Abstract
We analyzed viral nucleic acids in stool samples collected from 35 South Asian children with nonpolio acute flaccid paralysis (AFP). Sequence-independent reverse transcription and PCR amplification of capsid-protected, nuclease-resistant viral nucleic acids were followed by DNA sequencing and sequence similarity searches. Limited Sanger sequencing (35 to 240 subclones per sample) identified an average of 1.4 distinct eukaryotic viruses per sample, while pyrosequencing yielded 2.6 viruses per sample. In addition to bacteriophage and plant viruses, we detected known enteric viruses, including rotavirus, adenovirus, picobirnavirus, and human enterovirus species A (HEV-A) to HEV-C, as well as numerous other members of the Picornaviridae family, including parechovirus, Aichi virus, rhinovirus, and human cardiovirus. The viruses with the most divergent sequences relative to those of previously reported viruses included members of a novel Picornaviridae genus and four new viral species (members of the Dicistroviridae, Nodaviridae, and Circoviridae families and the Bocavirus genus). Samples from six healthy contacts of AFP patients were similarly analyzed and also contained numerous viruses, particularly HEV-C, including a potentially novel Enterovirus genotype. Determining the prevalences and pathogenicities of the novel genotypes, species, genera, and potential new viral families identified in this study in different demographic groups will require further studies with different demographic and patient groups, now facilitated by knowledge of these viral genomes.
Metagenomic analyses of human gut or stool specimens for bacterial (15, 22) and viral (7, 8, 13, 41) communities have revealed a large degree of microbial diversity. The use of sequence-independent amplification of viral nucleic acids (2, 19, 35, 38) for viral metagenomics avoids the potential limitations of traditional methods, including the failure of virus to replicate in cell cultures, unsuccessful PCR amplification or microarray hybridization due to high-level genetic divergence from known viruses, and the failure of antibodies to known viruses to cross-react. The application of viral metagenomics may also be useful for the study of diseases with unexplained etiologies possibly involving uncharacterized viruses or combinatorial viral infections (13, 21).
Acute flaccid paralysis (AFP), characterized by the rapid onset of asymmetric paralysis, can be caused by a variety of viral infections or coinfections (34). AFP in children under 15 years old is currently monitored in countries where poliovirus is still endemic, Pakistan, India, Afghanistan, and Nigeria, as part of the Global Polio Laboratory Network (9). Besides wild-type and revertant vaccine strains of polioviruses, several nonpolio enteroviruses, including human enterovirus species A (HEV-A) serotype EV71, have also been associated with AFP, linked to up to a third of AFP cases in children (6, 11, 16, 31, 32). In the United States, AFP is also observed in 10 to 41% of the estimated <1% of persons newly infected with West Nile virus who exhibit neurological symptoms (17, 30). Recent studies indicate that Chikungunya virus may also cause AFP in rare cases (33).
In this study, we utilized sequence-independent amplification of partially purified viral nucleic acids from stool samples obtained from South Asian children with nonpolio AFP. All samples had been tested previously by cell culture and found to be negative for poliovirus. Sequence data were obtained both by Sanger sequencing of subcloned DNA and by 454 pyrosequencing.
MATERIALS AND METHODS
Viral particle purification.
Stool samples were resuspended in Hanks' balanced salt solution (Gibco BRL) and vigorously vortexed. Five hundred microliters of supernatant from thrice-repeated 15,000 × g centrifugation using a tabletop microfuge was filtered through a 0.45-μm filter (Millipore) to remove eukaryotic- and bacterial-cell-sized particles. The filtrate was then treated with a cocktail of DNases (Turbo DNase from Ambion, Baseline-ZERO from Epicentre, and Benzonase from Novagen) and RNase (Fermentas) to digest non-particle-protected nucleic acid (20). Viral nucleic acids, protected from digestion within viral capsids, were then extracted using the QIAamp viral RNA extraction kit (Qiagen).
Sequence-independent amplification of viral nucleic acids.
Viral cDNA synthesis from extracted viral RNA/DNA was performed as described previously (38). Briefly, 100 pmol of a primer containing a fixed sequence followed by a randomized octomer at the 3′ end was used in a reverse transcription reaction with SuperScript III reverse transcriptase (Invitrogen). A single round of DNA synthesis was then performed using Klenow fragment polymerase (New England Biolabs) with an additional 50 pmol of the same primer. PCR amplification of nucleic acids was then performed using primers consisting of the fixed portions of the random primers (38). Primers used for downstream plasmid subcloning were based on primer K (GAC CAT CTA GCG ACC TCC ACN NNN NNN N) (35) or RA01 (GCC GGA GCT CTG CAG ATA TCN NNN NNN NNN) (19). For the 10 samples submitted for 454 pyrosequencing, the following primers were used in the PCR and corresponding primers containing an additional eight N residues at the 3′ end were used for priming during reverse transcription: 454-A, ATC GTC GTC GTA GGC TGC TC; 454-B, GTA TCG CTG GAC ACT GGA CC; 454-C, CGC ATT GGT CGG CAC TTG GT; 454-D, CGT AGA TAA GCG GTC GGC TC; 454-E, CAT CAC ATA GGC GTC CGC TG; 454-F, CGC AGG ACC TCT GAT ACA GG; 454-G, CGC ACT CGA CTC GTA ACA GG; 454-H, CGT CCA GGC ACA ATC CAG TC; 454-I, CCG AGG TTC AAG CGA GGT TG; and 454-J, ACG GTG TGT TAC CGA CGT CC. Either amplification products were subcloned into bacterial plasmids and Sanger sequenced, or PCR products were sequenced directly using a GS-FLX 454 pyrosequencing system (Roche). Briefly, PCR fragments were treated as sonicated bacterial DNA for 454 pyrosequencing, with polishing of the extremities of fragments and ligation of adaptors prior to emulsion PCR as recommended by the system manufacturer (Roche).
Data assembly and processing.
For 454 sequencing, primer tag signatures on the random PCR primers were used as identifier tags to assign sequences to the corresponding sample. Sequences were then automatically trimmed of the fixed primer sequences plus eight additional nucleotides at the 3′ end corresponding to the random N residue portion used in cDNA synthesis and Klenow extension. Trimmed sequences were assembled into contigs by using Sequencher software (Gene Codes), with a criterion of 95% identity or greater over 35 bp for 454 pyrosequenced products. Similarly, for cloned and sequenced products, primer sequences were removed from plasmid inserts and assembled by using Sequencher software (Gene Codes), with a criterion of 85% identity over 50 bp. The assembled contigs and singlet sequences were analyzed using tBLASTx. Sequences with tBLASTx E values lower than 10−3 were classified as either eukaryotic viral, bacteriophage, eukaryotic, bacterial, or other based on the match with the best E value. Those sequences with tBLASTx, BLASTn, and BLASTx E values greater than 10−3 for the best hit were deemed unclassifiable.
Sequencing of HEV VP1 region.
Nested consensus primers consisting of sequences within the VP3 and 2A genes of HEV-C were designed to amplify the complete VP1 gene. Primers were as follows (5′ to 3′): VP1_C_F1, GGBACNCAYRTNATHTGGGA; VP1_C_F2, GCNTGYMMNGAYTTYWSHGT; VP1_C_R1, GGRCACCANVMNCKNACATG; and VP1_C_R2, GYTTDGGYTTCATGTACACTC. Reverse transcriptase PCR conditions were as follows. Ten microliters of extracted viral RNA was incubated with 100 pmol of random hexamer oligonucleotide and 0.5 mM (each) deoxynucleoside triphosphates at 75°C for 5 min. Subsequently, 40 U of RNase inhibitor, 10 mM dithiothreitol, 1× first-strand extension buffer, and 200 U of SuperScript III reverse transcriptase (Invitrogen) were added to the mixture, and the mixture was incubated at 25°C for 5 min and then at 37°C for 1 h. For PCR, 5 μl of the reaction mixture described above was used in a total reaction volume of 50 μl containing 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1× manufacturer's buffer (New England Biolabs), 0.2 μM (each) primers, and 5 U of Taq polymerase (New England Biolabs). For the first round, the PCR cycle included 4 min of denaturation at 95°C; 30 cycles of 95°C for 45 s, 52°C for 1 min, and 72°C for 1 min; and final extension at 72°C for 7 min. One microliter of the first-round PCR product was used as a template in a second round of amplification under identical conditions, with appropriate second-round primers.
Phylogenetic analysis.
Reference HEV sequences were obtained from NCBI and edited for alignment using GeneDoc software. Three representative sequences from each serotype, when available, were used. Sequence alignments were generated using the CLUSTAL_W package with the default settings. Aligned sequences were trimmed to match the genomic regions of the sequences obtained in this study and used to generate phylogenetic trees in MEGA4 using either neighbor joining, maximum likelihood, or maximum parsimony with bootstrap values calculated from 1,000 replicates. Accession numbers of reference sequences used are as follows: HEV-A sequences, AY177911 and AY69748 to AY69761; HEV-B sequences, AF029859, AF083069, AF85363, AF105342, AF114383, AF231763, AF233852, AF241360, AF311939, AF317694, AF524867, AJ493062, AY167105, AY302539 to AY302560, AY556057, and AY556070; HEV-C sequences, AB205396, AF081308, AF081310, AF205396, AF465511 to AF465515, AF499635 to AF499643, AF546702, AY876912, and AY876913; and HEV-D sequences, AY426531, DQ201177, and EF107097.
Patient demographics and sample processing.
All samples previously tested negative for poliovirus by PCR analysis of cytopathogenic cell culture supernatants in L20M and RD cell lines (ATCC). Patient ages averaged 52 months, with a range of 1 month to 14.5 years. The patients included 24 boys and 11 girls. Control samples were collected from healthy contacts of children with AFP and were assigned identification numbers ending with C, e.g., 5006C.
Nucleotide sequence accession numbers.
High-quality sequences and contigs have been deposited in GenBank under accession numbers FI578338 through FI591728.
RESULTS
Sampling.
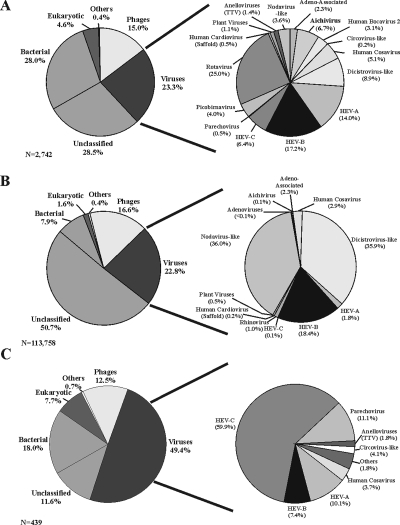
Sequence-independent amplification of viral nucleic acids was performed on 35 fecal samples from children diagnosed with AFP within 2 weeks prior to sampling. Clinical outcomes of the AFP cases ranged from full recovery to death (Table 1). Viral particles were first partially purified, and the nucleic acids were randomly amplified and subcloned into plasmids (see Materials and Methods). Between 35 and 240 subcloned fragments were sequenced from each sample. 454 pyrosequencing analysis of a subset of 10 samples was also performed, yielding between 3,715 and 25,516 high-quality sequences per sample. The average sequence length for subcloned products was 432 bp, while 454 pyrosequencing yielded an average of 201 bp. Unique sequences (singlets) or contigs were then analyzed by tBLASTx against the entries in the GenBank nonredundant database. Sequences with E values of ≤10−3 were categorized according to their closest BLAST match. Totals of 29 and 51% of sequences could not be classified using Sanger and 454 pyrosequencing, respectively, similar to levels from previous metagenomic studies of stool samples (7, 8, 13, 15, 22, 41). Both Sanger and 454 pyrosequencing yielded ∼23% eukaryotic viral sequences (Fig. 1A and B).
TABLE 1.
Sample summarya
| Subject | Sex | Age (mos) | Additional diagnosis | Outcome | No. of subclones analyzed by Sanger sequencing | No. of sequences obtained by 454 pyrosequencing | Eukaryotic virus(es) detected |
|---|---|---|---|---|---|---|---|
| AFP patients | |||||||
| 1449 | M | 96 | Unknown | Residual weakness | 49 | 14,713 | Cosavirus, nodavirus-like virus |
| 6278 | F | 30 | Hemiplegia | Residual weakness | 130 | 8,924 | None |
| 6341 | M | 4 | Hemiplegia | Full recovery | 57 | 5,456 | None |
| 6344 | M | 60 | Unknown | Full recovery | 102 | 23,477 | Cosavirus, dicistrovirus-like virus |
| 5727 | F | 24 | Other | Residual weakness | 130 | 3,715 | Tobacco green mosaic virus |
| 6178 | M | 36 | Hemiplegia | Death | 181 | 20,052 | Cosavirus, dicistrovirus-like virus, anellovirus (TTV), HEV-B, HEV-C |
| 6187 | M | 60 | Encephalitis/meningitis | Full recovery | 240 | 17,705 | Aichi virus, adenovirus |
| 5192 | M | 36 | Unknown | Full recovery | 59 | 4,019 | Adenovirus, Aichi virus, cosavirus |
| 5550 | M | 1 | NPE myelitis | Death | 52 | 8,276 | Adenovirus, cosavirus, HEV-B, HEV-C, cucumber mosaic virus, rhinovirus |
| 6572 | F | 48 | Unknown | Unknown | 129 | 25,516 | Cosavirus, HEV-C, HEV-B, human cardiovirus, plant virus |
| 1988 | M | 13 | Other | Full recovery | 45 | HEV-B | |
| 2111 | F | 120 | Hypokemia | Full recovery | 40 | HEV-A | |
| 2131 | M | 11 | Hypokemia | Full recovery | 36 | HEV-A, HEV-B | |
| 2178 | M | 30 | Unknown | Residual weakness | 50 | Adenovirus, HEV-C | |
| 2204 | M | 15 | Unknown | Residual weakness | 45 | HEV-A | |
| 2255 | F | 12 | Hypokemia | Full recovery | 38 | Parechovirus, picobirnavirus, pepper mottle virus | |
| 2263 | F | 24 | Hemiplegia | Residual weakness | 47 | Aichi virus, tobacco green mosaic virus | |
| 2291 | M | 122 | Other | Full recovery | 41 | Aichi virus | |
| 2295 | F | 26 | Traumatic neuritis | Full recovery | 39 | Aichi virus, picobirnavirus | |
| 2296 | M | 174 | Unknown | Death | 92 | Cosavirus, HEV-B | |
| 2299 | M | 96 | Hemiplegia | Residual weakness | 40 | HEV-A | |
| 5002 | F | 7 | Hypokemia | Full recovery | 58 | Rotavirus | |
| 5003 | M | 7 | Traumatic neuritis | Full recovery | 91 | Anellovirus (TTV), HEV-B, human cardiovirus, rotavirus | |
| 5004 | M | 72 | Unknown | Residual weakness | 43 | HEV-A | |
| 5005 | M | 96 | Other | Full recovery | 44 | HEV-A | |
| 5008 | M | 36 | Unknown | Full recovery | 35 | HEV-A | |
| 5034 | M | 84 | Unknown | Residual weakness | 110 | Cosavirus, HEV-C | |
| 5048 | M | 42 | Unknown | Unknown | 62 | None | |
| 5152 | M | 156 | Other | Full recovery | 41 | Human cardiovirus, unclassified Partitiviridae viruses | |
| 5222 | F | 60 | Unknown | Unknown | 88 | None | |
| 5510 | M | 36 | Unknown | Full recovery | 98 | Bocavirus, dicistrovirus-like virus, HEV-C | |
| 5551 | M | 8 | Encephalitis/meningitis | Residual weakness | 133 | Rotavirus | |
| 6197 | F | 23 | Encephalitis/meningitis | Death | 183 | HEV-B, HEV-C | |
| 6377 | M | 84 | Traumatic neuritis | Full recovery | 44 | None | |
| 6584 | F | 72 | Unknown | Unknown | 85 | None | |
| Contacts | |||||||
| 5006C | F | 24 | NA | NA | 83 | Aichi virus, cosavirus, circovirus-like virus, HEV-C | |
| 5045C | M | 25 | NA | NA | 86 | HEV-C, parechovirus | |
| 5044C | F | 24 | NA | NA | 70 | HEV-C, anellovirus (TTV) | |
| 5046C | F | 6 | NA | NA | 91 | Cosavirus, HEV-B, HEV-C, rhinovirus, anellovirus (TTV) | |
| 5047C | M | 11 | NA | NA | 47 | HEV-B, HEV-C | |
| 5048C | M | 48 | NA | NA | 62 | Cosavirus, HEV-A, HEV-B, HEV-C |
M, male; F, female; NA, not applicable; NPE, nonpolio enterovirus.
FIG. 1.
Sequence classification and distribution. (Left panels) Sequences with E values of ≤10−3 were classified as either eukaryotic, phage, bacterial, or viral based on the best tBLASTx score. Unclassified sequences are those which had E values of >10−3 by tBLASTx, BLASTn, and BLASTx analyses. Sequences which did not fall into set categories were designated “other” and included fungal and plasmid vector sequences. (Right panels) The subsets of sequences classified as viral were broken down further by family, genus, and species. Classification results for Sanger sequencing clones derived from 35 AFP patients (A), sequences generated by 454 pyrosequencing from a subset of 10 patients (B), and Sanger sequencing clones obtained from six healthy contacts (C) are shown. Others in the viral pie chart of healthy contacts consist of rhinovirus and Aichi virus.
Diversity of viruses and genetic composition.
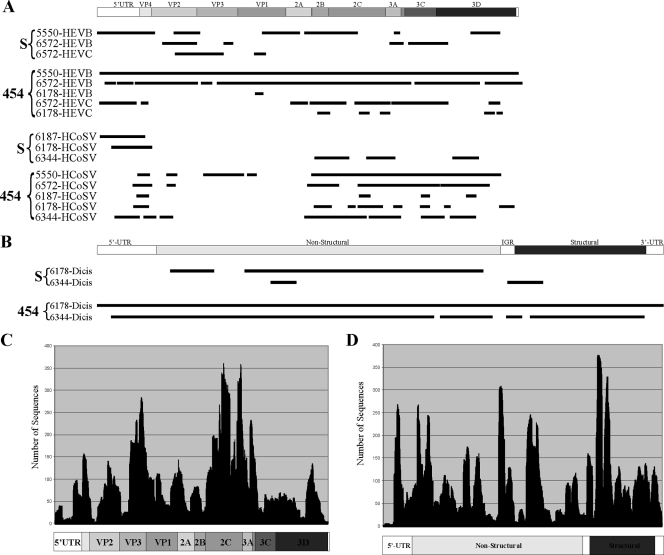
Multiple previously characterized and divergent novel viruses were detected. The majority of the known viral sequences were those of single-stranded RNA viruses belonging to the order Picornavirales. We also detected double-stranded RNA (dsRNA) viruses (picobirnavirus and rotavirus), single-stranded DNA viruses (Torque teno virus [TTV] anelloviruses and adeno-associated virus), and dsDNA viruses (adenoviruses) (Fig. 1). Highly divergent viruses with levels of amino acid sequence identity of less than 55% were described as being like the closest tBLASTx match (e.g., circovirus like or dicistrovirus like). Viral detection results from limited Sanger sequencing and 454 pyrosequencing of products from 10 samples were compared (Table 1). The identification of coinfection increased from an average of 1.4 (range, 0 to 4) eukaryotic viruses detected per sample using Sanger sequencing to 2.6 (range, 0 to 6) eukaryotic viruses detected per sample using 454 pyrosequencing. The fraction of each viral genome recovered increased from an average of 19% using Sanger sequencing to 41% using 454 pyrosequencing (Fig. 2A and B). 454 pyrosequencing enabled near-complete genome sequencing of several HEV strains, a novel nodavirus-like virus (unpublished results), and a novel dicistrovirus-like virus (unpublished results). For these near-complete genome sequences, the average depth of sequencing was 90 to 100×; however, it was highly variable (2 to 378×) and diminished near regions of known complex secondary RNA structure (Fig. 2C and D). Regions with overrepresented sequencing depth were often preceded by stretches of nucleotide residues exhibiting identity in 4 of 4 or 4 of 5 bases at the 3′ end of the fixed region of the primer preceding the random N octamer used for “random“ amplification (Fig. 2 and data not shown).
FIG. 2.
Viral genome coverage and depth of sequencing. (A) Lines below the graphical depiction of a generic picornavirus genome represent the locations of HEV-B, HEV-C, or human cosavirus (HCoSV) singlets or contigs acquired from different AFP patients by either Sanger sequencing (S) or 454 pyrosequencing (454). 5′-UTR, 5′ untranslated region; IGR, intergenomic region. (B) Lines below the graphical depiction of a generic dicistrovirus genome represent the locations of dicistrovirus (Dicis) singlets or contigs acquired from samples from two AFP patients by either Sanger sequencing or 454 pyrosequencing. (C and D) The depth of sequencing for each nucleotide position is shown for 454 pyrosequenced HEV-B from patient 5550 (C) and dicistrovirus from patient 6178 (D).
Healthy contact control samples.
Stool samples from six healthy children who had family contact with an AFP patient were also analyzed using Sanger sequencing. Unexpectedly, the fraction of viral sequences among total sequences more than doubled, increasing from 23 to 49%, compared to that from AFP patients (Fig. 1A and C). HEV-C composed the majority (60%) of the viral clones sequenced and was observed at a higher frequency in samples from healthy contacts (6 of 6) than in those from children with AFP (6 of 35) (P < 0.001). Different genotypes of HEV-C, including those of coxsackievirus A14 (CoxA14), CoxA13, and CoxA24 and poliovirus (Sabin 2 vaccine strain), were found. While several coxsackieviruses have been shown previously to correlate with enteric diseases, CoxA14, CoxA13, and CoxA24 have yet to be associated with illness (29). The poliovirus sequences identified here are likely derived from the ingestion of oral vaccine, as Pakistani children routinely received 10 or more doses.
Viral populations in each patient sample.
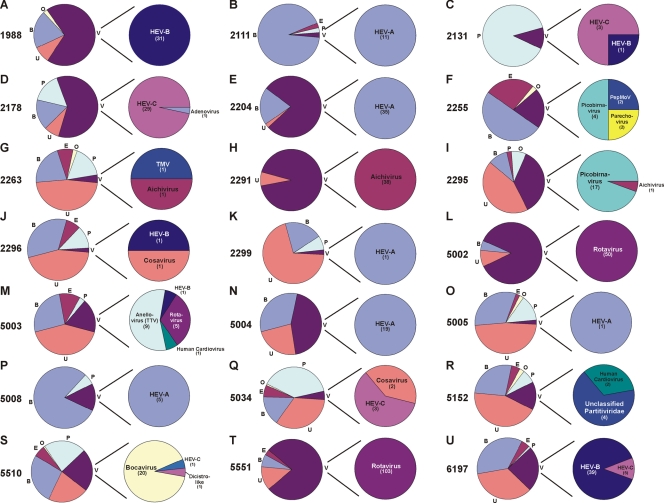
Within each patient sample, the percentages of viral and nonviral sequences varied greatly, consistent with previously published data (13). Eukaryotic viruses were detected in stool samples from 29 (83%) of 35 AFP patients. Based on tBLASTx classification, sequences from seven virus families, plus four novel virus groups, were detected. Of the 29 virus-positive samples, 17 contained at least one HEV, and 5 of these 17 samples appeared to be coinfected with two clearly distinguishable HEV species (Fig. 3 and 4). Targeted amplification from stool samples, using pan-HEV primers (39), indicated that 23 of these 35 tested patients were positive for HEV infection (20), 17 of which were also identified using viral metagenomics. Other known enteric viruses observed included adenovirus, picobirnavirus, rotavirus, Aichi virus, parechovirus, and rhinovirus, as well as assorted plant viruses of the families Partitiviridae and Tobamoviridae (Fig. 3 and 4; Table 1). In addition to previously identified viruses, several potentially novel viruses were observed. These viruses included one with weak identity (<35% amino acid identity) to the insect virus family Dicistroviridae (a member of the order Picornavirales; detected in samples from three AFP patients), viruses belonging to a novel candidate picornavirus genus named Cosavirus (present in eight samples) (20), a novel circovirus-like virus most closely related to porcine circovirus (<55% amino acid identity; present in a single patient sample), a novel human bocavirus (<80% amino acid identity; present in a single patient sample) (18), and a virus displaying weak amino acid identity (<33%) to a fish nodavirus (present in a single patient sample) (Fig. 3 and 4).
FIG. 3.
Sequence classification per patient. (Left pie charts) Sequences generated by subcloning and Sanger sequencing alone from samples from individual patients in which eukaryotic viral sequences were detected were categorized as bacterial (B), unclassified (U), phage (P), eukaryotic (E), viral (V), or other (O). (Right pie charts) Characterization of viral sequences by viral family or species. Values in parentheses are numbers of sequences detected. Virus abbreviations: TMV, tobacco mosaic virus; dicistro-like, dicistrovirus-like virus; and PepMoV, pepper mottle virus.
FIG. 4.
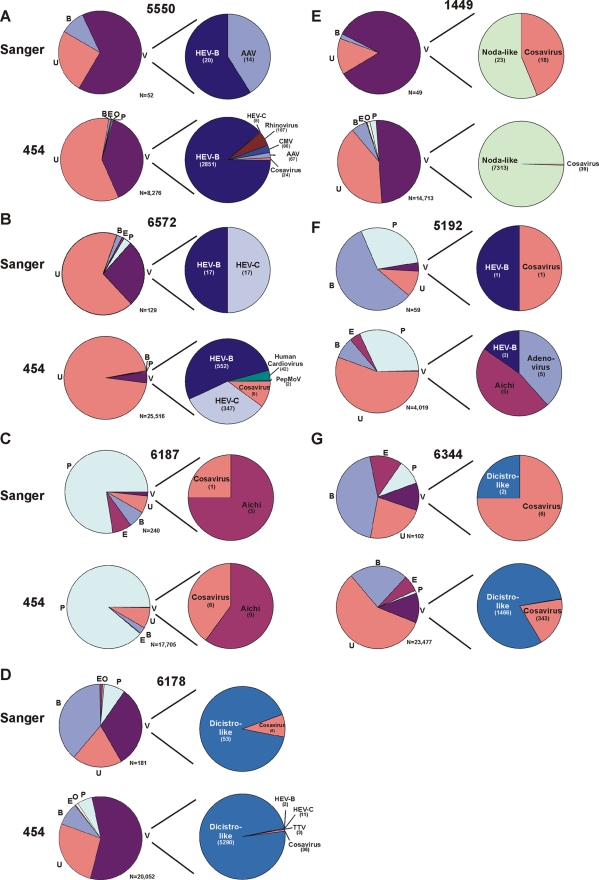
Comparison of sequences obtained by Sanger sequencing (upper charts) and pyrosequencing (lower charts). Pie charts are labeled as described in the legend to Fig. 3. In samples from patients 6278 and 6341, no recognized viral sequences were detected by either 454 pyrosequencing or Sanger sequencing (data not shown). Abbreviations: AAV, adeno-associated virus; CMV, cucumber mosaic virus; dicistro-like, dicistrovirus-like virus; noda-like, nodavirus-like virus; and PepMoV, pepper mottle virus. Results for the sample from patient 5727, which had a single tobacco green mosaic virus sequence, are not shown.
The classifications of sequences obtained from 10 individual samples analyzed by both Sanger sequencing and 454 pyrosequencing were compared (Fig. 4). In the majority of cases, the same viruses were found by both methods, although in 4 of 10 samples (those from patients 5192, 6178, 5550, and 6572), more viruses were found using pyrosequencing. The viral and nonviral sequence ratios determined and the specific viral species detected within the same samples by both methods were generally comparable, with some exceptions (e.g., the nodavirus-like virus/cosavirus ratio in the sample from 1449 and the adenovirus/other virus ratio in the sample from 5550).
Potentially new, divergent HEV serotypes.
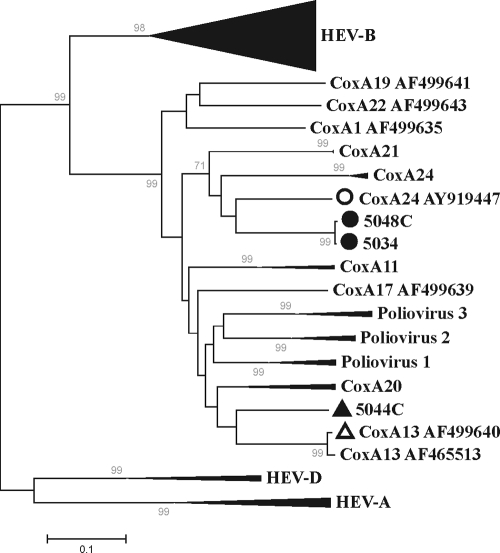
All HEV singlet sequences or contigs were examined individually by BLASTx and BLASTn similarity searches and by phylogenetic analyses (data not shown). Based on these analyses, three samples were recognized to contain the most divergent HEV sequences, thereby identifying these HEV variants as candidates for new HEV serotypes. HEV variants showing the same antibody neutralization profile (i.e., belonging to the same serotype) have previously been shown to carry VP1 proteins and genes with ≥88% amino acid identity and ≥75% nucleotide identity (25). Degenerate PCR primers flanking the VP1 gene were used to amplify VP1 sequences from the three samples with divergent HEV variants. VP1 genes and proteins from samples obtained from subjects 5034, 5044C, and 5048C exhibited 78, 75, and 75% nucleotide identity and 88, 87, and 83% amino acid identity, respectively, to the closest HEV sequences available in GenBank. Sequences from the samples from patient 5034 and healthy contact 5048C exhibited 98% nucleotide identity to each other, were collected in 2007 within 3 months of each other, and were both from the Punjab province of Pakistan. Phylogenetic analyses of amplified VP1 nucleotide sequences show that the sequences from the 5034 and 5048C samples represent deeply rooted CoxA24 serotypes, while the sequence from the 5044C sample may be divergent enough from preexisting genotypes to qualify as a prototype of a new HEV-C genotype (Fig. 5).
FIG. 5.
Unrooted neighbor-joining phylogenetic analysis showing relationships based on the alignment of divergent HEV VP1 gene nucleotide sequences from samples obtained from subjects 5044C, 5034, and 5048C. Filled symbols represent VP1 sequences amplified from samples from AFP patients or healthy contacts of AFP patients. Open symbols represent the corresponding closest BLASTn matches in GenBank. Collapsed branches represent at least three representative sequences from HEV species (HEV-A, HEV-B, and HEV-D) or serotypes (e.g., CoxA21).
DISCUSSION
In this study, we examined the viral nucleic acids in stool samples from 35 South Asian children with nonpolio AFP and 6 healthy contacts. Viruses were detected in 29 of 35 children with AFP and in all six healthy contacts. We also used 454 pyrosequencing and compared this technique to traditional shotgun subcloning and Sanger sequencing. Pyrosequencing provided superior genomic coverage (41% versus the 19% average coverage of all detected viral genomes) and more sensitive viral detection (average, 2.6 versus 1.4 viruses per patient sample) (Fig. 4). Consistent with the data in previous reports (40), the shorter reads associated with pyrosequencing nearly doubled the portion of unclassifiable sequences compared to the portion obtained by Sanger sequencing (51 versus 29%) (Fig. 1A and B). It appears that this increase in unclassifiable sequences is due primarily to a reduced ability to classify diverse bacterial sequences, resulting in a reduction in the portion of sequences classified as bacterial from 28% by Sanger sequencing to 7.9% by 454 pyrosequencing. Viral sequences accounted for ∼23% of the total, regardless of the sequencing method. Despite the problems associated with shorter sequence reads, pyrosequencing was superior in both viral detection and genome coverage for the 10 patients tested, at approximately the same financial cost. As pyrosequencing technology improves to generate longer sequence reads, it is likely to supplant Sanger shotgun sequencing as a method of viral identification and discovery.
Prior viral metagenomic studies of feces utilized Sanger shotgun sequencing of 532 (8), 4,600 (13), and 36,769 (41) plasmid subclones at the cost of analyzing fewer samples (1, 12, and 3, respectively). Two of these studies detected primarily plant viruses (41) or bacteriophage (8) (in this case, likely the result of focusing on dsDNA viruses), while the third study, using diarrhea samples, detected known viral pathogens as well as sequences divergent enough to potentially belong to two new viral species (astrovirus and nodavirus) (13, 14). In our study, the most common plant virus detected was pepper mild mottle virus, which has also been reported to occur at high frequencies in North American and Singaporean human stool samples (41). In addition to bacteriophage and plant viruses, we detected known pathogenic enteric viruses, including rotavirus, adenovirus, picobirnavirus, and numerous members of the Picornaviridae family, including parechovirus, Aichi virus, rhinovirus, cardioviruses, and HEV-A to HEV-C, as well as several new viral species. The high proportion of healthy children with viruses in their stool samples (six of six) (Table 1) underlines the often asymptomatic nature of many enteric viral infections whose clinical outcomes are likely dictated by a combination of viral and host genetics, active and passive immunity (i.e., maternal antibodies), and overall health (26).
Specific nested panenterovirus PCR primers detected HEV infection in 23 of 35 of the AFP cases (20), while 17 of 35 AFP samples exhibited at least one HEV sequence in the viral metagenomic analysis. Both metagenomic analysis and pan-HEV PCR detected HEV infection in all six healthy contacts. This correlation was less pronounced for members of the new candidate picornavirus genus Cosavirus than for HEV, as cosavirus sequences were found in only 9 of 41 samples from AFP patients and healthy contacts by shotgun sequencing, compared to 19 of these 41 by nested PCR (20). Similarly, human cardiovirus SAFV was found in 3 of 57 nonpolio AFP children using shotgun Sanger sequencing and in 9 of 57 patients using RT-nested PCR (5). It is possible that the cosavirus loads in stool samples are generally lower than HEV loads, thereby making detection using limited shotgun sequencing less likely. Indeed, in-depth 454 sequencing of 8,276 clones from the sample from patient 5550 and 25,516 clones from the sample from patient 6572 revealed the presence of cosaviruses missed by Sanger sequencing. These results indicate that while a wide range of distinct viruses (belonging to different and in some cases new viral species) can be detected using low-level Sanger subclone sequencing, the very high sensitivity of nested PCR stills allows more cases of presumably low-level infections with known viruses to be detected.
We detected at least five novel viruses or groups of viruses: a new human bocavirus (18), members of a new Picornaviridae genus (20), a new circovirus (unpublished results), a new nodavirus (unpublished results), and new discistroviruses (unpublished results). Sequences from divergent viruses that may represent new genotypes of enteroviruses, parechoviruses (23), cardioviruses (5), and picobirnaviruses (unpublished data) were also found. The novel nodavirus sequences were clearly distinguishable from the nodavirus sequences recently generated from diarrhea samples, overall exhibiting less than 41% amino acid identity to the previously generated sequences (13). The most diverse viral sequences detected and reported here belonged to the dicistrovirus-like category, in which polymerase and other enzymatic regions exhibited less than 35% amino acid identity to dicistrovirus sequences currently in GenBank. Dicistrovirus-like sequences were detected in samples from three patients, two of which, patients 6178 and 6344, were coinfected with members of the new Picornaviridae genus, Cosavirus. The dicistrovirus-like sequences exhibited 70 to 75% nucleotide identity to one another, a level of divergence otherwise seen among different species of dicistroviruses.
It remains to be determined which of these novel viruses are capable of replication in the human gut, as it is conceivable that some were consumed and their nucleic acids traveled through the digestive tract intact, as attested to by the detection of nucleic acids from plant viruses which have previously been shown to remain infectious (41).
Nodaviruses are small, single-stranded, bipartite RNA viruses that to date have been shown to naturally infect only insects and fish. Nodaviruses have been detected previously in human stool (13) and are semipermissive of replication in mammalian tissues (4, 12). Dicistroviruses have been shown to replicate and be pathogenic in insects (10, 37). The internal ribosomal entry site between the two cistronic segments can act as a powerful promoter in mammalian cells (28). However, reports of viral replication within mammalian cell lines are contradictory; one group has demonstrated the replication of a dicistrovirus, Taura syndrome virus, in human cell lines (3), while another has failed to reproduce Taura syndrome virus growth in mammalian cell lines (27). The ability of pathogenic porcine circovirus 2 to replicate in pigs is well established (1, 24, 36). Whether the circovirus detected in the sample from patient 5006 represents the first human circovirus or a circovirus from ingested meat remains unknown. In vitro replication as well as serological and larger epidemiological studies will be necessary to determine the range of host species tropisms and pathogenic potentials of these new viruses.
Three of the 35 AFP cases were fatal: the sample from child 5550, in which six distinguishable eukaryotic viruses (adenovirus, cosavirus, HEV-B, HEV-C, rhinovirus, and cucumber mosaic virus) were observed, exhibited the highest level of coinfection; patient 2296 was coinfected with HEV-B and a cosavirus; and patient 6178 exhibited coinfection with dicistrovirus and cosavirus. Stool from patient 6178 was likely to contain a high titer of dicistrovirus based on the large fraction of sequence from this virus derived by random amplification (Fig. 2B and D) and dilution end point PCR, which indicated a viral load of approximately 106 genome copies per ml of stool supernatant (data not shown). While cosaviruses were present in all three fatal cases, the difference in cosavirus prevalence among all AFP patients combined and healthy controls was not statistically significant (20). Since even clearly pathogenic picornaviruses, such as poliovirus, typically produce no clinical manifestations in 99 to 99.9% of infections (26), failure to detect a significant association with disease in this small cohort does not absolve cosaviruses, cardioviruses, or other new viruses of possible pathogenic roles.
In summary, we have used limited Sanger sequencing of stool samples from children with AFP to detect both known and novel viruses. By increasing the depth of the nucleic acid sampling using 454 pyrosequencing, we detected more viruses likely present at lower viral loads. These studies provide a framework for further studies that can be applied to numerous cases of AFP reported by the Global Polio Laboratory Network; of the 700,000 cases reported since 1997, only ∼6.5% have been attributed to poliovirus and 15 to 30% have been attributed to nonpolio enteroviruses (9). PCR studies of stool and tissue samples from subjects of different ages and geographic origins, both with and without diseases, as well as serological testing, will be required to determine the epidemiology and pathogenicity of these new viruses. The numerous known and new viruses in stool samples from developing countries, a likely result of limited access to adequate sanitary conditions resulting in frequent enteric infections, also indicates that such samples provide readily accessible material for further viral discovery.
Acknowledgments
We thank Michael P. Busch and the Blood Systems Research Institute for sustained support. We also thank Hope Biswas for statistical assistance and Shahzad Shaukat, Salmaan Sharif, Muhammad Masroor Alam, and Mehar Angez for sample collection and poliovirus testing.
This research was supported by NHLBI grant R01HL083254 to E.L.D.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clarke, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 103-10. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T., S. U. Emerson, R. E. Engle, R. H. Purcell, and J. Bukh. 18 September 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA 9811609-11614. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audelo-del-Valle, J., O. Clement-Mellado, A. Magana-Hernandez, A. Flisser, F. Montiel-Aguirre, and B. Briseno-Garcia. 2003. Infection of cultured human and monkey cell lines with extract of penaeid shrimp infected with Taura syndrome virus. Emerg. Infect. Dis. 9265-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A. 1992. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 662335-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blinkova, O., A. Kapoor, J. Victoria, M. Jones, N. Wolfe, A. Naeem, S. Shaukat, S. Sharif, M. M. Alam, M. Angez, S. Zaidi, and E. L. Delwart. 2009. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J. Virol. 834631-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolanaki, E., C. Kottaridi, E. Dedepsidis, Z. Kyriakopoulou, V. Pliaka, A. Pratti, S. Levidiotou-Stefanou, and P. Markoulatos. 14 February 2008. Direct extraction and molecular characterization of enteroviruses genomes from human faecal samples. Mol. Cell. Probes 22156-161. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 7.Breitbart, M., M. Haynes, S. Kelley, F. Angly, R. A. Edwards, B. Felts, J. M. Mahaffy, J. Mueller, J. Nulton, S. Rayhawk, B. Rodriguez-Brito, P. Salamon, and F. Rohwer. 1 May 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159367-373. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 1856220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2007. Laboratory surveillance for wild and vaccine-derived polioviruses—worldwide, January 2006-June 2007. MMWR Morb. Mortal. Wkly. Rep. 56965-969. [PubMed] [Google Scholar]
- 10.Cox-Foster, D. L., S. Conlan, E. C. Holmes, G. Palacios, J. D. Evans, N. A. Moran, P. L. Quan, T. Briese, M. Hornig, D. M. Geiser, V. Martinson, D. vanEngelsdorp, A. L. Kalkstein, A. Drysdale, J. Hui, J. Zhai, L. Cui, S. K. Hutchison, J. F. Simons, M. Egholm, J. S. Pettis, and W. I. Lipkin. 6 September 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318283-287. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 11.da Silva, E. E., M. T. Winkler, and M. A. Pallansch. 1996. Role of enterovirus 71 in acute flaccid paralysis after the eradication of poliovirus in Brazil. Emerg. Infect. Dis. 2231-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delsert, C., N. Morin, and M. Comps. 1997. Fish nodavirus lytic cycle and semipermissive expression in mammalian and fish cell cultures. J. Virol. 715673-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkbeiner, S. R., A. F. Allred, P. I. Tarr, E. J. Klein, C. D. Kirkwood, and D. Wang. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkbeiner, S. R., C. D. Kirkwood, and D. Wang. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 3121355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward, J. C., S. M. Gillespie, K. M. Kaplan, R. Packer, M. Pallansch, S. Plotkin, and L. B. Schonberger. 1989. Outbreak of poliomyelitis-like paralysis associated with enterovirus 71. Pediatr. Infect. Dis. J. 8611-616. [DOI] [PubMed] [Google Scholar]
- 17.Jean, C. M., S. Honarmand, J. K. Louie, and C. A. Glaser. 2007. Risk factors for West Nile virus neuroinvasive disease, California, 2005. Emerg. Infect. Dis. 131918-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapoor, A., E. Slikas, P. Simmonds, T. Chieochansin, A. Naeem, S. Shaukat, M. M. Alam, S. Sharif, M. Angez, S. Zaidi, and E. Delwart. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor, A., J. Victoria, P. Simmonds, C. Wang, R. W. Shafer, R. Nims, O. Nielsen, and E. Delwart. 2008. A highly divergent picornavirus in a marine mammal. J. Virol. 82311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor, A., J. G. Victoria, P. Simmonds, T. Chieochansin, E. Slikas, A. Naeem, S. Shaukat, S. Salmaan, A. M. Muhammad, M. Angez, C. Wang, R. W. Shafer, Z. Sohail, and E. L. Delwart. 2008. A highly diversified and prevalent new Picornaviridae genus in the stool of South Asian children. Proc. Natl. Acad. Sci. USA 10520482-20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kistler, A. L., D. R. Webster, S. Rouskin, V. Magrini, J. J. Credle, D. P. Schnurr, H. A. Boushey, E. R. Mardis, H. Li, and J. L. DeRisi. 2007. Genome-wide diversity and selective pressure in the human rhinovirus. Virol. J. 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 22 May 2008. Evolution of mammals and their gut microbes. Science 3201647-1651. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, L., J. Victoria, A. Kapoor, A. Naeem, S. Shaukat, S. Sharif, M. Masroor, M. Angez, S. Zaidi, and E. Delwart. 28 January 2009. Genomic characterization of a novel human parechovirus type. Emerg. Infect. Dis. 15288-291. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morozov, I., T. Sirinarumitr, S. D. Sorden, P. G. Halbur, M. K. Morgan, K. J. Yoon, and P. S. Paul. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 362535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 731941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallansch, M., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-776. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Pantoja, C. R., S. A. Navarro, J. Naranjo, D. V. Lightner, and C. P. Gerba. 2004. Nonsusceptibility of primate cells to Taura syndrome virus. Emerg. Infect. Dis. 102106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfingsten, J. S., and J. S. Kieft. 30 May 2008. RNA structure-based ribosome recruitment: lessons from the Dicistroviridae intergenic region IRESes. RNA 141255-1263. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 21230-38. [DOI] [PubMed] [Google Scholar]
- 30.Saad, M., S. Youssef, D. Kirschke, M. Shubair, D. Haddadin, J. Myers, and J. Moorman. 6 November 2004. Acute flaccid paralysis: the spectrum of a newly recognized complication of West Nile virus infection. J. Infect. 51120-127. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 31.Saeed, M., S. Z. Zaidi, A. Naeem, M. Masroor, S. Sharif, S. Shaukat, M. Angez, and A. Khan. 2007. Epidemiology and clinical findings associated with enteroviral acute flaccid paralysis in Pakistan. BMC Infect. Dis. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoja, Z. O., H. Tabatabie, S. Shahmahmoudi, and R. Nategh. 2007. Comparison of cell culture with RT-PCR for enterovirus detection in stool specimens from patients with acute flaccid paralysis. J. Clin. Lab. Anal. 21232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, S. S., S. P. Manimunda, A. P. Sugunan, Sahina, and P. Vijayachari. 2008. Four cases of acute flaccid paralysis associated with chikungunya virus infection. Epidemiol. Infect. 1361277-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solomon, T., and H. Willison. 2003. Infectious causes of acute flaccid paralysis. Curr. Opin. Infect. Dis. 16375-381. [DOI] [PubMed] [Google Scholar]
- 35.Stang, A., K. Korn, O. Wildner, and K. Uberla. 2005. Characterization of virus isolates by particle-associated nucleic acid PCR. J. Clin. Microbiol. 43716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischer, I., W. Mields, D. Wolff, M. Vagt, and W. Griem. 1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91271-276. [DOI] [PubMed] [Google Scholar]
- 37.Van Munster, M., A. M. Dullemans, M. Verbeek, J. F. Van Den Heuvel, A. Clerivet, and F. Van Der Wilk. 2002. Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae. J. Gen. Virol. 833131-3138. [DOI] [PubMed] [Google Scholar]
- 38.Victoria, J. G., A. Kapoor, K. Dupuis, D. P. Schnurr, and E. L. Delwart. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 4e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch, J. B., K. McGowan, B. Searle, J. Gillon, L. M. Jarvis, and P. Simmonds. 2001. Detection of enterovirus viraemia in blood donors. Vox Sang. 80211-215. [DOI] [PubMed] [Google Scholar]
- 40.Wommack, K. E., J. Bhavsar, and J. Ravel. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 741453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]