylidene difluoride membrane, and immunoblotted using rabbit polyclonal antibody K67.3 (37, 38) or rabbit polyclonal antibody 2638 that was raised against the DBD of EBNA1. Immunoblots were visualized using a horseradish peroxidase-conjugated secondary antibody, followed by chemiluminescent detection.
Luciferase reporter assays.
Reporter assays were performed as described earlier (1, 20). Briefly, 500 ng of reporter plasmid 53 or 1033 was cotransfected with 10 μg of the effector plasmid, and 500 ng of the cytomegalovirus-EGFP expression plasmid. Cells were harvested 48 h posttransfection, and a fraction (1/10) was analyzed by flow cytometry to determine the percentage of live-transfected cells. Cytometry was performed using a Becton-Dickinson FACSCalibur instrument and analyzed using CellQuest software. For luciferase assays, cells were lysed at 2 × 107 cells/ml in reporter lysis buffer and analyzed for luminescence using a luciferase assay system (Promega, Madison, WI) and a Zylux FB15 tube luminometer. Data were normalized using the percentage of live-transfected (GFP-positive) cells observed in each transfection.
Indirect immunofluorescence microscopy and image deconvolution.
C33a cells transfected with wild-type EBNA1 or derivatives [S78D, S78A, S78T, 2×LR1, 2×UR1, ΔATH1, 1×Hook(ΔATH1), and 3×Hook(ΔATH1)] were plated on type 1 coverslips and grown to 30 to 50% confluence. Cells were washed with phosphate-buffered saline (PBS), fixed with 3.5% formaldehyde (prepared in PBS) for 10 min at room temperature, and then blocked and permeabilized in PBS containing 3% bovine serum albumin and 0.5% Triton X-100 for 10 min at room temperature. The permeabilized cells were incubated with either antibody K67.3 or 2638 (1:1,000 dilution in PBS), followed by a secondary antibody conjugated to Alexa Fluor 488. Cells were counterstained with Hoechst 33342 (1:5,000 dilution in PBS) for 1 min at room temperature and mounted onto glass slides with Prolong Gold antifade mounting medium. Images were obtained using an inverted Zeiss AxioVision AX10 microscope, with a 100× (numerical aperture, 1.35) objective without any optical enhancement with an AxioCam MRm camera. Z-stacks of at least 15 200-nm optical sections were obtained and deconvolved using AxioVision software, version 4.6.3. All images were deconvolved using the constrained iterative algorithm.
Pharmacological modulation of PKA.
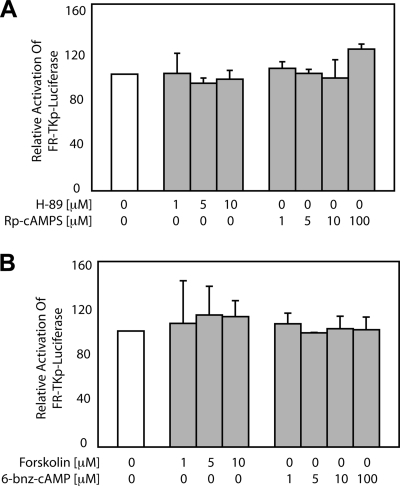
Forskolin (PKA activator), 6-benzoyl-cAMP (6-bnz-cAMP; PKA agonist), H-89 (PKA inhibitor), and Rp-cAMPS (PKA antagonist) were added 6 h posttransfection at the indicated concentrations (Fig. 2). Cells were incubated with the indicated agent (Fig. 2) for an additional 42 h, following which the fraction of live-transfected cells was measured by flow cytometry, and the expression of luciferase was measured by luminescence.
FIG. 2.
Pharmacologic modulators of PKA activity do not affect transactivation by EBNA1. C33a cells were transfected with an EBNA1 expression plasmid and FR-TKp-luciferase reporter plasmid prior to treatment with PKA inhibitors H-89 and Rp-cAMPS (A) and with PKA activators forskolin and 6-bnz-cAMP (B) at the indicated concentrations for 48 h prior to luciferase assay. At this time, EBNA1 transactivates FR-TKp-luciferase approximately 80-fold over the DBD alone, and this value was set to 100%. The luciferase values observed after treatment are expressed as a fraction of 100% and were not altered significantly by either set of conditions. The data represent the means ± standard deviations from five independent experiments. Treated samples did not vary statistically from the untreated control (P > 0.05).
Statistical analysis.
All the statistical analysis, where indicated, was performed using MSTAT, version 5 (N. Drinkwater, McArdle Laboratory for Cancer Research, University of Wisconsin Medical School). A Wilcoxon rank sum test was used for pairwise comparisons.
RESULTS
UR1 contains a conserved canonical PKA recognition site.
A derivative of EBNA1 containing aa 40 to 89, termed LR1, fused to the DBD of EBNA1 activates transcription at levels comparable to wild-type EBNA1 (23). LR1 contains two conserved features: (i) a GR repeat that functions as an AT-hook and (ii) UR1, corresponding to aa 65 to 89 of EBNA1 (23). While it has been shown previously that a UR1-deleted derivative of EBNA1 is impaired in its ability to transactivate, the mechanism by which UR1 enables EBNA1 to transactivate is unknown (23). A portion of UR1 (aa 75 to 85) is highly conserved in the EBNA1 orthologs of other gamma herpesviruses (Fig. 1). This sequence closely resembles one-half of a C4 zinc finger in several eukaryotic zinc-binding proteins (26). The conserved region contains a dicysteine motif that is flanked at one end by a canonical recognition site for cAMP-dependent PKA, namely, K/R-K/R-X-S/T, corresponding to Ser78 of EBNA1. Ser78 is also identified by the predictive algorithm pkaPS as a strong candidate site for PKA phosphorylation (32). Depending on the transactivator, PKA phosphorylation either increases transactivation or down-modulates it (16, 28, 33, 41, 47). Because Ser78 lies in a domain of EBNA1 required for transactivation, we examined whether modulating PKA activity altered transactivation by EBNA1.
FIG. 1.
The UR1 region of EBNA1 is highly conserved in the EBNA1 orthologs of EBV-like gammaherpesviruses. The region of aa 65 to 89 of EBNA1 from EBV strain B95-8 (accession no. YP_401677) are aligned with the corresponding region from the EBNA1 orthologs encoded by Cercopithecine herpesvirus 15 (accession no. YP_067973), cynomolgus EBV (accession no. BAB03281), and Cercopithecine herpesvirus 12 (accession no. AAA66373). In all four EBNA1 proteins, aa 75 to 85 are highly conserved, as indicated by the box, and also share identity with half of a C4 zinc finger found in the catalytic subunit of DNA polymerase δ from several eukaryotes (accession nos. NP_001087694, NP_001034899, XP_001641357, and XP_002008314).
The effect of PKA on transactivation by EBNA1.
PKA exists as an inactive holoenzyme with catalytic (C) and regulatory (R) subunits. When the R subunit binds cAMP, it dissociates from the C subunit, which then phosphorylates target proteins (40). H-89 is competitive inhibitor of the active site in the C subunit (10, 29), while the antagonist Rp-cAMPS binds the R subunit and prevents its dissociation from the C subunit (18). H-89 and Rp-cAMPS were used to determine whether inhibiting PKA affected EBNA1's ability to transactivate in C33a epithelial cells and BJAB Burkitt's lymphoma cells. C33a cells cotransfected with the EBNA1-expression plasmid and the FR-TKp-luciferase reporter plasmid were exposed to vehicle alone or the indicated concentrations of H-89 and Rp-cAMPS for 48 h (Fig. 2A), followed by a luciferase assay. In the absence of any pharmacologic modulator, EBNA1 transactivates FR-TKp-luciferase approximately 80-fold in C33a cells relative to the DBD alone, a value set to be 100% in Fig. 2A. Neither H-89 nor Rp-cAMPS had any effect on EBNA1's ability to transactivate FR-TKp-luciferase in C33a cells (Fig. 2A). Similar results were observed with an oriP-BamHI-Cp-luciferase reporter in BJAB cells, indicating that this result is not altered in a different cell type or with a different promoter (data not shown). Treatment with lower concentrations of the antagonists or for shorter times (15 or 24 h) also had no effect on transactivation (data not shown). To determine the effect of activating PKA, transfected cells were treated with forskolin or 6-bnz-cAMP. Forskolin causes a rise in the intracellular levels of cAMP (13), while 6-bnz-cAMP associates with the R subunit and causes it to dissociate from the C subunit (11). As shown in Fig. 2B, neither forskolin nor 6-bnz-cAMP had any effect on the ability of EBNA1 to activate transcription from the FR-TKp-luciferase reporter in C33a cells. Similar observations were made in BJAB cells using the oriP-BamHI-Cp-luciferase reporter plasmid (data not shown). These results indicate that although the UR1 domain of EBNA1 contains a conserved PKA recognition site, PKA does not affect EBNA1's ability to transactivate.
Ser78 is not regulated by cellular kinases or phosphatases.
Ser78 is conserved in the UR1 domain of EBNA1 orthologs, and deletion of UR1 decreases EBNA1's ability to transactivate. Therefore, site-directed mutagenesis was used to determine whether other cellular kinases or phosphatases might regulate EBNA1 through Ser78. For this, Ser78 was altered to alanine (S78A), aspartic acid (S78D), or threonine (S78T) (Fig. 3A). These substitutions were chosen because alanine structurally resembles an unphosphorylated serine, aspartic acid mimics a phosphorylated serine in charge and structure, and several serine kinases also phosphorylate threonine (15, 27, 42). Wild-type EBNA1 and these derivatives were expressed at similar levels in transfected C33a cells (Fig. 3B). In reporter assays, wild-type EBNA1 and EBNA1 with the mutation S78D, S78A, or S78T transactivated FR-TKp-luciferase to similar levels in C33a cells (Fig. 3C) and oriP-BamHI-Cp-luciferase in BJAB cells (data not shown). These results clearly indicate that although serine 78 is conserved in EBNA1 and its orthologs, the phosphorylation status of this residue does not regulate the ability of EBNA1 to transactivate.
FIG. 3.
Mutation of the conserved PKA recognition site in UR1 does not affect transactivation by EBNA1. (A) The wild-type version of EBNA1 used in these studies contains five copies of the Gly-Gly-Ala repeat and is depicted schematically, indicating domains of EBNA1. The sequence of the conserved portion of UR1 is shown with serine 78 indicated in bold. The sequence of the S78D, S78A, and S78T substitution mutations constructed for this study are also shown with the substituted amino acids shown in bold. (B) Immunoblot of 5 × 105 C33a cells transfected with expression plasmids for EBNA1, DBD, S78D, S78A, and S78T. Proteins were detected using the K67.3 or 2638 antibodies against the DBD of EBNA1 and were visualized by chemiluminescence. The migration of prestained markers of known molecular sizes are indicated by the arrowheads. (C) C33a cells were transfected with an expression plasmid for EBNA1, S78D, S78A, or S78T along with an FR-TKp-luciferase reporter. Assays for luciferase activity were performed at 48 h posttransfection. At this time, EBNA1 transactivates FR-TKp-luciferase approximately 80-fold over the DBD alone, and this value was set to 100%. Data from three independent transfections indicate that replacing the conserved serine in UR1 with alanine, aspartic acid, or threonine does not significantly affect transactivation (P > 0.05 for pairwise comparisons with EBNA1).
UR1 and ATH1 contribute to transactivation by EBNA1.
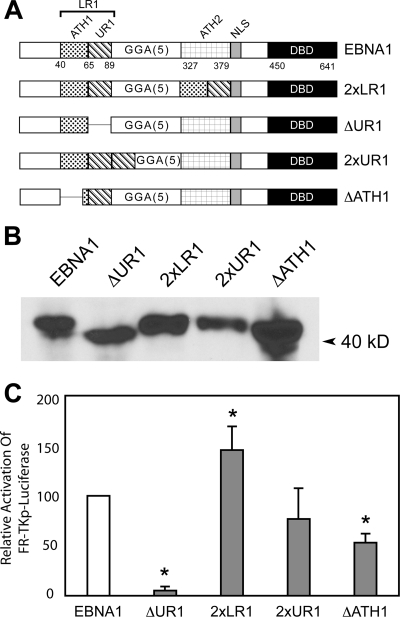
An EBNA1 derivative containing 2×LR1 activates transcription better than wild-type EBNA1 (23). This derivative of EBNA1 contains two copies of UR1 and two copies of the AT-hook motif within LR1 (ATH1), which closely resembles the AT-hook present in architectural transcription factors such as HMGA1a (37, 38). The derivatives of EBNA1 shown in Fig. 4A were constructed to determine the contributions of UR1 and ATH1 to EBNA1's ability to transactivate. The first two of these, 2×LR1 and ΔUR1, correspond to previously described derivatives of EBNA1 (23, 30), with the sole difference being that this version of ΔUR1 has a smaller deletion (aa 71 to 88). This change does not affect the previously described transactivation properties of this mutant (Fig. 4C and data not shown). The third derivative of EBNA1 contains a duplication of only UR1 (2×UR1) while the last derivative contains a deletion of ATH1 (ΔATH1). All four derivatives were expressed similarly in transfected C33a cells as determined by immunoblotting (Fig. 4B). The ability of these proteins to activate transcription was evaluated in C33a cells and BJAB cells using the FR-TKp-luciferase and oriP-BamHI-Cp-luciferase reporter plasmids, respectively. As observed previously (23), 2×LR1 was more active than wild-type EBNA1 in its ability to transactivate FR-TKp-luciferase. Similarly, ΔUR1 was severely impaired in its ability to activate transcription (Fig. 4C). However, in contrast to 2×LR1, 2×UR1 did not activate transcription at levels higher than wild-type EBNA1 (23), suggesting that both ATH1 and UR1 contribute to the increased transactivation observed with 2×LR1. This conclusion is further supported by the results obtained with the ΔATH1 mutant. ΔATH1 activated transcription of FR-TKp-luciferase approximately 50% as well as wild-type EBNA1. Similar results were obtained with the oriP-BamHI-Cp-luciferase reporter in BJAB cells, indicating that the requirement for ATH1 is not cell type or promoter specific (data not shown). Because ATH1 is one of two regions used by EBNA1 to retain FR-containing plasmids (38), such as the transcription reporter plasmids used here, the decreased reporter activity observed with ΔATH1 could reflect either a decrease in transcription or an inefficient retention of the reporter plasmid in transfected cells. To distinguish between these possibilities, ΔATH1 was tested for its ability to activate transcription from an integrated FR-dependent luciferase reporter constructed in BJAB cells (23).
FIG. 4.
Both the UR1 and ATH1 domains within LR1 contribute to transactivation of an episomal FR-TKp-luciferase reporter in C33a cells. (A) A schematic representation of wild-type EBNA1, the previously described 2×LR1, ΔUR1 derivatives, and the 2×UR1 and ΔATH1 derivatives used in this study. 2×LR1 replaces aa 327 to 377 of EBNA1 with a second copy of aa 40 to 89. In ΔUR1 aa 71 to 88 are deleted. 2×UR1 contains a second copy of aa 65 to 89 inserted after UR1 while the GR repeat from aa 40 to 54 is deleted in ΔATH1. (B) Expression of wild-type EBNA1, ΔUR1, 2×LR1, 2×UR1, and ΔATH1 in transfected C33a cells. Proteins from 5 × 105 transfected cells were visualized as described in the Materials and Methods section, and the migration of a prestained marker is indicated by the arrowhead. (C) C33a cells were transfected with an expression plasmid for EBNA1, ΔUR1, 2×LR1, 2×UR1, or ΔATH1 proteins along with an FR-TKp-luciferase reporter and assayed for luciferase activity 48 h posttransfection. At this time, EBNA1 transactivates FR-TKp-luciferase approximately 80-fold over the DBD alone, and this value was set to 100%. In three independent transfections 2×LR1 was observed to transactivate the reporter at greater levels than wild-type EBNA1 while ΔUR1 was impaired in transactivation. Transactivation by 2×UR1 could not be distinguished from wild-type EBNA1. ΔATH1 transactivated the reporter approximately 50% as well as wild-type EBNA1. Transactivation that differs significantly from that observed with wild-type EBNA1 (P < 0.05) is indicated by the asterisk.
Optimal transactivation by EBNA1 requires an AT-hook motif in LR1.
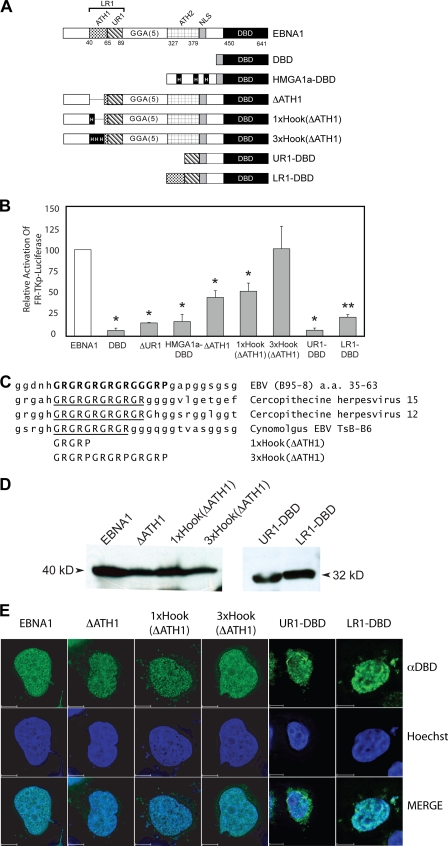
The reporter cell line BJAB/FR-TK-luciferase that contains an integrated FR-TKp-luciferase reporter has been used to discriminate between EBNA1's ability to transactivate and its ability to retain transcription reporter plasmids in transfected cells (23). To elucidate whether ATH1 facilitates EBNA1's ability to transactivate, BJAB/FR-TK-luciferase cells were electroporated with expression plasmids for EBNA1 and the following derivatives: DBD, HMGA1a-DBD, ΔUR1, or ΔATH1 (Fig. 5A). In these cells, as has been observed previously, wild-type EBNA1 activated transcription approximately 15-fold more than the DBD alone (3, 37). This value was set as 100%, and the transactivation by each EBNA1 derivative is expressed as a fraction of that value. As observed previously, HMGA1a-DBD and ΔUR1 were significantly impaired in their ability to activate transcription from this integrated reporter (Fig. 5B) (23, 37). ΔATH1 transactivated the integrated reporter approximately 45% as well as wild-type EBNA1, a decrease that is similar to that observed with episomal reporter plasmids in C33a and BJAB cells. This result confirms that in addition to UR1, the AT-hook motif within LR1 also contributes to EBNA1's ability to transactivate. However, as indicated by the result with HMGA1a-DBD, AT-hooks are not sufficient for EBNA1 to transactivate.
FIG. 5.
EBNA1's AT-hook domains significantly augment transactivation of an integrated FR-TK-luciferase reporter in BJAB cells. (A) A schematic depiction of wild-type EBNA1 and EBNA1 derivatives expressed in BJAB/FR-TK-luciferase cells. The DBD contains a fusion to EBNA1's nuclear localization signal (NLS) to aa 450 to 641. In ΔUR1, aa 71 to 88 are deleted. HMGA1a-DBD is a fusion of the architectural transcription factor HMGA1a to DBD. The three AT-hooks within HMGA1a are boxed and indicated by a capital H. In ΔATH1, the GR repeat in LR1 from aa 40 to 54 is deleted. The sequence deleted in ΔATH1 is replaced by a single AT-hook from HMGA1a in 1×Hook(ΔATH1) or three copies of this AT-hook in 3×Hook(ΔATH1). UR1-DBD contains a fusion of aa 59 to 89 of EBNA1 to aa 379 to 641. LR1-DBD contains a fusion of aa 40 to 89 of EBNA1 to aa 379 to 641. (B) Expression plasmids for the proteins described above were electroporated into BJAB/FR-TK-luciferase cells along with a cytomegalovirus-EGFP expression plasmid, as described in Materials and Methods. Luciferase activity in extracts was measured 48 h posttransfection and corrected for transfection efficiency, which ranged from 20% to 40%. Wild-type EBNA1 transactivated the reporter approximately 15-fold over the DBD alone, and this value was normalized to 100%. The relative induction observed with the EBNA1 derivatives is expressed as a fraction of 100%. Transactivation that differed significantly from that observed with wild-type EBNA1 (P < 0.05) is indicated by the asterisk. ΔATH1 and 1×Hook(ΔATH1) were similarly impaired in their transactivation of this reporter. In contrast, transactivation by 3×Hook(ΔATH1) could not be distinguished from wild-type EBNA1. (C) Sequence alignment of the GR repeat in LR1 aligned with the corresponding sequence from the indicated EBNA1 orthologs. The substituted sequences in the 3×Hook(ΔATH1) and 3×Hook(ΔATH1) are shown aligned to the GR repeat of the EBNA1 proteins. (D) Expression of EBNA1, ΔATH1, 1×Hook(ΔATH1), 3×Hook(ΔATH1), UR1-DBD, and LR1-DBD in electroporated BJAB/FR-Tk-luciferase cells. Protein from 5 × 105 electroporated cells was detected and visualized as described in Materials and Methods. Standards of known molecular sizes are indicated adjacent to the arrowheads. (E) Expression of EBNA1, ΔATH1, 1×Hook(ΔATH1), 3×Hook(ΔATH1), UR1-DBD, and LR1-DBD in C33a cells evaluated by indirect immunofluorescence. Transfected cells were grown on coverslips and processed for indirect immunofluorescence as described in Materials and Methods. The anti-DBD (αDBD) antibody was visualized with Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G, and nuclei were counterstained with Hoechst 33342. Staining by the anti-DBD and Hoechst stain is indicated in the merged image of the two. At least 15 z-sections of 200 nm were deconvolved for each image. The scale bar represents a distance of 5 μm.
ATH1 is conserved in EBNA1 orthologs (Fig. 5C) and is approximately three times longer than any one of the prototypic AT-hooks in HMGA1a. ATH1 is predicted to bind longer stretches of AT-rich DNA than a single AT-hook of HMGA1a, based on the structure of an AT-hook bound to DNA (22). To determine whether the removal of ATH1 could be compensated by the addition of an AT-hook from HMGA1a, two additional derivatives of EBNA1 were constructed, 1×Hook(ΔATH1) and 3 ×Hook(ΔATH1) (Fig. 5A). For 1 × Hook(ΔATH1), a single AT-hook motif from HMGA1a was inserted in lieu of the ATH1 deletion, whereas for 3×Hook(ΔATH1), the sequences deleted in ΔATH1 were replaced by three copies of the HMGA1a AT-hook motif juxtaposed to each other (Fig. 5C). As shown in Fig. 5B, although 1×Hook(ΔATH1) transactivated the integrated FR-TK-luciferase reporter at slightly higher levels than ΔATH1, it still displayed impaired function relative to wild-type EBNA1. In contrast, 3×Hook(ΔATH1) activated transcription in a manner that was statistically indistinguishable from wild-type EBNA1 (Fig. 5B). In addition to ATH1, EBNA1 contains a second AT-hook, termed ATH2, between aa 327 to 377. All the EBNA1 derivatives described above contain an unmodified ATH2. It has been shown previously that an EBNA1 derivative lacking LR1 but with two copies of ATH2 activates transcription from episomal reporters but not the integrated reporter employed here (23). While this result indicates that ATH2 is insufficient to transactivate in the absence of LR1, it remains possible that ATH2 contributes to transactivation. Further, because all the EBNA1 derivatives used here contain ATH2, it is also possible that the wild-type AT-hook present in ATH2 compensates for the absence of ATH1. To examine a role for ATH2, we studied two additional derivatives of EBNA1, namely, LR1-DBD and UR1-DBD, also shown schematically in Fig. 5A. Both derivatives were impaired significantly in their ability to activate transcription (Fig. 5B). LR1-DBD activated transcription to approximately 20% of the level of EBNA1 while UR1-DBD activated transcription to approximately the same level as DBD alone (about 7% of EBNA1) (23). It should be noted that when tested with an episomal reporter plasmid, UR1-DBD activated transcription twice as well as DBD alone while transactivation by LR1-DBD relative to EBNA1 was similar with integrated and episomal reporters (data not shown). LR1-DBD activated transcription three times as well as UR1-DBD, a difference that was found to be statistically significant. While this threefold increase is greater than the difference between EBNA1 and ΔATH1, it should be interpreted with the knowledge that LR1-DBD is significantly impaired in function relative to EBNA1. Indeed, comparing the results obtained with EBNA1, LR1-DBD, and 2×LR1 suggests that transactivation by EBNA1 is maximal when there are two sets of AT-hooks in addition to UR1. The 1×Hook(ΔATH1), 3×Hook(ΔATH1), UR1-DBD, and LR1-DBD proteins were expressed at levels similar to wild-type EBNA1 and ΔATH1, as demonstrated by several immunoblot assays (Fig. 5D). Finally, indirect immunofluorescence examination of the localization of wild-type EBNA1, ΔATH1, 1×Hook(ΔATH1), 3×Hook(ΔATH1), UR1-DBD, and LR1-DBD in C33a cells indicated that the decrease in function observed for ΔATH1, 1×Hook(ΔATH1), and LR1-DBD could not be attributed to deficiencies in nuclear localization (Fig. 5E). In contrast to all the other derivatives of EBNA1 shown here, UR1-DBD stained both the nucleus and the cytoplasm (Fig. 5E). In addition, cell fractionation experiments indicated that UR1-DBD is easily extracted from the nucleus in comparison to LR1-DBD (data not shown). These results confirm our previous conclusion that both ATH1 and UR1 domains are required for EBNA1 to transactivate optimally. In addition, it is likely that for wild-type levels of transactivation, two sets of AT-hooks are required in EBNA1.
DISCUSSION
Although the ability of FR-bound EBNA1 to activate transcription from EBV and heterologous promoters is well documented (34, 35, 43), the mechanism underlying this ability is as yet unclear. Studies with EBNA1 derivatives have revealed that a fusion of LR1 to the DBD activates transcription comparably to wild-type EBNA1 (23). In this study, the importance of LR1 in transactivation was confirmed when a portion of LR1, termed UR1, was deleted from EBNA1. The resulting protein, ΔUR1, is significantly impaired in its ability to transactivate without affecting other functions of EBNA1 (23). This finding is corroborated by the observation that the addition of four copies of UR1 to the transactivation-incompetent chimera protein HMGA1a-DBD creates a transactivation-competent protein (3). Only a portion of UR1 is conserved in EBNA1 orthologs from other gamma herpesviruses (Fig. 1). This portion, KRPSCIGCKG, shares sequence identity with half of a C4 zinc finger domain (26) from the catalytic subunit of DNA polymerase δ and includes a potential phosphorylation site for PKA. Our results indicate that although this potential phosphorylation site in UR1 is conserved, PKA does not modulate EBNA1's ability to transactivate. In addition, mutagenesis studies indicate that the phosphorylation status of this conserved serine residue does not affect transactivation by EBNA1.
In addition to UR1, LR1 contains a GR repeat that functions as an AT-hook (37, 38) and is therefore termed ATH1. ATH1 is similar in sequence to the prototypic AT-hook motifs in the architectural transcription factor, HMGA1a, and also associates specifically with AT-rich DNA (37, 38). ATH1 and a second GR repeat from aa 327 to 377 of EBNA1, termed ATH2, are used by EBNA1 to tether viral genomes or FR-containing plasmids to cellular chromosomes and thereby facilitate their retention and partitioning. EBNA1 derivatives in which either ATH1 or ATH2 is deleted (ΔATH1 and LR1-DBD) have a diminished capacity to transactivate from both episomal reporter plasmids and a chromosomally integrated reporter gene. The latter result indicates that decreases in episomal reporter expression observed with ΔATH1 and LR1-DBD proteins reflect decreases in transcription and not a defect in retention of reporter plasmids in transfected cells. For ATH1, it is likely that the number of GR repeats correlates with its function in transactivation. EBNA1 and its orthologs contain at least five GR repeats in ATH1. A single AT-hook motif from HMGA1a contains only two GR repeats, and when substituted for ATH1, it does not rescue the defect in transactivation. In contrast, when three copies of the HMGA1a AT-hook motif were substituted for ATH1, transactivation was equal to that observed with wild-type EBNA1.
We conjecture three possible ways by which AT-hooks may contribute to EBNA1's ability to transactivate on the basis of observations made with HMGA1a. HMGA1a displays a punctate pattern on condensed mitotic chromosomes and a granulated pattern in interphase nuclei (4, 14). Colocalization studies indicate that a fraction of the HMGA1a colocalizes with RNA polymerase II and is released under the same nuclease conditions that release RNA polymerase II (4, 17). On this basis it is claimed that some of the HMGA1a in cells is localized to actively transcribed chromatin (14, 17). Consistent with this interpretation, microinjection of HMGA1a into single-cell mouse embryos induces early transcription (6). The localization of EBNA1 on mitotic chromosomes is strikingly similar to that of HMGA1a (19, 31, 37). In addition, EBNA1 also displays a granulated pattern similar to HMGA1a in interphase nuclei (37, 38), possibly indicating preferred sites of association on chromosomal DNA. Derivatives of EBNA1 that contain only either ATH1 or ATH2 continue to display the same localization on condensed chromosomes and the granulated appearance in interphase nuclei as HMGA1a-DBD (37, 38). In our first model, we hypothesized that, like HMGA1a (14, 17), AT-hooks localize EBNA1 to actively transcribed chromatin in interphase nuclei, and transcription coactivators enriched at such sites may be recruited by another domain of EBNA1, such as UR1, to EBV genomes tethered to the DBD of EBNA1.
Our other two hypotheses are influenced by the two different mechanisms by which HMGA1a activates transcription. First, at the β-globin promoter, HMGA1a binds AT-rich sequences at promoter proximal and distal sites, loops intervening sequences by self-association, and thereby brings a distal enhancer close to the promoter (5). Second, for other promoters like the beta interferon promoter (24, 45) or the HPV-18 early promoter (7), HMGA1a functions as an architectural transcription factor. Using its AT-hooks, HMGA1a associates with AT-rich regions in the promoter and then bends adjacent sequences to form a structure, termed a transcription enhanceosome. Other proteins bound to DNA sequences within the enhanceosome structure recruit coactivators such as p300/CBP to these promoters and thereby activate transcription (8, 46). A unique characteristic of HMGA1a within enhanceosomes is that DNA phasing affects its function. Insertions or deletions of 5 or 6 bp decrease transactivation by 50% or more at the beta interferon and HPV-18 early promoters while insertions or deletions of 10 bp have no effect on transactivation (7, 24).
It is possible that EBNA1 transactivates by either mechanism. Regions upstream of some EBV promoters, such as the BamHI-Cp, have several long AT-rich sequences. We postulate that the AT-hooks of FR-bound EBNA1 associates with these sequences and forms an enhanceosome, in which context coactivators are recruited to the promoter, perhaps through interactions with UR1. It is of interest that, as with HMGA1a, changes in the phase of DNA between adjacent EBNA1 binding sites affects the ability of EBNA1 to transactivate by a magnitude similar to that observed with HMGA1a (20). It is also possible that looping between FR and the promoter is used to concentrate a large amount of EBNA1 at the promoter and thereby increase the local availability of transactivation domains. Both enhanceosome formation and looping between FR and the promoter are dependent on EBNA1's association with sequences close to BamHI-Cp. A recent chromatin immunoprecipitation analysis of EBNA1 across the first 60 kb of the EBV genome has revealed that in addition to its cognate binding sites in the FR, dyad symmetry element, and Qp, EBNA1 also associates with multiple sequences upstream of BamHI-Cp (12). Greater association was observed in cells displaying a latency III transcription profile in which BamHI-Cp is active than in cells displaying a latency I transcription profile in which this promoter is not used (12). We propose that EBNA1 associates with sequences upstream of BamHI-Cp through its AT-hooks, based on their capacity to bind AT-rich DNA and the presence of AT-rich sequences upstream of BamHI-Cp. Delineating the sequences bound by EBNA1 upstream of BamHI-Cp, along with the mechanism of association, will greatly elucidate how EBNA1 transactivates and will identify the contributions made to transactivation by UR1 and EBNA1's AT-hooks. Because transactivation by EBNA1 is necessary for EBV to immortalize naïve B cells, identifying the mechanism of transactivation will provide a basis to systematically devise anti-EBV therapeutics that block this essential function.
Acknowledgments
We thank Bill Sugden for providing the BJAB/FR-TK-luciferase cell-line and expression plasmids for 2×LR1 and LR1-DBD. We thank Tim Foster and Paolo Rodriguez for their experimental suggestions and Ben Kelly for critiquing the manuscript.
This research was supported by NIH awards P02RR021970 (to S.K. through A. Ochoa) and R01CA112564 (A.A.). G.S., A.H.Z., S.K., and A.A. gratefully acknowledge support from the Stanley S. Scott Cancer Center at Louisiana State University Health Sciences Center. S.A. is a graduate student in the Microbiology graduate program at Louisiana State University Health Sciences Center.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Aiyar, A., and B. Sugden. 1998. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J. Biol. Chem. 27333073-33081. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar, A., Y. Xiang, and J. Leis. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 57177-191. [DOI] [PubMed] [Google Scholar]
- 3.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. USA 10314188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amirand, C., A. Viari, J. P. Ballini, H. Rezaei, N. Beaujean, D. Jullien, E. Kas, and P. Debey. 1998. Three distinct sub-nuclear populations of HMG-I protein of different properties revealed by co-localization image analysis. J. Cell Sci. 1113551-3561. [DOI] [PubMed] [Google Scholar]
- 5.Bagga, R., S. Michalowski, R. Sabnis, J. D. Griffith, and B. M. Emerson. 2000. HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res. 282541-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaujean, N., C. Bouniol-Baly, C. Monod, K. Kissa, D. Jullien, N. Aulner, C. Amirand, P. Debey, and E. Kas. 2000. Induction of early transcription in one-cell mouse embryos by microinjection of the nonhistone chromosomal protein HMG-I. Dev. Biol. 221337-354. [DOI] [PubMed] [Google Scholar]
- 7.Bouallaga, I., S. Massicard, M. Yaniv, and F. Thierry. 2000. An enhanceosome containing the Jun B/Fra-2 heterodimer and the HMG-I(Y) architectural protein controls HPV 18 transcription. EMBO Rep. 1422-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouallaga, I., S. Teissier, M. Yaniv, and F. Thierry. 2003. HMG-I(Y) and the CBP/p300 coactivator are essential for human papillomavirus type 18 enhanceosome transcriptional activity. Mol. Cell. Biol. 232329-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin, M. 2001. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26152-153. [DOI] [PubMed] [Google Scholar]
- 10.Chijiwa, T., A. Mishima, M. Hagiwara, M. Sano, K. Hayashi, T. Inoue, K. Naito, T. Toshioka, and H. Hidaka. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 2655267-5272. [PubMed] [Google Scholar]
- 11.Christensen, A. E., F. Selheim, J. de Rooij, S. Dremier, F. Schwede, K. K. Dao, A. Martinez, C. Maenhaut, J. L. Bos, H. G. Genieser, and S. O. Doskeland. 2003. cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 27835394-35402. [DOI] [PubMed] [Google Scholar]
- 12.Day, L., C. M. Chau, M. Nebozhyn, A. J. Rennekamp, M. Showe, and P. M. Lieberman. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 816389-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza, N. J., A. N. Dohadwalla, and J. Reden. 1983. Forskolin: a labdane diterpenoid with antihypertensive, positive inotropic, platelet aggregation inhibitory, and adenylate cyclase activating properties. Med. Res. Rev. 3201-219. [DOI] [PubMed] [Google Scholar]
- 14.Disney, J. E., K. R. Johnson, N. S. Magnuson, S. R. Sylvester, and R. Reeves. 1989. High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J. Cell Biol. 1091975-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egelhoff, T. T., R. J. Lee, and J. A. Spudich. 1993. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75363-371. [DOI] [PubMed] [Google Scholar]
- 16.Fiol, C. J., J. S. Williams, C. H. Chou, Q. M. Wang, P. J. Roach, and O. M. Andrisani. 1994. A secondary phosphorylation of CREB341 at Ser129 is required for the cAMP-mediated control of gene expression. A role for glycogen synthase kinase-3 in the control of gene expression. J. Biol. Chem. 26932187-32193. [PubMed] [Google Scholar]
- 17.Giancotti, V., A. Bandiera, C. Sindici, L. Perissin, and C. Crane-Robinson. 1996. Calcium-dependent ADP-ribosylation of high-mobility-group I (HMGI) proteins. Biochem. J. 317865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjertsen, B. T., G. Mellgren, A. Otten, E. Maronde, H. G. Genieser, B. Jastorff, O. K. Vintermyr, G. S. McKnight, and S. O. Doskeland. 1995. Novel (Rp)-cAMPS analogs as tools for inhibition of cAMP-kinase in cell culture. Basal cAMP-kinase activity modulates interleukin-1 beta action. J. Biol. Chem. 27020599-20607. [DOI] [PubMed] [Google Scholar]
- 19.Grogan, E. A., W. P. Summers, S. Dowling, D. Shedd, L. Gradoville, and G. Miller. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc. Natl. Acad. Sci. USA 807650-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebner, C., J. Lasanen, S. Battle, and A. Aiyar. 2003. The spacing between adjacent binding sites in the family of repeats affects the functions of Epstein-Barr nuclear antigen 1 in transcription activation and stable plasmid maintenance. Virology 311263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 981865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huth, J. R., C. A. Bewley, M. S. Nissen, J. N. Evans, R. Reeves, A. M. Gronenborn, and G. M. Clore. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4657-665. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 236901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1119-129. [DOI] [PubMed] [Google Scholar]
- 25.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 711766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna, S. S., I. Majumdar, and N. V. Grishin. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin, L. R., and M. J. Zoller. 1990. Association of catalytic and regulatory subunits of cyclic AMP-dependent protein kinase requires a negatively charged side group at a conserved threonine. Mol. Cell. Biol. 101066-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, G., J. A. Harton, X. Zhu, and J. P. Ting. 2001. Downregulation of CIITA function by protein kinase a (PKA)-mediated phosphorylation: mechanism of prostaglandin E, cyclic AMP, and PKA inhibition of class II major histocompatibility complex expression in monocytic lines. Mol. Cell. Biol. 214626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lochner, A., and J. A. Moolman. 2006. The many faces of H89: a review. Cardiovasc. Drug Rev. 24261-274. [DOI] [PubMed] [Google Scholar]
- 30.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 193349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 734385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuberger, G., G. Schneider, and F. Eisenhaber. 2007. pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol. Direct 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols, M., F. Weih, W. Schmid, C. DeVack, E. Kowenz-Leutz, B. Luckow, M. Boshart, and G. Schutz. 1992. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP induced gene transcription. EMBO J. 113337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puglielli, M. T., M. Woisetschlaeger, and S. H. Speck. 1996. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 705758-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reisman, D., and B. Sugden. 1986. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 63838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 37.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 7711767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 7811487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinitz, M., and G. Klein. 1975. Comparison between growth characteristics of an Epstein-Barr virus (EBV)-genome-negative lymphoma line and its EBV-converted subline in vitro. Proc. Natl. Acad. Sci. USA 723518-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor, S. S., C. Kim, C. Y. Cheng, S. H. Brown, J. Wu, and N. Kannan. 2008. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim. Biophys. Acta 178416-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tratner, I., R. Ofir, and I. M. Verma. 1992. Alteration of a cyclic AMP-dependent protein kinase phosphorylation site in the c-Fos protein augments its transforming potential. Mol. Cell. Biol. 12998-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldmann, R., P. I. Hanson, and H. Schulman. 1990. Multifunctional Ca2+/calmodulin-dependent protein kinase made Ca2+ independent for functional studies. Biochemistry 291679-1684. [DOI] [PubMed] [Google Scholar]
- 43.Yates, J. L., S. M. Camiolo, S. Ali, and A. Ying. 1996. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology 2221-13. [DOI] [PubMed] [Google Scholar]
- 44.Yee, C., I. Krishnan-Hewlett, C. C. Baker, R. Schlegel, and P. M. Howley. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am. J. Pathol. 119361-366. [PMC free article] [PubMed] [Google Scholar]
- 45.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J. 183074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 9613108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1661-671. [DOI] [PubMed] [Google Scholar]